Abstract
Objective
To investigate the distribution and patterns of pfcrt and pfmdr1 polymorphisms in Plasmodium falciparum (P. falciparum) isolates collected from the malaria endemic area of Thailand along Thai-Myanmar border.
Methods
Dried blood spot samples were collected from 172 falciparum malaria patients prior received treatment. The samples were extracted using chelex to obtain parasite DNA. PCR-RFLP was employed to detect pfcrt mutation at codons 76, 220, 271, 326, 356 and 371, and the pfmdr1 mutation at codon 86. Pfmdr1 gene copy number was determined by SYBR Green I real-time PCR.
Results
Mutant alleles of pfcrt and wild type allele of pfmdr1 were found in almost all samples. Pfmdr1 gene copy number in isolates collected from all areas ranged from 1.0 to 5.0 copies and proportion of isolates carrying>1 gene copies was 38.1%. The distribution and patterns of pfcrt and pfmdr1 mutations were similar in P. falciparum isolates from all areas. However, significant differences in both number of pfmdr1 copies and prevalence of isolates carrying>1 gene copies were observed among isolates collected from different areas. The median pfmdr1 copy number in P. falciparum collected from Kanchanaburi and Mae Hongson were 2.5 and 2.0, respectively and more than half of the isolates carried>1 gene copies.
Conclusions
The observation of pfmdr1 wild type and increasing of gene copy number may suggest declining of artesunate-mefloquine treatment efficacy in P. falciparum isolates in this border area.
Keywords: Plasmodium falciparum, Multidrug resistance, Pfcrt, Pfmdr1, Gene mutation, Gene copy number
1. Introduction
Malaria is one of the major infectious diseases that causes a number of deaths in tropical and subtropical countries. In Thailand, the mortality rate had raised to 36 per 100 000 population in 1958, but continuously declined after the launch of malaria control program[1],[2]. Most of the affected populations are those who reside in/near forests and hilly areas along the international borders. The highest incidence has been reported from areas bordering Thai-Myanmar, followed by Thai-Malaysia, Thai-Cambodia and Thai-Laos PDR. A serious problem that limits the effectiveness of malaria control program of the country is the emergence and spread of multidrug resistance Plasmodium falciparum (P. falciparum)[3],[4]. To deal with the situation, the artemisinin-based combination therapy, a three-day artesunate-mefloquine combination is currently being used as first-line treatment of multidrug resistance P. falciparum according to recommendation of World Health Organization[4].
The gold standard for monitoring antimalarial drug efficacy mainly relies on in vivo investigation with supplemented information of in vitro parasite susceptibility. In recent years, attempt has been made to apply valid molecular markers of antimalarial drug resistance to predict treatment outcome following treatment with an antimalarial drug regimen[5]. The two candidate malarial parasite genes, P. falciparum chloroquine resistant transporter (pfcrt) and P. falciparum multidrug resistance 1 (pfmdr1) that express the transport proteins on the plasma membrane of the parasite's food vacuole pfcrt and pfmdr1, respectively, have been confirmed to link with resistance of the parasite to antimalarial drugs[6]. Mutation of pfcrt associated with chloroquine resistance in P. falciparum and distinct genotype polymorphisms depends on its origination. Most of P. falciparum isolates collected from Thailand carry pfcrt mutations at codons K76T, A220S, Q271E, N326S and R371I[7]. The mutation at codon 86 of pfmdr1 (86Y) related with chlolroquine resistance, while pfmdr1 wild type at the same codon (N86) including increased pfmdr1 gene copy number linked to resistance of the parasite to mefloquine and artesunate[8]–[10]. The aim of the present study was to investigate the distribution and patterns of pfcrt and pfmdr1 polymorphisms in P. falciparum isolates collected from the malaria endemic area of Thailand along Thai-Myanmar border.
2. Materials and methods
2.1. Sample collection and DNA extraction
A total of 172 P. falciparum-infected dried blood spot samples were collected prior to treatment, from patients with acute uncomplicated P. falciparum malaria during 2009-2010 from the four malaria endemic areas along Thai-Myanmar border of Thailand, i.e., Mae Hongson (MH, 41 samples), Tak (TK, 82 samples), Kanchanaburi (KN, 6 samples) and Ranong (RN, 43 samples) provinces (Figure 1). Genomic DNA was extracted from each sample using chelex resin according to the previously described method[11].
Figure 1. Map of Thailand presenting the four malaria endemic areas along the Thai-Myanmar border.
2.2. Determination of pfcrt and pfmdr1 single nucleotide polymorphisms
PCR-RFLP was employed to detect pfcrt mutation at codons 76, 220, 271, 326, 356 and 371[12] and pfmdr1 mutation at codon 86[13]. DNA of P. falciparum laboratory clones G112 and K1 served as control for chloroquine sensitive and chloroquine-resistant genotype, respectively.
2.3. Determination of pfmdr1 gene amplification
Pfmdr1 gene copy number in all samples was investigated by SYBR Green I real-time PCR[14]. DNA of 3D7 (1 pfmdr1 copy number) and Dd2 (4 pfmdr1 copy number) P. falciparum laboratory clones provided by Professor Dr. Steven A. Ward (School of Tropical Medicine, Liverpool, UK) were used as the internal control. The copy number was determined by relative quantification between pfmdr1 (target gene) and pfβ-actin (reference gene, an endogenous house-keeping gene which carries only a single copy) that was calculated using the comparative Ct method (also known as the 2−ΔΔCt method).
2.4. Statistical analysis
Qualitative variables were summarized as proportions and percentages, while quantitative variables were presented as median (95% CI). Differences among qualitative variables were determined using Chi-square test. Differences among quantitative variables were determined using Kruskal Wallis test. The statistical significance level was set at α=0.05 for all tests (SPSS version 17; SPSS, Chicago, Illinois, USA).
3. Results
3.1. Pfcrt and pfmdr1 single nucleotide polymorphisms
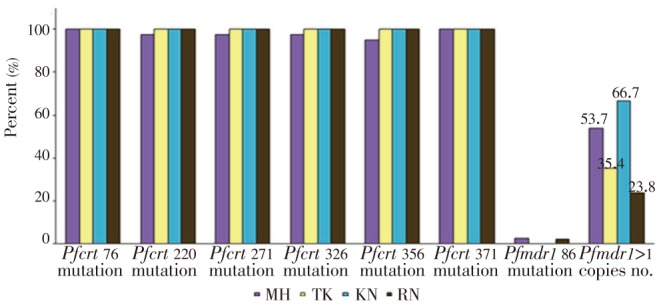
Mutations of the two candidate P. falciparum resistance genes, i.e., seven codons of pfcrt (K76T, A220S, Q271E, A326S, I356T and R371I), one codon of pfmdr1 (N86Y) were successful investigated in 172 P. falciparum isolates. The distribution and patterns of pfcrt and pfmdr1 mutations were similar in P. falciparum isolates collected from the four different endemic areas (Figure 2). All isolates carried mutant allele of pfcrt at codons 76 and 371. The prevalence of mutations at codons 220, 271 and 326 was 99.4%, by only one isolate collected from Mae Hongson was wild type. The mutation at codon 356 was 98.8% of which wild type was observed in two isolates collected from Mae Hongson. In contrast, the pfmdr1 mutation at codon 86 was identified in only two isolates (1.2%) which each from Mae Hongson and Ranong.
Figure 2. Proportions of pfcrt and pfmdr1 polymorphisms in P. falciparum isolates from the four study areas.
3.2. Pfmdr1 gene amplification
Pfmdr1 copy number of 168 evaluable P. falciparum isolates collected from different areas has been summarized in Figure 2. The copy number ranged from 1.0 to 5.0 copies [median (95% CI)=1.0 (1.0-1.0) copies] and proportions of isolates carrying 1 and>1 gene copy number were 61.9% and 38.1%, respectively. Significant differences in both the number of gene copies (P=0.003) and prevalence of isolates carrying>1 gene copies (P=0.017) were observed among isolates collected from different areas. The median copy number and prevalence of isolates carrying more than one copy number were highest in samples from Kanchanaburi (median copy number=2.5, prevalence of isolates carrying >1 gene copies=66.7%), followed by Mae Hongson (median copy number=2.0, prevalence of isolates carrying>1 gene copies=53.7%), Tak (median copy number=1.0, prevalence of isolates carrying>1 gene copies=35.4%) and Ranong (median copy number 1.0, prevalence of isolates carrying>1 gene copies=23.8%).
4. Discussion
Information on distribution and patterns of antimalarial drug resistance is essential for implementation of effective malaria control program and disease surveillance in all malaria endemic areas of the world including Thailand. Early detection of occurrence and spreading of antimalarial drug resistance would be greatly enhanced by the application of valid antimalarial resistance molecular markers. Among the candidate genes investigated to date, pfcrt mutation is widely acceptable as a reliable marker of chloroquine resistance, while pfmdr1 wild type including number of gene copies strong correlate to mefloquine and artesunate resistance in P. falciparum[15]–[17]. Despite the fact that chloroquine was withdrawn from treatment of falciparum malaria in Thailand for many decades, pfcrt mutation at least at the codon 76 was conserved in all P. falciparum isolates. This observation could be explained by a result of natural selection against chloroquine pressure which has been maintained in this area as chloroquine has been used as the first line treatment of Plasmodium vivax infection for more than six decades[4],[18]. Furthermore the drug has always been prescribed in some case with P. falciparum infection due to misdiagnosis of P. falciparum with Plasmodium vivax, thus unintentionally exposing the P. falciparum parasite to subtherapeutic concentration of chloroquine[19],[20].
Mefloquine was introduced for clinical use in Thailand since 1983, and since then, it has effectively select resistance genotype particularly the pfmdr1 codon 86 wild type (N86)[21],[22]. Extensive use of mefloquine in area along Thai-Myanmar border provided significant impact on the intensity and patterns of P. falciparum drug resistance genotypes, the pfmdr1 mutation at codon 86 is gradually replaced by wild type (N86) genotype[21]. Prevalence of pfmdr1 mutation at codon 86 in Tak and Kanchanaburi during 2001-2003 was 2.0% and 5.8%, respectively[23], and it was completely cleared from these areas in 2009-2010. Nevertheless, a number of isolates were found to carry mutant allele of pfmdr1 codon 86, 1 isolate each from Mae Hongson and Ranong. This decreasing trend of pfmdr1 mutation at codon 86 also observed in isolates collected from the Thai-Cambodia border by the prevalence of pfmdr1 mutation at codon 86 was reduced from 11.4% during 1988-1993 to 4.8% in 2003[7].
A three-day combination regimen of artesunate-mefloquine has been adopted as first-line treatment of acute uncomplicated P. falciparum in Thailand since 2005[2]. The observation of good correlation between pfmdr1 and declining of artesunate and mefloquine sensitivity supports results from the current study that both pfmdr1 wild type at codon 86 and amplification of pfmdr1 could apply as reliable tools for prediction of mefloquine and artesunate resistance[15],[24].
With regards to pfmdr1 copy number, the increase of copy number found in some P. falciparum isolates in this study correlates well with the observation of decline in sensitivity of P. falciparum to artesunate and mefloquine and therefore, the chance of recrudescence/treatment failure following treatment with this artemisinin-based combination therapy regimen[25],[26]. Application of the median copy number and prevalence of isolates carrying more than one gene copies can sort out the provinces with relatively high possibility of treatment failure and require close monitoring for drug resistance as Kanchanaburi, followed by Mae Hongson, Tak and Ranong, respectively.
As mefloquine resistance had occurred in these areas before the implementation of artesunate-mefloquine combination, emergence of P. falciparum resistance to artesunate is alarming and of greater concern since artesunate-mefloquine combination regimen is being used as first-line treatment for falciparum malaria in Thailand. In recent years, evidence of confirmed emerging resistance to artemisinins has been gathered from clinical studies in western Cambodia as well as the bordering areas between Thailand and Myanmar[27]–[30]. Furthermore, the observation of pfmdr1 wild type and increasing of gene copy number may suggest declining of artesunate-mefloquine treatment efficacy in P. falciparum isolates in this border area. Continued monitoring of distribution and patterns of these two candidate molecular markers of antimalarial drug resistance is essential for adjusting the policy of malarial control program of the country.
Acknowledgments
This work was supported by the National Research University Project (NRU) of Thailand (Grant No. 10/2555) and Thammasat University (Grant for student No. 33/2555).
Comments
Background
Malaria is usually found in area along Thai-international border. Several antimalarial drugs were used in malaria treatment due to spreading of multidrug resistance P. falciparum. The drug resistance P. falciparum developed by gene mutation and/or gene amplification. However, alteration of prevalence of polymorphisms always occurs after changed treatment policy.
Research frontiers
This work investigated prevalence of pfcrt and pfmdr1 polymorphisms in P. falciparum collected from 4 provinces of malaria endemic area along Thai-Myanmar border during 2009–2010 using PCR-RFLP and real-time PCR. The prevalence of gene mutation and amplification including median copy number were compared among the sample collection areas.
Related reports
Food vacuole is a location for heam degradation. Drug accumulation in food vacuole correlates to treatment efficacy of chloquine, mefloquine and may also artesunate. Mutations of the drug transport proteins lead to resistance. Previous publications confirmed relationship between pfcrt mutations and chloroquine resistance while pfmdr1 mutation at codon 86 and gene amplification contribute to declining of mefloquine and artesunate efficacy.
Innovations and breakthroughs
Using of pfcrt and pfmdr1 polymorphims reveal P. falciparum resistance situation to chloroquine and artesunate-mefloquine in Kanchanaburi, Mae Hongson, Tak and Ranong during 2009-2010. Even the four provinces located at the same border, the prevalence of isolates carried more than one pfmdr1 copy number including median copy number which linked to artesunate and mefloquine resistance, are significantly different among the areas that means to different degree of drug resistance.
Applications
Investigation of antimalarial resistance molecular markers of chloroquine including artesunate-mefloquine combination, pfcrt and pfmdr1, respectively can apply for monitoring of emerging of the antimalarial drug resistance and further adapt to estimate treatment efficacy of artesunate-mefloquine combination regimen indirectly instead of in vivo study. In addition, the information also useful in processes of treatment policy designs.
Peer review
This study is very interesting and applicable that established prevalence of pfcrt and pfmdr1 polymorphims in provinces along Thai- Myanmar border after chloroquine withdrawal and estimately 15 years of artesunate and mefloquine usage. Application of well known drug resistance molecular markers; pfcrt and pfmdr1 could be applied for surveillance of chloroquine, artesunate and mefloquine resistance progression among the four provinces. Furthermore, the degree of resistance estimated from this study is advantage to classify requirement of intensive monitoring in each location.
Footnotes
Foundation Project: Supported by the National Research University Project (NRU) of Thailand (Grant No. 10/2555) and Thammasat University (Grant for student No. 33/2555).
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Aungkulanon S, McCarron M, Lertiendumrong J, Olsen S, Bundhamcharoen K. Infectious disease mortality rates, Thailand, 1958–2009. Emerg Infect Dis. 2012;18(11):1794–1801. doi: 10.3201/eid1811.120637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Na-Bangchang K, Congpuong K. Current malaria status and distribution of drug resistance in East and Southeast Asia with special focus to Thailand. Tohoku J Exp Med. 2007;211(2):99–113. doi: 10.1620/tjem.211.99. [DOI] [PubMed] [Google Scholar]
- 3.Kaewpitoon S, Rujiragul R, Namwichaisirikul N, Churproong S, Ueng-arporn N, Matrakool L, et al. Malaria in Thailand 2007–2011. Srinagarind Med J. 2012;27(Suppl):118–120. [Google Scholar]
- 4.Na-Bangchang K, Karbwang J. Current status of malaria chemotherapy and the role of pharmacology in antimalarial drug research and development. Fundam Clin Pharmacol. 2009;23:387–409. doi: 10.1111/j.1472-8206.2009.00709.x. [DOI] [PubMed] [Google Scholar]
- 5.Marfurt J, Müller I, Sie A, Oa O, Reeder J, Smith T, et al. The usefulness of twenty-four molecular markers in predicting treatment outcome with combination therapy of amodiaquine plus sulphadoxine-pyrimethamine against falciparum malaria in Papua New Guinea. Malar J. 2008;7:61. doi: 10.1186/1475-2875-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez C, Dave A, Stein W, Lanzer M. Transporters as mediators of drug resistance in Plasmodium falciparum. Int J Parasitol. 2010;40(10):1109–1118. doi: 10.1016/j.ijpara.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Mungthin M, Suwandittakul N, Chaijaroenkul W, Rungsrihirunrat K, Harnyuttanakorn P, Seugorn A, et al. The patterns of mutation and amplification of Plasmodium falciparum pfcrt and pfmdr1 genes in Thailand during the year 1988 to 2003. Parasitol Res. 2010;107(3):539–545. doi: 10.1007/s00436-010-1887-x. [DOI] [PubMed] [Google Scholar]
- 8.Eyase F, Akala H, Ingasia L, Cheruiyot A, Omondi A, Okudo C, et al. The role of Pfmdr1 and Pfcrt in changing chloroquine, amodiaquine, mefloquine and lumefantrine susceptibility in western-Kenya P. falciparum samples during 2008–2011. PLoS One. 2013;8(5):64299. doi: 10.1371/journal.pone.0064299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim P, Alker A, Khim N, Shah N, Incardona S, Doung S, et al. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar J. 2009;8:11–17. doi: 10.1186/1475-2875-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witkowski B, Iriart X, Soh P, Menard S, Alvarez M, Naneix-Laroche V, et al. Pfmdr1 amplification associated with clinical resistance to mefloquine in West Africa: implications for efficacy of artemisinin combination therapies. J Clin Microbiol. 2010;48(10):3797–3799. doi: 10.1128/JCM.01057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wooden J, Gould E, Paull A, Sibley C. Plasmodium falciparum: a simple polymerase chain reaction method for differentiating strains. Exp Parasitol. 1992;75(2):207–212. doi: 10.1016/0014-4894(92)90180-i. [DOI] [PubMed] [Google Scholar]
- 12.Fidock D, Nomura T, Talley A, Cooper R, Dzekunov S, Ferdig M, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein Pfcrt and evidence for their role in chloroquine resistance. Mol Cell. 2000;6(4):861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djimdé A, Doumbo O, Cortese J, Kayentao K, Doumbo S, Diourté Y, et al. A molecular marker for chloroquine resistant falciparum malaria. N Engl J Med. 2001;334(4):257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira I, Rosario V, Cravo P. Real-time quantitative PCR with SYBR Green I detection for estimating copy numbers of nine drug resistance candidate genes in Plasmodium falciparum. Malar J. 2006;5:1. doi: 10.1186/1475-2875-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaijaroenkul W, Wisedpanichkij R, Na-Bangchang K. Monitoring of in vitro susceptibilities and molecular markers of resistance of Plasmodium falciparum isolates from Thai-Myanmar border to chloroquine, quinine, mefloquine and artesunate. Acta Trop. 2010;113(2):190–194. doi: 10.1016/j.actatropica.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Mehlotra R, Mattera G, Bockarie M, Maguire J, Baird J, Sharma Y, et al. Discordant patterns of genetic variation at two chloroquine resistance loci in worldwide populations of the malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 2008;52(6):2212–2222. doi: 10.1128/AAC.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinayak S, Alam M, Sem R, Shah N, Susanti A, Lim P, et al. Multiple genetic backgrounds of the amplified Plasmodium falciparum multidrug resistance (pfmdr1) gene and selective sweep of 184F mutation in Cambodia. J Infect Dis. 2010;201(10):1551–1560. doi: 10.1086/651949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang F, Tang L, Yang H, Zhou S, Liu H, Li J, et al. Molecular epidemiology of drug resistance markers of Plasmodium falciparum in Yunnan Province, China. Malar J. 2012;11:243. doi: 10.1186/1475-2875-11-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown T, Smith L, Oo E, Shawng K, Lee T, Sullivan D, et al. Molecular surveillance for drug-resistant Plasmodium falciparum in clinical and subclinical populations from three border regions of Burma/Myanmar: cross-sectional data and a systematic review of resistance studies. Malar J. 2012;19:333. doi: 10.1186/1475-2875-11-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barber B, William T, Grigg M, Yeo T, Anstey N. Limitations of microscopy to differentiate Plasmodium species in a region co-endemic for Plasmodium falciparum, Plasmodium vivax and Plasmodium knowlesi. Malar J. 2013;12:8. doi: 10.1186/1475-2875-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lekana-Douki J, Dinzouna Boutamba S, Zatra R, Zang Edou S, Ekomy H, Bisvigou U, et al. Increased prevalence of the Plasmodium falciparum Pfmdr1 86N genotype among field isolates from Franceville, Gabon after replacement of chloroquine by artemether-lumefantrine and artesunate-mefloquine. Infect Genet Evol. 2011;11(2):512–517. doi: 10.1016/j.meegid.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Wongsrichanalai C, Meshnick S. Declining artesunate-mefloquine efficacy against falciparum malaria on the Cambodia-Thailand border. Emerg Infect Dis. 2008;14(5):716–719. doi: 10.3201/eid1405.071601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Congpuong K, Na Bangchang K, Mungthin M, Bualombai P, Wernsdorfer W. Molecular epidemiology of drug resistance markers of Plasmodium falciparum malaria in Thailand. Trop Med Int Health. 2005;10(8):717–722. doi: 10.1111/j.1365-3156.2005.01450.x. [DOI] [PubMed] [Google Scholar]
- 24.Zatra R, Lekana-douki J, Lekoulou F, Bisvigou U, Ngoungou E, Ndouo F. In vitro antimalarial susceptibility and molecular markers of drug resistance in Franceville, Gabon. BMC Infect Dis. 2012;12:307. doi: 10.1186/1471-2334-12-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muhamad P, Phompradit P, Sornjai W, Maensathian T, Chaijaroenkul W, Rueangweerayut R, et al. Polymorphisms of molecular markers of antimalarial drug resistance and relationship with artesunate-mefloquine combination therapy in patients with uncomplicated Plasmodium falciparum malaria in Thailand. Am J Trop Med Hyg. 2011;85(3):568–572. doi: 10.4269/ajtmh.2011.11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phompradit P, Wisedpanichkij R, Muhamad P, Chaijaroenkul W, Na-Bangchang K. Molecular analysis of pfatp6 and pfmdr1 polymorphisms and their association with in vitro sensitivity in Plasmodium falciparum isolates from the Thai-Myanmar border. Acta Trop. 2011;120(1–2):130–135. doi: 10.1016/j.actatropica.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Noedl H, Se Y, Schaecher K, Smith B, Socheat D, Fukuda M. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359(24):2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 28.Noedl H, Se Y, Sriwichai S, Schaecher K, Teja-Isavadharm P, Smith B, et al. Artemisinin resistance in Cambodia: a clinical trial designed to address an emerging problem in Southeast Asia. Clin Infect Dis. 2010;51(11):82–89. doi: 10.1086/657120. [DOI] [PubMed] [Google Scholar]
- 29.Noedl H, Socheat D, Satimai W. Artemisinin-resistant malaria in Asia. N Engl J Med. 2009;361(5):540–541. doi: 10.1056/NEJMc0900231. [DOI] [PubMed] [Google Scholar]
- 30.Satimai W, Sudathip P, Vijaykadga S, Khamsiriwatchara A, Sawang S, Potithavoranan T, et al. Artemisinin resistance containment project in Thailand. II: Responses to mefloquine-artesunate combination therapy among falciparum malaria patients in provinces bordering Cambodia. Malar J. 2012;11:300. doi: 10.1186/1475-2875-11-300. [DOI] [PMC free article] [PubMed] [Google Scholar]


