Abstract
Objective
To isolate and identify Bacillus subtilis (B. subtilis) from soil and to characterize and partially purify the bacteriocin. To evaluate the antimicrobial activity against four diabetic foot ulcer bacterial pathogens.
Methods
Genotypic identification was done based on Bergey's manual of systemic bacteriology. Antimicrobial susceptibility test was done by Kirby-Bauer disc diffusion method. Colonies were identified by colony morphology and biochemical characterization and also compared with MTCC 121 strain. Further identification was done by 16S rRNA sequencing. Inhibitory activities of partially purified bacteriocin on all the DFU isolates were done by agar well diffusion method. The strain was identified to produce bacteriocin by stab overlay assay. Bacteriocin was extracted by organic solvent extraction using chloroform, further purified by HPLC and physical, and chemical characterization was performed.
Results
The four isolates showed high level of resistance to amoxyclav and sensitivity to ciprofloxacin. HPLC purification revealed that the extracts are bacteriocin. The phylogenetic tree analysis results showed that the isolate was 99% related to B. subtilis BSF01. The results reveled activity to all the four isolates and high level of activity was seen in case of Klebsiella sp.
Conclusions
Partially purified bacteriocin was found to have antimicrobial activity against the four diabetic foot ulcer bacterial pathogens, which can thus be applied as a better drug molecule on further studies. The strain B. subtilis are found to be safe for use and these antimicrobial peptides can be used as an antimicrobial in humans to treat DFU bacterial pathogens.
Keywords: 16S rRNA, Antimicrobial activity, Micrococcus leuteus, HPLC, Physicochemical characterization
1. Introduction
Diabetic patients who develop foot ulcers are at more risk of dying prematurely than those without the complication. Those patients with foot ulcers and diabetes showed more cardiovascular risk factors, such as high blood pressure, and were more likely to die from cardiovascular causes. Approximately half of the additional mortalities were due to cardiovascular disease, such as heart attack or stroke[1]. Mostly these diabetes related foot ulcers occur in the presence of peripheral sensory neuropathy, foot deformity and/or trauma, with peripheral arterial disease and infection being further complicating factors that prevent or delay ulcer healing[2]. Most often these ulcers are polymicrobial. Both gram positive and gram negative organisms are seen. Several gram positive bacterias such as Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus sp., Enterococcus sp. (mostly Enterococcus faecalis) and Corynebacterium sp. inhabitate gram negative bacteria that account for DFU are Proteus sp., Pseudomonas sp., Enterobacteriaceae such as E. coli, Klebsiella, Citrobacter and Enterobacter[3],[4]. To date, no topical anti-infective agent has been proven to be effective for treating DFI[5].
Bacteriocins are unique and assorted group of molecules produced by living organisms of all types and considered to be part of the host innate immunity[6]. Thus they are promising candidates for the treatment of various skin infections[5]. Ribosomally synthesized antimicrobial polypeptides, bacteriocins are usually inhibitory only to strains closely related to the producing bacteria. Bacteriocins from gram positive bacteria are often membrane-permeabilizing cationic peptides with fewer than 60 amino acid residues[7],[8]. Clinical antimicrobial used of RAMPs is because they are active against antibiotic resistant isolates and have limited natural bacterial resistance, which are proven to kill bacteria in animal model etc[9]. Bacteriocins are of different groups. Small peptides that undergo extensive post-translational modification to produce the active peptides example nisin are grouped as Class I bacteriocins. Heat stable, low molecular weight, membrane active peptides are grouped as Class II bacteriocins. Members of Class III are large heat labile proteins, and a fourth class (complex bacteriocin) has also been suggested, requiring nonprotein moieties for activity.
Bacteriocins produced by gram positive bacteria have been largely studied and also biochemically and genetically characterized. Bacteriocin activity is very specific. Due to difference in cell wall composition, the activity spectra by gram positive bacteria is wider when compare to gram negative bacteria[10]. The genus Bacillus encompasses a number of bacteriocinogenic species, such as Bacillus subtilis (B. subtilis) which produces subtilin and subtilosin, Bacillus coagulans which produces coagulin and Bacillus megatirium which produces megacin. Bacillus thuringiensis is widely used in agriculture for the control of many insect pathogens[11]. Because of the safety use in humans, probiotic strain B. subtilis is of great interest[12]. It is found that subtilin and subtilosin from B. subtilis are active against many strains of gram positive bacteria and these are the only bacteriocins of Bacillus sp. to be characterized at the amino acid and DNA sequence level[13]. The present study was designed to isolate and identify B. subtilis from soil and to characterize and partially purify the bacteriocin.
2. Materials and methods
2.1. Isolation and identification of diabetic foot ulcer bacterial pathogens
Wound swabs of diabetic foot ulcer patients were collected from Ideal Clinic, Coimbatore and were processed by standard microbiological analysis. Genotypic identification was done based on Bergey's manual of systemic bacteriology.
2.2. Antimicrobial susceptibility testing
To analyze the antibiogram of the isolates, antimicrobial susceptibility test was done by Kirby-Bauer disc diffusion method. The antibiotic discs that are currently used in diabetic foot ulcer treatment were used, amoxyclave 30 µg, ceftriaxone 30 µg, ceftazidime 30 µg, cephalothin 30 µg, ampicillin 25 µg, co-trimoxazole 25 µg, ciprofloxacin 30 µg, chloramphenicol 30 µg, gentamycin 30 µg and Amikacin µg.
2.3. Isolation of Bacillus from soil
Approximately 1 g of soil sample collected was placed in 5 mL distilled water and vortexed vigorously to dissolve the particles. The soil sample was boiled for 10 min in water bath and allowed to cool. Ten-fold, one hundred-fold and one thousand-fold dilution of the re-suspended soil samples were made and 0.1 mL from each sample including the undiluted solution was plated on nutrient agar plates. The plates were incubated overnight at 37 °C[11]. After incubation, the colonies were identified by colony morphology and biochemical characterization and also compared with MTCC 121 strain.
2.4. Identification and confirmation of bacterial strain
Further identification was done by 16S rRNA sequencing. PCR was used to amplify the 16S ribosomal DNA and was determined by direct sequencing. Total DNA was isolated by using SoluteReady® genomic DNA purification kit. Agarose gel (2%) were used to isolate 16S rRNA. 16S rRNA was separated by gel electrophoresis on gel made with 2% acrylamide and bis-acrylamide. The confirmed strain was then screened for bacteriocin production.
2.5. Detection of the strain to produce bacteriocin
2.5.1. Stab overlay assay
The 24 h culture of B. subtilis was stabbed on nutrient agar plate and incubated at 37 °C for 24 h. After incubation, the colonies on the stabbed area were scrapped and the plate was exposed to chloroform vapors for 15 min. Then soft agar was prepared and 24 h old indicator organism (Micrococcus luteus) was added and vortexed and was poured on stabbed plate and incubated for 24 h at 37 °C. After incubation, plates were examined for zone of inhibition around the stabbed area[12].
2.6. Bacteriocin extraction and purification
Nutrient broth was seeded with B. subtilis and incubated at 37 °C at 150 r/min for 24 h. The sample was centrifuged at 10 000 r/min for 20 min, and the supernatant was collected. The culture supernatant (100 mL) was stirred vigorously with chloroform (100 mL) and transferred in a separating funnel. The interface layer between the aqueous and organic phase, which contain bacteriocin was harvested, and the residual chloroform was eliminated by speed vacuum[14].
2.7. Purification by HPLC
The purification of the bacteriocin was carried out by using HPLC. This analytical technique has been shown to be extremely valuable for the analysis of these peptide antibiotics, since peptide antibiotics are generally resistant to different organic solvents used as mobile phase and the high pressure employed through the chromatographic process. Here, the chromatographic process was carried with C18 column. The mobile phase was water/trifluoroacetic acid as eluent A and acetonitril/trifluoroacetic acid as eluent B. The flow rate was 1 mL/min. The peaks were measured by UV-photo diode array detector.
2.8. Sensitivity to temperature, pH and enzymes
To determine the effect of temperature on bacteriocin activity, aliquots of partially purified bacteriocin were incubated at various temperatures (40, 60, 80 and 100 °C) for 30 min. The residual bacteriocin activity was determined by agar well diffusion method.
Effect of pH on bacteriocin was determined by adjusting the pH of the partially purified bacteriocin with diluting HCl and NaOH as 3.0, 4.0, 9.0 and 10.0 and incubated for 2 h at 37 °C, and all samples were then readjusted to neutral pH and then tested for antimicrobial activity.
For enzyme stability, partially purified bacteriocin was treated for 2 h with enzyme lysozyme at a final concentration of 1 mg/mL. Bacteriocin activity was analyzed by agar well diffusion method against indicator organism. In all the three cases, untreated partially purified bacteriocin served as control.
2.9. Molecular weight determination by SDS-PAGE
The molecular weight of the isolated bacteriocin was identified by performing SDS-PAGE using 12% gel. Following electrophoresis which was conducted at 50 V for 40 min, the gel was stained with coomassic brilliant blue.
2.10. Antimicrobial activity of partially purified bacteriocin on DFU bacterial isolates
Inhibitory activities of partially purified bacteriocin on all the DFU isolates were done by agar well diffusion method.
3. Results
3.1. Isolation and identification of DFU bacterial pathogen
Most of the samples processed were polymicrobial on standard microbiological analysis. The organisms were identified as Pseudomonas sp., Staphylococcus sp., Klebsiella sp. and Proteus sp.
3.2. Antimicrobial susceptibility testing
The four isolates showed high level of resistance to amoxyclave and sensitivity to ciprofloxacin.
3.3. Isolation and confirmation of B. subtilis
B. subtilis was isolated from soil. The strain was identified by morphological biochemical, and species level identification which was done by 16S r RNA sequencing. The sequence length was found to be 34, 5′GCC CAA CTA AAT GAT GGC AAC TAA AAT CAA GGG T3′. The phylogenetic tree analysis results showed that the isolate was 99% related to B. subtilis BSF01 (Figure 1, 2 and Table 1).
Figure 1. BLAST hits on the query sequence for B. subtilis.
Figure 2. BLAS tree for B. subtilis.
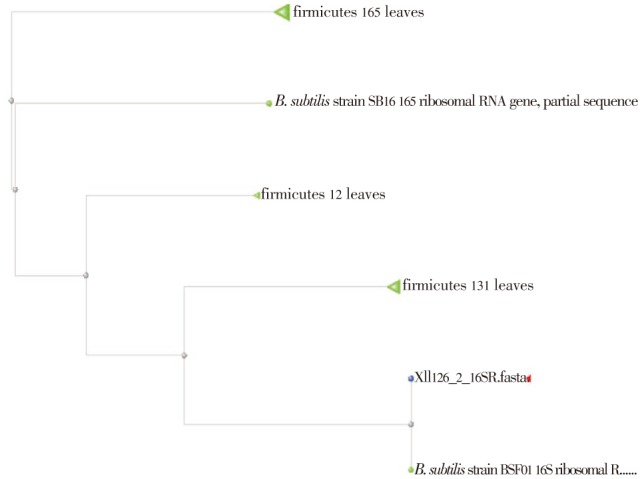
Table 1. Accession description.
| Accession number | Organism | Maximum score | Total score | Query coverage% | E value | Maximum identity % |
| JF706263.1 | B. subtilis strain BSF01 | 67.9 | 67.9 | 100 | 2e-12 | 100 |
| HM133949.1 | B. subtilis strain BGSm254 | 60.0 | 60.0 | 100 | 5e-10 | 97 |
| JN215487.1 | B. subtilis strain MB 2 | 60.0 | 60.0 | 100 | 5e-10 | 97 |
3.4. Screening of the isolate for bacteriocin production
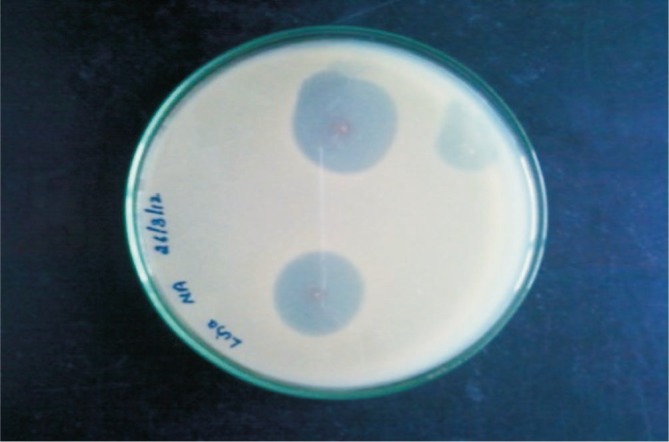
The ability to the isolate for bacteriocin production by stab overlay assay used Micrococcus luteus as the indicator organism. It exhibited antibacterial activity producing a clear zone of inhibition around the indicator strain (Figure 3).
Figure 3. Stab overlay assay plate showed a clear zone of inhibition of indicator strain.
3.5. Bacteriocin production and purification
Bacteriocin production was carried out in nutrient broth at 37 °C and incubated for 24 h. The sample was centrifuged and the supernatant was collected and mixed with chloroform for the extraction of bacteriocin in a separating funnel and mixed well for 30 min. The intermediate layer was collected and further purification was done by HPLC. The peaks revealed that the extracts are bacteriocin. The peaks obtained had a height of 139 619 and 19 797 with Rf values of 2.082 and 112.486 respectively.
3.6. Effect of various temperature, pH and enzymes
The effect of various temperatures on partially purified bacteriocin was investigated. Bacteriocin activity was not affected in all temperature ranging from 40–100 °C. pH stability was observed in both acidic and basic pH. High level of activity was seen in acidic pH. On treatment of partially purified bacteriocin with lysozyme, it showed reduced level of activity (Table 1).
3.7. Molecular weight determination
The molecular weight of the isolated bacteriocin was analyzed by 12% SDS-PAGE with bovine serum albumin as protein marker. The band revealed the molecular weight of the isolated bacteriocin got on comparison with the standard protein marker to be 6.4 kDa.
3.8. Antimicrobial activity of partially purified bacteriocin on DFU bacterial pathogens
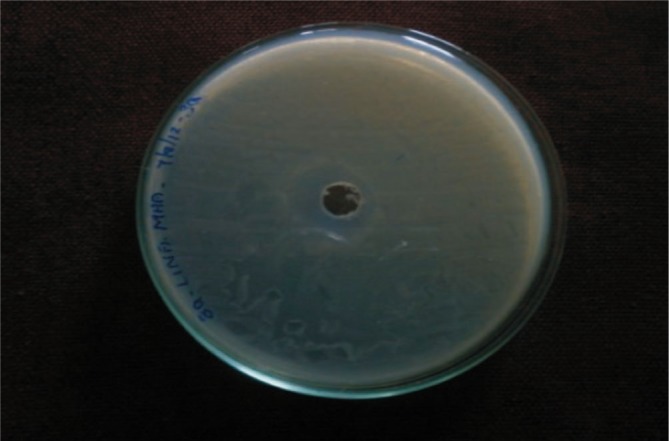
To determine the antimicrobial activity of partially purified bacteriocin, agar well diffusion method was adopted and the results revealed activity to all the four isolates. High level of activity was seen in case of Klebsiella sp. (Figure 4).
Figure 4. Antimicrobial activity of partially purified bacteriocin against Klebsiella sp.
4. Discussion
Multifactorial process is involved in diabetic foot ulceration. The factors are neuropathy, abnormal foot biomechanics and peripheral arterial disease. Infection occurs following the traumatic injury with introduction of bacteria. The diabetic ulcer infections are polymicrobial, harboring anaerobic organisms synergistically with aerobes. In superficial wounds, aerobic bacterias are predominant pathogens. Anaerobic organisms are found in deeper wounds. Failure to recognize and control of the infectious process may have devastating consequences, such as limb amputation, sepsis and mortality[15].
Multidrug resistant organisms were common in hospitalized patients with diabetic foot ulcers. High level of resistance was seen with normally used antibiotics like oxacillin, vancomycin. Imipenem, piperacillin/tazobactam and cefoperazone/sulbactam were the agents which were most effective against gram negative organisms. While vancomycin was effective against the gram positive organisms. Infection with these organisms may limit the choice of the antibiotic treatment and lead to worse outcomes[16]. In the present study, polymicrobial nature of the DFU was evident and they were also found to be multidrug resistant. Since there is an increasing rate of multidrug resistant organisms, there is a need for continuous surveillance to provide the basis of the empirical therapy and to reduce the risk of the complications.
Bacteriocins are bacterially produced peptides that are active against other bacteria and against which the producer has a specific immunity mechanism. They are produced by all major lineages of bacteria and archaea, and constitute heterogeneous group of peptides with respect to size, structure, mode of action, antimicrobial potency, immunity mechanisms and target cell receptors. Bacteriocins may function as colonizing peptides, facilitating the introduction and/or dominance of a producer into an already occupied niche. Alternatively, bacteriocins may act as antimicrobial or killing peptides, directly inhibiting competing strains or pathogens. Lastly, bacteriocins may function as signaling peptides, either signaling other bacteria through quorum sensing and bacterial cross talk within microbial communities or signaling cells of the host immune system[17]. In recent years, groups of antibacterial protein produced by gram positive bacteria have attracted great interest in their potential use as food preservative[18]. In the present study, B. subtilis was the organism of interest.
Bacillus which is a soil inhabitant bacterium is found to have the ability to produce several antimicrobial compounds, which were also found to be safe to use. In the present study, Bacillus was isolated from soil and was confirmed as B. subtilis by 16 S rRNA which serves as an effective tool in genotypic identification of organisms. The screening for bacteriocin production was done by stab overlay assay. B. subtilis was outstanding in the genus Bacillus with regards to its potential to produce so many different antibiotics[19]. High antibacterial activity of the solvent extracted bacteriocin was seen in the study by applied chloroform extraction to recover lacidin, pediocin, nisin, bacilli and subtilin. On analyzing the phsico chemical characterization, the partially purified bacteriocin showed a wide range of activity towards various temperature, pH, enzymes and chemicals. The partially purified bacteriocin showed antimicrobial activity towards all the bacterial isolates where a high level of activity was found against Klebsiella sp. a capsulated bacteria. This proves that the bacteriocin from B. subtilis have high antimicrobial activity.
The findings of the present study lead us to conclude that the bacteriocin producing Bacillus strains could easily be isolated from soil. The bacteriocin activity of the isolate has been made clear with stab overlay assay. Production of bacteriocin was done in nutrient broth and extraction of bacteriocin was well achieved by using chloroform as the organic solvent. Since bacteriocin and the strain B. subtilis are found to be safe for use, these antimicrobial peptides can be used as an antimicrobial in humans to treat DFU bacterial pathogens.
Acknowledgments
We extend our sincere thanks to Ideal Clinic Coimbatore, Synergy Scientific Services, Chennai and South Indian Textile Association, Coimbatore. The supporting agency: CSIR, Head HRDG, New Delhi (Grant No: 27/0237/10 EMR II).
Comments
Background
Bacteriocin is an important source of chemical compound to control bacterial pathogen. This compound is very specific being an alternative to common antibiotics.
Research frontiers
The present paper aimed the discovery a new bacteriocin. However, the methodology and the results are not clear and confuse. A restructuration is vital to publish the present manuscript.
Related reports
The results are not clear, being impossible related it with other reposts. The discussion is very poor too.
Innovations and breakthroughs
The new bacteriocin should be very well applied to control bacterial pathogen that are described as antibiotic resistant.
Applications
Control of bacterial human pathogen.
Peer review
The research is important to medical control of ulcer disease, being alternative to control antibiotic resistant bacterial pathogen.
Footnotes
Foundation Project: CSIR, Head HRDG, New Delhi (Grant No. 27/0237/10 EMR II)
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Brownrigg JR, Davey J, Holt PJ, Davis WA, Thompson MM, Ray KK, et al. The association of ulceration of the foot with cardiovascular and all-cause mortality in patients with diabetes: a meta-analysis. Diabetologia. 2012;55:2906–2912. doi: 10.1007/s00125-012-2673-3. [DOI] [PubMed] [Google Scholar]
- 2.Bergin SM, Gurr JM, Allard BP, Holland EL, Horsley MW, Kamp MC, et al. Australian diabetes foot network: management of diabetes-related foot ulceration-a clinical update. Med J Aust. 2012;197:226–229. doi: 10.5694/mja11.10347. [DOI] [PubMed] [Google Scholar]
- 3.Gontcharova V, Youn E, Sun Y, Wolcott RD, Dowd SE. A comparison of bacterial composition in diabetic ulcers and contralateral intact skin. Open Microbiol J. 2010;4:8–19. doi: 10.2174/1874285801004010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banashankari GS, Rudresh HK, Harsha AH. Prevalence of gram negative bacteria in diabetic foot-A clinico microbiological study. Al Ameen J Med Sci. 2012;5:224–232. [Google Scholar]
- 5.Lipsky BA, Holroyd KJ, Zasloff M. Topical versus systemic antimicrobial therapy for treating mildly infected diabetic foot ulcers: a randomized, controlled, double-blinded, multicenter trial of pexiganam cream. Clin Infect Dis. 2008;47:1537–1545. doi: 10.1086/593185. [DOI] [PubMed] [Google Scholar]
- 6.Smajs D, Micenková L, Smarda J, Vrba M, Sevčíková A, Vališová Z, et al. Bacteriocin synthesis in uropathogenic and commensal Escherichia coli: colicin E1 is a potential virulence factor. BMC Microbiol. 2010;10:288–292. doi: 10.1186/1471-2180-10-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jack RW, Tagg JR, Ray B. Bacteriocin of gram positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klaenhammer TR. Genetics of bacteriocin produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–85. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 9.Pag U, Oedenkoven M, Papo N, Oren Z, Shai Y, Sahl HG. In vitro activity and mode of action of diastereometric antimicrobial peptides against bacterial clinical isolates. J Antimicrob Chemother. 2004;53:230–239. doi: 10.1093/jac/dkh083. [DOI] [PubMed] [Google Scholar]
- 10.Ansari A, Aman A, Siddiqui NN, Iqbal S, Ali ul Qader S. Bacteriocin (BAC-IB17): Screening, isolation and production from Bacillus subtilis KIBGE IB-17. Pak J Pharm Sci. 2012;25:195–201. [PubMed] [Google Scholar]
- 11.Okulate MA. Antimicrobial activity of bioactive compounds produced by Bacillus species. A final report of microbial diversity course 2009. [Online] Available from: http://courses.mbl.edu/microbialdiversity/research_projects/research_projects_docs/FinalReports_2009/BolajiOkulate_FinalReport.pdf. [Accessed on 27 August, 2013].
- 12.Pinchuk IV, Bressollier P, Verneuil B, Fenet B, Sorokulova IB, Mégraud F, et al. In vitro anti-helicobacter pylori activity of the probiotic strain Bacillus subtilis 3 is due to secretion of antibiotics. Antimicrob Agents Chemother. 2001;45:3156–3161. doi: 10.1128/AAC.45.11.3156-3161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Marrec C, Hyronimus B, Bressollier P, Verneuil B, Urdaci MC. Biochemical and genetic characterization of coagulin, a new antillisterial bacteriocin in the pediocin family of bacteriocins, produced by Bacillus coagulans. App Environ Microbiol. 2000;66:5213–5220. doi: 10.1128/aem.66.12.5213-5220.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mezaini A, Chihib NE, Bouras AD, Nedjar-Arroume N, Hornez JP. Antibacterial activity of some lactic acid bacteria isolated from an Algerian dairy product. J Environ Public Health. 2009 doi: 10.1155/2009/678495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banoo S, Shubha DS, Shashidhar V, Venkatesha D. Bacterial and clinical profile of diabetic foot patients. Ann Trop Med Public Health. 2012;5:69–73. [Google Scholar]
- 16.Mohanasoundaram KM. The microbiological profile of diabetic foot infections. J Clin Diagn Res. 2012;6:409–412. [Google Scholar]
- 17.Alleson D, Paul D, Paul R, Colin H. Bacteriocin production: a probiotic trait? J Appl Environ Microbiol. 2012;78:1–6. doi: 10.1128/AEM.05576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shelar SS, Warang SS, Mane SP, Sutar RL, Ghosh JS. Characterization of bacteriocin produced by Bacillus atrophaeus strain JS-2. Int J Biol Chem. 2012;6:10–16. [Google Scholar]
- 19.Hammami I, Jaouadi B, Ben Bacha A, Rebai A, Bejar S, Nesme X, et al. Bacillus subtilis bacteriocin Bac 14B with a broad inhibitory spectrum: purification, amino acid sequence analysis, and physicochemical characterization. Biotechnol Bioprocess Eng. 2012;17:41–49. [Google Scholar]


