Abstract
We report the expression, purification, liposome reconstitution and functional validation of uniformly 13C and 15N isotope labeled KcsA, a bacterial potassium channel that has high homology with mammalian channels, for solid-state NMR studies. The expression and purification is optimized for an average yield of ~ 35–40 milligrams per liter of M9 media in a time-efficient way. The protein purity is confirmed by gel electrophoresis and the protein concentration is quantified by UV-Vis absorption spectroscopy. Protocols to efficiently reconstitute KcsA into liposomes are also presented. The presence of liposomes is confirmed by cryo-electron microscopy images and the effect of magic angle spinning on liposome packing is shown. High-resolution solid-state NMR spectra of uniformly isotope labeled KcsA in these liposomes reveal that our protocol yields to a very homogenous KcsA sample with high signal to noise and several well-resolved residues in NMR spectra. Electrophysiology of our samples before and after solid-state NMR show that channel function and selectivity remain intact after the solid-state NMR.
Keywords: Isotopic Labeling, Membrane proteins, reconstitution, solid-state NMR, liposomes
Introduction
Potassium channels are highly conserved intrinsic membrane proteins that selectively conduct potassium ions across cell membranes near the rate of free diffusion. They are implicated in a variety of physiological functions including osmoregulation, muscle contraction and the generation of synaptic action potentials, and are consequently key targets for many diseases like cardiac arrhythmia and hypoglycemia[1]. KcsA is a 160 amino-acid potassium channel isolated from the soil bacterium S. lividans and was the first potassium channel to have a solved crystal structure. It is a homotetrameric protein with two transmembrane helices per monomer and an ion-conducting pore in the center [2,3]. Due to its experimental convenience, high sequence homology and structural similarity to key regions of mammalian potassium channels, it has become a model system for studies of the biophysical details of ion-conduction and gating by a variety of different methods including x-ray crystallography[3,4], electrophysiology[5,6] and NMR[7–9].
Solid state NMR is particularly suited to study membrane proteins because it does not require that the protein be soluble in water, and can be used to study protein systems in a native bilayer environment. NMR can offer atom-specific insights into protein structure and dynamics and can also be used to study ligand binding and conformational exchange. However studies by NMR typically require milligram quantities of isotope labeled proteins, often difficult to achieve for membrane proteins due to their low expression and toxicity to cells. Furthermore, in order to get high-resolution spectra of uniformly 13C and 15N labeled proteins, some degree of local sample homogeneity is necessary in order to efficiently resolve peaks in the spectrum.
In this paper we describe protocols and methods for the efficient expression, purification of 13C and 15N labeled KcsA and subsequent reconstitution of this protein into lipid bilayers. KcsA is a well-studied protein that was first expressed and reconstituted over a decade ago [10,11] and has been successfully expressed on many occasions, however the yields for the protein were typically less than 10mg/L of media, especially for isotope enriched preparations. Previous work in our group led to yields of 6–8 milligrams of U-13C-15N labeled KcsA per liter of M9 enriched media using a PQE60 plasmid [8]. The group of Shimada has reported yields of 2 milligrams of deuterated KcsA per liter of the C-terminal deletion construct [12], the group of Miller original reported a yield of 1–3 mg/liter [13] and the group of Nimigean report yields of 5–10 milligrams per liter of LB for WT-KcsA using a Pask90 vector system. Several groups have also reported NMR quantities of KcsA [7,14]. We report significant improvements to the protocol, which now yields 35–40 milligrams of KcsA per liter of M9 enriched media (after a 4X concentration relative to LB or 9–10 mg/liter of initial LB culture). Our protocol has been tested several times and is tailored to reliably yield tens of milligrams of protein, and homogenous, functional samples that are a necessary prerequisite for NMR studies of this important system. The composition of the minimum media used can be easily modified to incorporate different isotope labeling schemes. We show cryo-EM images of the reconstituted liposomes to validate the bilayer structure and high-resolution solid-state NMR spectra of the reconstituted protein. These are important pre-requisites for site-specific structural and dynamic studies of this system.
Materials and Methods
Plasmids
We tried two plasmids encoding KcsA (primary accession number P0A334 in the Uniprot/Swiss-Prot databank). Plasmid 1 was a gift from the group of Rod Mackinnon at Rockefeller University. The plasmid encodes the KcsA sequence with a mutation at the second residue (P2A) and a deletion of the last two carboxyl terminal arginines. The gene was engineered such that the hexa-histidine tag was included at the carboxyl terminus of the protein and was cloned for over-expression into a PQE-60 vector. This plasmid was sequenced used the primer 5’ TGGTCAAACTGCTGCTCG from Invitrogen. Plasmid 2 was a gift from the group of Crina Nimigean at Weill Cornell Medical School. This plasmid encodes full-length wildtype KcsA with an N-terminal hexa-histidine tag. The gene was cloned into the PASK90 vector system. Plasmid 2 does not have the P2A mutation or any arginine deletions. The plasmid was sequenced using the primer 5’ GCTTGCTCGTCT GGTTAAGTTG also from Invitrogen. We found that isotope labeled protein expression using plasmid 2 was better than plasmid 1, but the same protocol with small modifications can be used with both plasmids.
Protein Expression
The two plasmids require different cell-lines for optimal expression. Plasmid 1 was optimal with E. coli M15 cells and plasmid 2 was optimal with E. coli JM83 cells. The difference between these two cell lines is that JM83 is auxotrophic for proline, so proline needs to be supplemented in the minimum media for the cells to survive. The PQE-60/M15 expression system is induced with isopropyl-1-thio-beta-D-galactopyranoside (IPTG) and the plasmid encodes both ampicillin and kanamycin resistance. The PASK90/JM-83 system is induced with anhydrotetracyclin (aTC) and the plasmid contains only ampicillin resistance. The protocols are otherwise identical.
Isotopic protein expression was always started with a fresh transformation of the plasmid into competent cells prepared using the standard protocol described in the Qiagen manual. The most common reasons for failed transformations were old competent cells, a badly calibrated heat-bath or a heat-shock that was too long. Competent cells were prepared and stored at −80°C for 6 months for optimal yield. After this period a significant decrease was observed in the overall protein yield. The transformed cells were grown on LB (Luria Broth)-Agar plates with the requisite antibiotic (100 µg/L Ampicillin and 25 µg/L Kanamycin for plasmid 1 and 100 µg/ml Ampicillin for plasmid 2). The plates were incubated at 37°C for 6–8 hours.
Four single separate colonies were picked. Each one was transferred into a separate 10 ml LB pre-culture with the appropriate antibiotic. The pre-culture was incubated at 37°C and 250 rpm shaking for 3–4 hours or until the OD600 is ~0.5. The preculture was then transferred into 4 flasks with 4 liters of LB and incubated at same conditions of temperature and shaking. OD600 (optical density at 600 nm) was monitored every hour for 3–4 hours or until it reached ~0.9. The cells were then harvested by centrifugation at 5700g for 20 mins at 4°C and resuspended in 1L of M9 minimum media [15] (recipe in Appendix) supplemented with 3 g of 13C-labelled glucose, 0.5 g of 15-labelled NH4Cl, 10 ml of 13C-15N labeled Bioexpress (Cambridge Isotopes # CGM-1000-CDN-50S) and 1 g proline in the case of plasmid 2. Bioexpress has been shown previously to improve expression of isotope labeled proteins [16]. Due to the 4X concentration from LB to M9, the OD600 of the M9 solution is typically much higher than 2 and it needs to be diluted in order to be measured by UV. Cells were allowed to grow for 1 hour in the M9 media and then protein expression was induced using 1 mM IPTG for plasmid 1 or 1 mM aTC (from Cayman Chemicals #10009542) for plasmid 2. After induction, the cells were allowed to grow for 12–14 hours (overnight) at 37°C and then harvested by centrifugation at 5700g for 20 mins at 4°C and stored at −80°C. Typically 22–25 g of cells were harvested per liter of M9.
Protein Purification
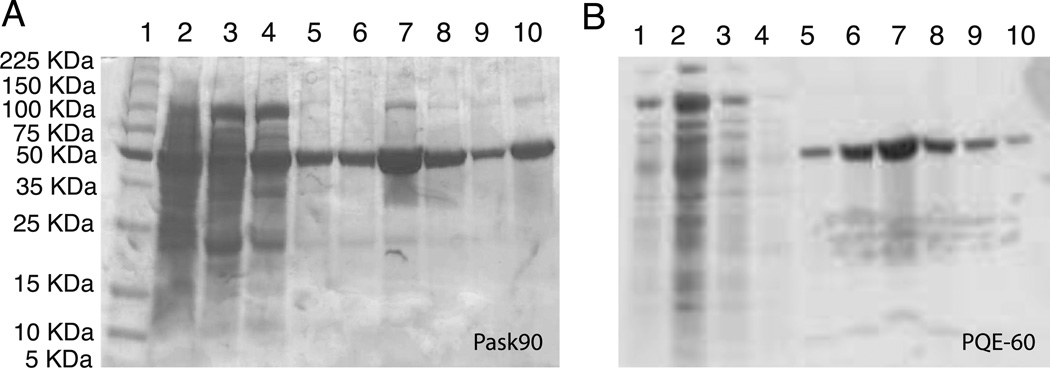
The cells were thawed by leaving them in an ice-water bath for 30 mins and then lysed using 2–3 passages through a French Press at 12,000 psi. After lysis the cells acquire a distinctly darker color, which indicates that the lysing process was successful. Approximately 1 g of Decyl-β-Maltopyranoside (DM, from Affymetrix #D310S) was added for every 10 g of cells and the lysate is incubated on a shaker at 4°C for at least 8 hours to extract the protein. The Unlysed cells and membranes were pelleted by centrifugation at 30,000g for 45 mins at 4°C. The bright yellow supernatant was carefully pipetted out and loaded onto a Nickel column pre-equilibrated with 50 mM Tris, 150 mM KCl, 10 mM DM, pH 7.5 buffer for purification. After loading, the column was washed with at least 3 column volumes of buffer containing 50 mM imidazole to remove non-specific binding. The washing was stopped when the yellow colored protein was no longer visible in the flow through. KcsA was then eluted with 150 mM imidazole and checked using gel electrophoresis. A typical acrylamide gel is shown in Figure 1A. KcsA is stable as a tetramer in SDS and observed as a clean band on an acrylamide gel. The imidazole buffer was replaced with a buffer containing 50 mM Tris, 150 mM KCl, 2 mM DM using an Amicon concentrator with a 10,000 KDa molecular weight cutoff. The final concentration of DM is much higher than 10 mM because DM micelles do not penetrate the filter well. The concentrated protein solution was then dialyzed against a 5 mM DM solution for 12 hours at room temperature (25 C) to reduce the detergent concentration to a known value. This is an important step for the lipid reconstitution and also for the accurate determination of protein concentrations because excess detergent leads to baseline errors in the UV spectra due to scattering. The KcsA yield was quantified by UV absorption at 280 nm using the published extinction coefficient[17] of 33570 M−1 cm−1 for the monomer. A typical UV spectrum of pure KcsA solubilized in DM is shown in Figure 1B. Using this protocol we are routinely able to make ~ 40 mg of U-[13C]-[15N]-KcsA per liter of M9 or 10 mg of KcsA per liter of LB. KcsA is stable as a tetramer when stored at a 1–2 mg/ml concentration in 10 mM DM at 4°C for 3–4 months. For longer periods it was reconstituted and stored in liposomes at −20°C.
Figure 1.
SDS-PAGE analysis of KcsA during purification is shown. Panel A shows results from Nickle column on the lysate from pask90 expression and panel B shows results using the PQE-60 expression system. The band around 50 KDa is the KcsA tetramer. Weak bands at ~100 KDa and ~20 KDa correspond to octamers and monomers respectively. Panel A lanes: 1-marker 2-cell lysate supernatent, 3,4 – consecutive 50 mM imidazole washes, 5,6 - 100 mM imidazole elution 7– 150 mM imidazole elution, 8- 200 mM imidazole elution, 9- 250 mM imidazole elution, 10- pure KcsA standard. Panel B lanes: 1,3,4- 50 mM imidazole washes, 2-cell lysate supernatant, 5–6 – 100 mM imidazole elution, 7 – 150 mM imidazole elution, 8– 200 mM imidazole elution, 9–10 – 250mM imidazole elution.
Reconstitution into liposomes and Cryo-EM
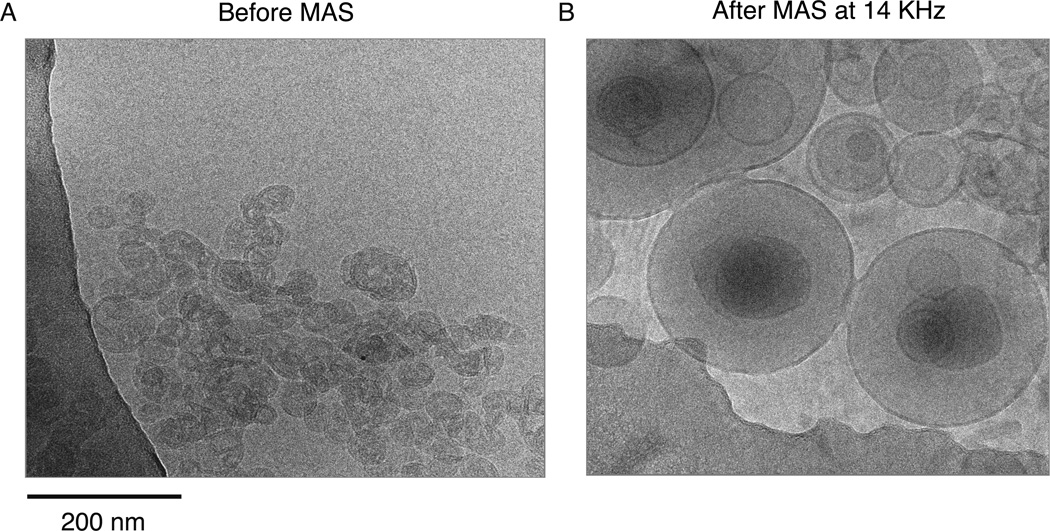
The U-[13C]-[15N]-KcsA was reconstituted into 9:1 DOPE/DOPS liposomes. DOPE (18:1 PE, 1,2-Dioleoyl-sn-Glycero-3-Phosphoethanolamine, Avanti # 850725) has a zwitterionic headgroup and DOPS (18:1 PS, 1,2-Dioleoyl-sn-Glycero-3-[Phospho-L-Serine], Avanti # 840035) has an anionic headgroup. Bacterial membranes are known to be significantly anionic and anionic lipids are known to be important for KcsA function [18,19]. DOPE/DOPS mixtures were prepared with a lipid: lipid weight ratio of 9:1. The chloroform was evaporated under nitrogen and the lipids were resuspended in 50 mM KCl, 50 mM Tris, 0.01 mM sodium azide, 10 mM DM, pH=7.5 buffer to a total lipid concentration of 10 mg/ml. The lipid solution was then added to the desired amount of KcsA in DM such that the protein: lipid weight ratio was 1:1, this leads to a protein: lipid molecular ratio of ~ 1:100 and ensures that there is a layer of lipids surrounding each tetramer of KcsA. The mixture was diluted ~5–10X to around the critical micelle concentration of DM (1.8 mM) with a 50 mM Tris/150 mM KCl buffer at pH=7.5 and dialyzed against a 50 mM Tris/150 KCl buffer at pH=7.5 using a dialysis membrane with a MW cut-off of 12,000–14,000 KDa. The KcsA-DOPE/DOPS proteoliposomes precipitate within a few hours and are visible as a cloudy white precipitate inside the dialysis tube. The kinetics of detergent dialysis are significantly slower above the CMC. We found that over 80% of total pellet yield is obtained within 36 hours of dialysis if the initial concentration is around the CMC, whereas it takes over 72 hours to obtain the same yield if the initial concentration is not controlled. Typically 3–4 buffer exchanges were necessary to get optimal yields. The liposome pellet was then harvested by spinning at 5500g for 30 mins. UV-Vis spectra of the supernatant were acquired to check for any residual protein, which was typically negligible. Pellet volumes for 10 mg lipid, and 10 mg protein were typically ~ 50–70 µl due to excess water in the liposomes. The pellets were then subject to a −80°C overnight freeze-thaw cycle to remove excess bulk water, which reduced the volume to ~ 30–40 µl. This step also made the pellet more manageable to pack into NMR rotors. The presence of liposomes was checked using cryo-electron Microscopy imaging of a proteoliposome suspension diluted to ~ 50 µg/ml with deionized buffer and imaged at the New York Structural Biology Center.
Solid-state NMR characterization
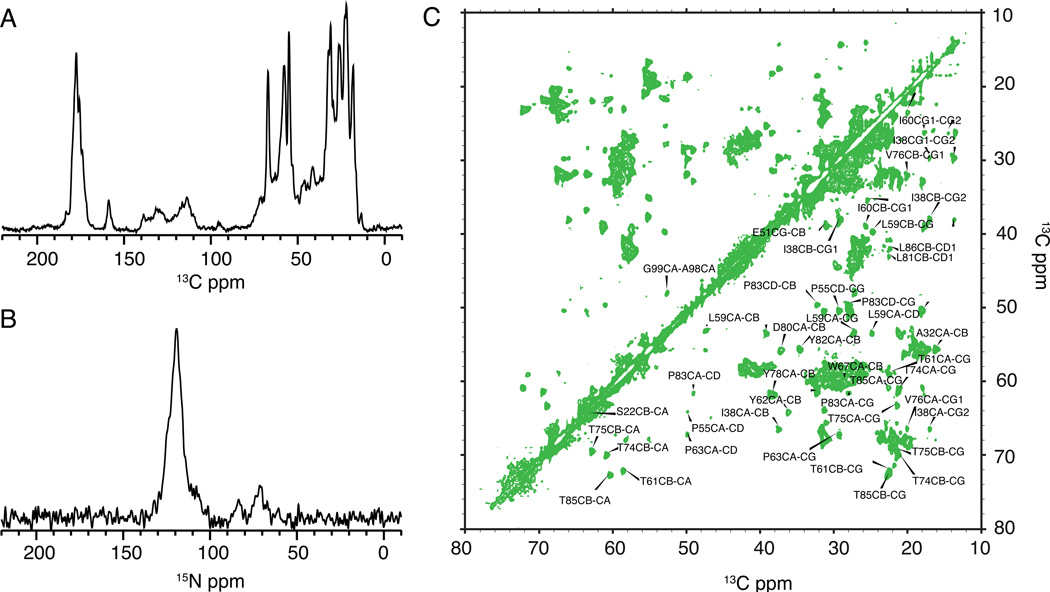
The liposome pellets were centrifuged into 3.2 mm or 4 mm Bruker rotors. Typically 20–30 µl of sample were packed into the rotors, which corresponds to ~ 10 mg of KcsA. 13C and 15N 1D spectra were measured on a Bruker 750 MHz instrument with a triple channel probe. Typically an MAS rate of 12–14 KHz was optimal for the 4 mm probes and an MAS rate of 20 KHz was optimal for the 3.2 mm probes. The temperature of the cooling gas was ~−30 °C which corresponds to a sample temperature of ~ 0–10 °C. 1D 13C and 15N cross-polarization (CP) spectra were used as diagnostics of a good sample. Typical 90° pulse lengths for 1H were ~2.5µs. 13C and 15N 90° pulse lengths were ~ 5µs. 80–85 KHz of SPINAL64[20] decoupling was used on the 1H channel during acquisition with a 3 s recycle delay. Typical 13C and 15N 1D CP spectra of KcsA are shown in Figure 3. High-resolution 13C-13C correlation experiments using the 2D DARR [21] sequence and a 15 ms mixing time show narrow lines for several resonances. Data were acquired for 10 ms in each dimension. 13C chemical shifts were referenced using the downfield adamantane line set to 40.48 ppm. 15N chemical shifts are referenced indirectly relative to 13C using the relative gyromagnetic ratios of 13C to 15N reported by the Biological Magnetic Resonance bank.[22
Figure 3.
Solid-state NMR spectra of ~ 10 mg of uniformly 13C-15N labeled KcsA at 50 mM K+, pH 7.5 measured on a 750 MHz instrument are shown. Panel A shows a C1D CP spectrum acquired with 32 scans acquired at an MAS of 12 KHz and a sample temperature of 0–10 C. Panel B shows a N1D CP spectrum acquired with 128 scans at an MAS of 14 KHz and the same temperature. Panel C shows the Cα-Cβ region of a 13C-13C correlation spectrum of KcsA using the DARR mixing sequence and 15ms mixing. The spectra show well-ordered narrow lineshapes for several resonances
Electrophysiology
Liposome vesicles were prepared by mixing 10 mg/ml of chloroform-solubilized lipids (POPE and POPG; Avanti Polar Lipids) in a 3:1 ratio. The chloroform was removed via evaporation under a nitrogen gas stream. To ensure that all chloroform was removed the lipids were resuspended in one volume of pentane (Sigma), which was subsequently removed via evaporation under a nitrogen gas stream. The lipids were then solubilized in swelling buffer (400 mM KCl, 20 mM Tris, pH 7.5) and 34 mM CHAPS detergent (Affymetrix) and sonicated for several minutes. KcsA samples from solid-state NMR experiments were resuspended in buffer B (100 mM KCl, 20 mM Tris, 5 mM DM, pH 7.5) and reconstituted into liposomes at a protein-to-lipid ratio of 10 µg of protein per mg of lipid. Detergent was removed by running the protein/lipid mixture over a hand-packed G50 fine (GE) gel filtration column in swelling buffer and 100 µl liposome aliquots were flash-frozen in liquid nitrogen and stored at −80°C. Before being used for lipid bilayer recordings, liposomes were thawed and sonicated for 5 sec. Lipid bilayers were formed from a 10 mg/ml POPE: POPG (3:1) mix resuspended in n-decane (Sigma) on a 50-µm partition (transparency slide, IKON) appended with vacuum grease to a larger hole separating two horizontal chambers containing the recording solutions. Since KcsA is a pH gated channel, which opens at low intracellular pH, by placing pH 4 recording solution (70 mM KCl, 30 mM KOH, and 10 mM succinate) in the grounded lower chamber and pH 7 recording solution (70 mM KCl, 30 mM KOH, 10 mM MOPS) in the upper chamber, we made recordings from those channels that were inserted with the extracellular domain in the lower chamber. Liposomes were applied to bilayer in 1 µl aliquots and the insertion of channels into the bilayer was monitored electrically. Currents were recorded in Clampex 10 under continuous mode with an Axopatch 200 A, digitized with a Digidata 1320 (Molecular Devices), sampled at 20 kHz and filtered at 2 kHz. Single-channel current amplitudes were measured by hand using Clampfit 10 (Molecular Devices) and verified using amplitude histograms.
Results and Discussion
Several strategies were used to increase the yield of U-[13C]-[15N] labeled KcsA. By switching from plasmid #1 to plasmid #2 we were able to go from ~6–8 mg/L of M9 to 10–11 mg/L of M9. We then optimized the induction time for protein expression. For the PQE-60/M15 expression system we found that maximum yield (~4–5 mg/L of M9) could be obtained using an induction period of ~3–4 hours and the yield did not change if the induction period was raised. For the PASK-90 vector system, a 12 hour induction period increased the yield from ~10 to ~18 mg/L compared to a 3–4 hour period. We also found that the use of a small amount (10 ml per liter) of 13C-15N Bioexpress increased the yields U-[13C]-[15N]-KcsA to ~ 40 mg/L of M9 using PASK90/JM83 system. This strategy that has been reported previously [16]. A different strategy to increase the yield is to increase the concentration of the initial transfer of cells. We found that while an increase in cell weight between a 2 → 1 to a 4 → 1 transfer is nearly linear, the increase between a 4 → 1 and a 10 → 1 transfer, was not linear, so it was more time-efficient to use the 4 → 1 transfer [15,23]. Typically we harvested ~ 3–3.5 g of cells per liter of LB and transferred 12–14 g of cells into 1 liter of M9. We note that these protocols are valid for wildtype KcsA. Site-specific mutations can affect the yield and the sample homogeneity significantly and must be optimized on a case-by-case basis using this protocol as a starting point.
During purification, the maximum elution for KcsA off the Ni-CAM column was observed at ~ 150 mM imidazole. The KcsA tetramer solubilized in SDS typically runs lower than its expected molecular weight on an SDS page and is seen as a clean band a little above the 50 KDa protein marker. A typical SDS-Page analysis of KcsA is shown in Figure 1A. The characteristic UV absorbance spectrum of KcsA between 320 nm and 250 nm was used to quantify the amount of protein. In order to measure the protein concentration accurately, the detergent concentration must be between 5–10 mM in order to minimize light scattering by the micelles, which leads to an elevated baseline. The UV spectrum of KcsA shows a strong absorbance at 275 and 280 nm and a weaker shoulder at ~ 290 nm. An absorbance of 1 corresponds to ~0.5 mg/ml of the KcsA monomer.
In order to validate the presence of lipid bilayers in the liposome sample, we conducted Cryo-electron microscopy imaging of a dilute liposome pellet. We imaged the pellet before and after solid-state magic-angle spinning and saw that magic angle spinning at 14 KHz changes the single unilamellar vesicles to large multilamellar vesicles as shown in Figure 2. To the best of our knowledge the multilamellar vesicles do not affect the quality of solid-state NMR spectra for the membrane embedded regions of KcsA. We have several cryo-EM images of different NMR samples spun for different lengths of time and they all show similar multilamellar structure. Given that during MAS we spin at speeds of 5–20 KHz i.e 300,000 rpm to 1,200,000 rpm, and that these speeds are achieved within a few minutes of inserting the sample into the magnet, we suspect that the transition to multilamellar vesicles occurs very soon after the MAS is started.
Figure 2.
Cryo-EM images of KcsA proteoliposomes in DOPE/DOPS lipid membranes are shown. In (A) an image of the sample before magic angle spinning shows predominantly small, unilamellar vesicles. Panel (B) shows an aliquot of the sample after magic angle spinning at 14 KHz for several days. The spinning causes these the vesicles to be converted into large multilamellar vesicles, but the bilayer remains intact.
Typical 13C and 15N solid-state NMR spectra of KcsA in liposomes are shown in Figure 3A and 3B. A 13C-13C 2D correlation spectrum is shown in Figure 3C to illustrate the sharp peaks (typically < 100 Hz in the 13C dimension) for individual resonances that can be assigned using the higher dimensional spectra. Spectra were collected at 50 mM K+ and neutral pH. Under these conditions we observe only one set of chemical shifts for each residue in KcsA, indicating that our preparation yields a homogenous and symmetric tetramer. The chemical shifts are reproducible to within 0.2 ppm between different preparations and we have used this protocol several times to produce high quality, high signal: noise 13C-15N labeled KcsA for solid-state NMR studies described elsewhere[24,25].
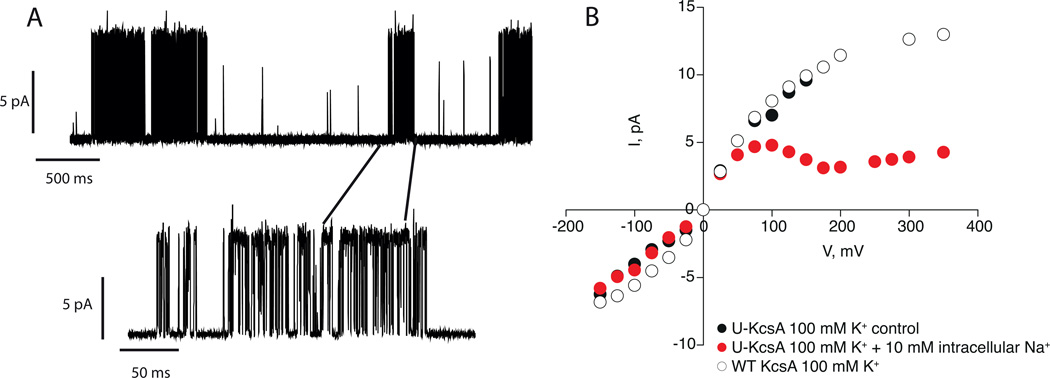
In order to ensure that our protocols yielded stable, functioning channels, we used electrophysiology to measure current-voltage relationships and single channel current traces of KcsA before and after 1 continuous week of solid-state NMR experiments. The results, shown in Figure 4, prove that our preparation yields K+ selective channels that open at a low pH of 4, close at neutral pH and exhibit a current-voltage curves that are characteristic for KcsA.[6,26] Our measurements however, do not reveal how much of the sample is active and if a certain fraction of the sample has been rendered inactive during the experiments. Further work is necessary to clarify this point.
Figure 4.
Electrophysiology results on Uniformly 13C-15N labeled KcsA (U-KcsA). Channels were reconstituted into 3POPE: 1POPG liposomes as described in the paper. Currents were recorded in symmetrical 100 mM K+. In panel A, a single channel trace of U-KcsA at 100 mV is shown. Traces were low pass filtered at 500 Hz. In Panel B the Current-voltage curve for U-KcsA (black circles. −175 mV to 175 mV) and WT KcsA (open circles, −175 mV to 375 mV) is shown. The current-voltage curve is also measured in the presence of 10 mM intracellular Na+ (red circles, −175 mV to 375 mV) to confirm selectivity. The data show that our protocols yield selective and functional K+ channels.
The protocols that we present here document the complete procedure to prepare a membrane protein sample for solid-state NMR studies starting from the plasmid preparation to the lipid reconstitution and NMR spectra. The reproducibility of the protocol is key to make reliable samples to answer biologically important questions about this system. We expect that as more groups begin to study membrane proteins by solid-state NMR, these protocols may be used as a starting point to design and optimize the biochemistry for different systems.
Supplementary Material
Highlights.
-
➢
Protocols to express 40mg/Lit of pure 15N and 13C labeled KcsA, a bacterial K+ channel for NMR studies are reported.
-
➢
Optimized protocols to reconstitute membrane proteins into liposomes for solid-state NMR are shown.
-
➢
Cryo-electron microscopy of liposomes and single-channel measurements of KcsA validate channel function.
-
➢
These methods yield reliable samples for high-resolution NMR spectra of KcsA for biophysical studies.
Acknowledgments
The authors would like to thank Ruben Diaz Avaloz at the New York Structural Biology center for his help with the cryo-EM measurements. We also thank all members of the McDermott and Nimigean groups for help and advice. Work in the McDermott Lab was supported by grants from the National Institutes of Health NIH P41 GM66354 and NIH R01 GM88724 to AEM. AEM is a member of the New York Structural Biology Center where the NMR data was acquired. The center is a STAR center supported by the New York State Offce of Science, Technology and Academic Research. BJW was supported by NIH NRSA F32 087908. Work in the Nimigean lab was supported by NIH grant R01GM088352 and the Irma T. Hirschl trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tang XD, Santarelli LC, Heinemann SH, Hoshi T. Metabolic regulation of potassium channels. ANNUAL REVIEW OF PHYSIOLOGY. 2004;66:131–159. doi: 10.1146/annurev.physiol.66.041002.142720. [DOI] [PubMed] [Google Scholar]
- 2.Doyle D, Cabral J, Pfuetzner R, Kuo A, Gulbis J. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y, Morais-Cabral JH, Kaufman A, Mackinnon R. Chemistry of ion coordination and hydration revealed by a K channel Fab complex at 2.0A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 4.Cuello LG, Jogini V, Cortes DM, Pan AC, Gagnon DG, Dalmas O, et al. Structural basis for the coupling between activation and inactivation gates in K+ channels. Nature. 2010;466:272–275. doi: 10.1038/nature09136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson AN, Posson DJ, Parsa PV, Nimigean CM. Molecular mechanism of pH sensing in KcsA potassium channels. Proc. Nat. Acad. Sci. 2008;105:6900–6905. doi: 10.1073/pnas.0800873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LeMasurier M, Heginbotham L, Miller C. KcsA: it’s a potassium channel. Journal of General Physiology. 2001;118:303–314. doi: 10.1085/jgp.118.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chill JH, Louis JM, Miller C, Bax A. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Protein Sci. 2006;15:684–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhate MP, Wylie BJ, Tian L, McDermott AE. Conformational dynamics in the selectivity filter of KcsA monitored by solid-state NMR. J. Mol. Bio. 2010;401:155–166. doi: 10.1016/j.jmb.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker K a, Tzitzilonis C, Kwiatkowski W, Choe S, Riek R. Conformational dynamics of the KcsA potassium channel governs gating properties. Nat Struct Mol Biol. 2007;14:1089–1095. doi: 10.1038/nsmb1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrempf H, Schmidt O, Kummerlen R, Steinkamp T, Wagner R. A prokaryotic potassium ion channel with two predicted transmembrane segments from streptomyces lividans. EMBO Journal. 1995;14:5170–5178. doi: 10.1002/j.1460-2075.1995.tb00201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heginbotham L, Kolmakova-Partensky L, Miller C. Functional reconstitution of a prokaryotic K+ channel. Journal of General Physiology. 1998;111:741–749. doi: 10.1085/jgp.111.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeuchi K, Takahashi H, Kawano S, Shimada I. Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. The Journal of Biological Chemistry. 2007;282:15179–15186. doi: 10.1074/jbc.M608264200. [DOI] [PubMed] [Google Scholar]
- 13.Heginbotham L, Odessey E, Miller C. Tetrameric stoichiometry of a prokaryotic K+ channel. Biochemistry. 1997;36:10335–10342. doi: 10.1021/bi970988i. [DOI] [PubMed] [Google Scholar]
- 14.Schneider R, Ader C, Lange A, Giller K, Hornig S, Pongs O, et al. Solid-state NMR spectroscopy applied to a chimeric potassium channel in lipid bilayers. Journal of the American Chemical Society. 2008;130:7427–7435. doi: 10.1021/ja800190c. [DOI] [PubMed] [Google Scholar]
- 15.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. Journal of Biomolecular NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Berthold DA, Frericks HL, Gennis RB, Rienstra CM. Partial 13C and 15N chemical-shift assignments of the disulfide-bond-forming enzyme DsbB by 3D magic-angle spinning NMR spectroscopy. ChemBioChem. 2007;8:434–442. doi: 10.1002/cbic.200600484. [DOI] [PubMed] [Google Scholar]
- 17.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Analytical Biochemistry. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 18.Marius P, Zagnoni M, Sandison ME, East JM, Morgan H, Lee AG. Binding of anionic lipids to at least three nonannular sites on the potassium channel KcsA is required for channel opening. Biophysical Journal. 2008;94:1689–1698. doi: 10.1529/biophysj.107.117507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Dalen A, Hegger S, Killian JA, de Kruijff B. Influence of lipids on membrane assembly and stability of the potassium channel KcsA. FEBS Letters. 2002;525:33–38. doi: 10.1016/s0014-5793(02)03061-2. [DOI] [PubMed] [Google Scholar]
- 20.Fung BM, Khitrin AK, Ermolaev K. An Improved Broadband Decoupling Sequence for Liquid Crystals and Solids . Journal of Magnetic Resonance. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 21.Takegoshi K, Nakamura S, Terao T. C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 22.Http://www.bmrb.wisc.edu, Biological Magnetic Resonance Data Bank, (n.d.)
- 23.Williams JC, McDermott a E. Dynamics of the flexible loop of triosephosphate isomerase: the loop motion is not ligand gated. Biochemistry. 1995;34:8309–8319. doi: 10.1021/bi00026a012. [DOI] [PubMed] [Google Scholar]
- 24.Bhate MP, Wylie BJ, Tian L, McDermott AE. Conformational Dynamics in the Selectivity Filter of KcsA in Response to Potassium Ion Concentration. Mol. Bio. J. 2010;401:155–166. doi: 10.1016/j.jmb.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhate MP, McDermott AE. Protonation state of E71 in KcsA and its role for channel collapse and inactivation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:15265–15270. doi: 10.1073/pnas.1211900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nimigean C, Miller C. Na+ block and permeation in K+ channel of known structure. Journal of General Physiology. 2002;120:323–325. doi: 10.1085/jgp.20028614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.