Abstract
Promoters of many developmentally regulated genes have a bivalent mark of H3K27me3 and H3K4me3 in embryonic stem cells state, which is proposed to confer precise temporal activation upon differentiation. Although Polycomb repressive complex 2 (PRC2) is known to implement H3K27me3, the COMPASS family member responsible for H3K4me3 at bivalently-marked promoters was previously unknown. Here, we identify Mll2 (KMT2b) as the enzyme responsible for H3K4me3 on bivalently-marked promoters in embryonic stem cells. Although H3K4me3 at bivalent genes is proposed to prime future activation, we did not detect a substantial defect in rapid transcriptional induction after retinoic acid treatment in Mll2 depleted cells. Our identification of the Mll2 complex as the COMPASS family member responsible for implementing H3K4me3 at bivalent promoters provides an opportunity to reevaluate and experimentally test models for the function of bivalency in the embryonic stem cell state and in differentiation.
INTRODUCTION
Determining the mechanisms coordinating the precise spatio-temporal transcriptional programs driving organismal development is a long-standing interest in biology. Recent genome-wide studies of pluripotent cells suggest that developmentally regulated changes in chromatin structure play a pivotal role in this process. Embryonic stem cells reflect an early stage of mammalian development and possess a unique chromatin landscape in which “bivalent” domains of trimethylated histone H3 lysine 4 (H3K4me3) and lysine 27 (H3K27me3) mark key lineage-specific genes1,2. Histone H3K4me3 and H3K27me3 are normally associated with active and repressed genes respectively, however, the promoters of the genes carrying both of these marks simultaneously, “bivalent promoters”, are transcribed at very low levels and are considered to be poised for activation by developmental signals3,4.
A striking example of bivalent chromatin is at the Hox gene clusters, which encode developmental transcription factors that are conserved from insects to mammals5. In mammalian embryonic stem cells, the Hox clusters carry particularly large bivalent domains spanning hundreds of kilobases, suggesting an important role for these histone modifications and their associated complexes in Hox expression2,5. Studies in Drosophila and vertebrates established the H3K27 trimethylase, Polycomb repressive complex 2 (PRC2), as a critical negative regulator of Hox genes6–8. However, the methylase responsible for H3K4me3 at bivalent chromatin has not been previously identified due to a presumed redundancy among the multiple COMPASS-like complexes that implement this mark in mammals9. There is only one H3K4 methylase Set1 (COMPASS) in yeast, while in Drosophila cells this activity is divided within three COMPASS-like enzymes: Set1, Trithorax (Trx), and Trithorax-related (Trr)10 (Fig. 1a). Mammals have an extended set of six complexes representing duplications of the Drosophila enzymes: Set1A and Set1B (Set1-like), which are the main H3K4 trimethylases10,11, Mll1 and Mll2 (Trx-like), and Mll3 and Mll4 (Trr-like) which were recently identified as enhancer monomethylases12 (Fig. 1a). The mammalian Trx branch of COMPASS, consisting of Mll1 and Mll2, are both essential for mouse embryonic development13–15, however, neither factor has been previously linked to bivalent chromatin in embryonic stem cells.
Figure 1.
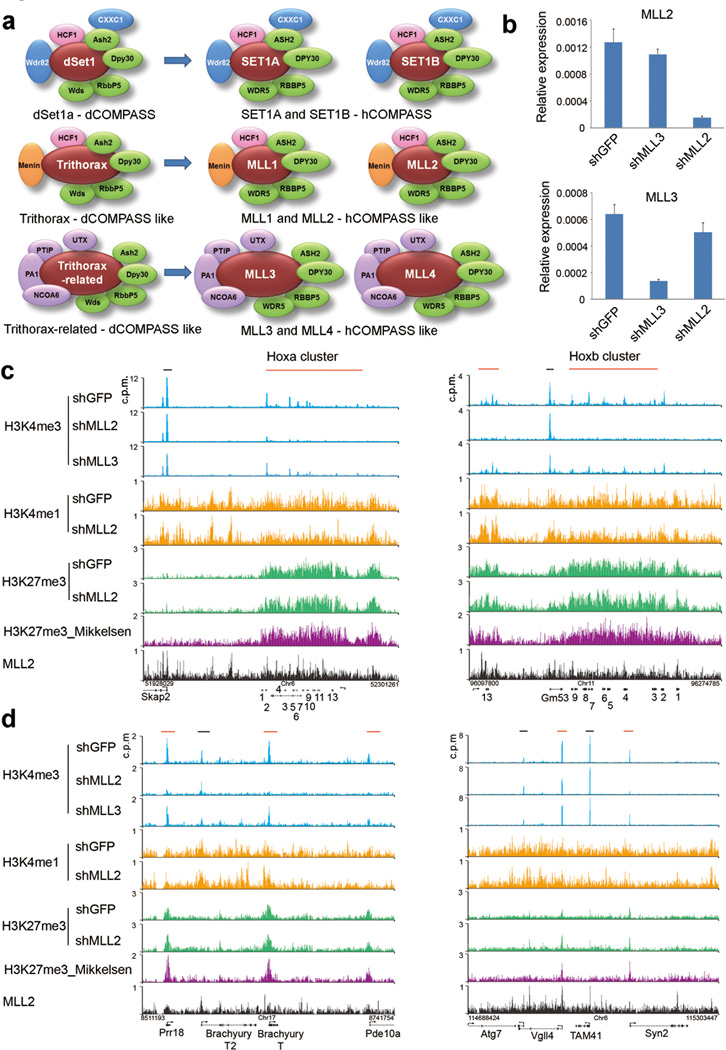
Mll2 is required for the trimethylation of Histone H3 lysine 4 at bivalent homeotic genes. (a) The COMPASS family of H3K4 methylases in Drosophila (3 members on the left) and mammals (6 members, on the right). Subunits common to all COMPASS family members from yeast to human are shown in green; complex-specific subunits are shown in blue, orange and purple. (b) Mll2 and Mll3 mRNA levels after RNAi-mediated knockdown in mouse embryonic stem cells with shRNAs targeting GFP (shGFP), Mll3 (shMll3) or Mll2 (shMll2). Expression was determined by qRT-PCR and is shown relative to Actin (Actb). Results are shown as means and s.d. (n=2 technical replicates, representative of three biological replicate experiments). (c) ChIP-seq track file examples of H3K4me3 at mouse Homeobox (Hox) gene clusters. Red and black bars above the tracks indicate bivalent and non-bivalent regions, respectively. H3K27me3 data from Mikkelsen et al.4 is shown for comparison (purple). (d) ChIP-seq track examples of bivalent and non-bivalent chromatin in control and Mll2 shRNA-treated cells. Bivalently marked genes such as Prr18, Brachyury (T), Vgll4 and Syn2 are shown with red bars above the tracks as in (c).
Here we set out to gain insight into the mechanism by which Mll2 contributes to embryonic stem cell function and find that it is required for the implementation of H3K4me3 at bivalent promoters, providing an opportunity to experimentally test the functional relevance of the bivalent chromatin state.
RESULTS
Mll2 is required for H3K4me3 at Homeobox gene clusters
We investigated the bulk level of H3K4me1, H3K4me2, and H3K4me3 by immunoblotting and genome-wide H3K4me3 occupancy by chromatin immunoprecipitation coupled with high throughput DNA sequencing (ChIP-seq) in mouse embryonic stem cells depleted for Mll2. For comparison, we also performed RNAi-mediated knockdown of Mll3. Although the expression of Mll2 and Mll3 were each decreased by more than 80% (Fig. 1b), we detected no change in the overall bulk levels of H3K4 methylation for either knockdown condition by Western blotting (Supplementary Fig. 1a). This is consistent with earlier studies that members of the MLL family do not appear to individually mediate bulk changes in H3K4 di- and trimethylation11,16.
To determine whether Mll2 or Mll3 regulates H3K4 methylation at specific genomic loci, we mapped H3K4me3 in control and Mll2 or Mll3-depleted mouse embryonic stem cells using ChIP-seq. Although H3K4me3 at the stem cell pluripotency genes Nanog and Oct4 was unaffected by knockdown of either factor (Supplementary Fig. 1b), we found that H3K4me3 is broadly reduced across all four Hox clusters in Mll2-knockdown mouse embryonic stem cells (Fig. 1c and Supplementary Fig. 1c). This contrasts with the results from embryonic fibroblasts, which demonstrated that implementation of H3K4 trimethylation at the Hox genes is shared by both Mll1 and Mll217. Although Hox genes are marked by both H3K4me3 and H3K27me3 in embryonic stem cells, H3K27me3 was largely unchanged at these loci after Mll2 depletion (Fig. 1c and Supplementary Fig. 1c). Outside of Hox loci, we find other bivalent genes that neighbor non-bivalently marked genes, with the bivalently-marked genes such as Brachyury (T) losing H3K4me3 in the absence of Mll2, while at many active genes such as Brachyury the second (T2), H3K4me3 remained unchanged (Fig. 1d and Supplementary Fig. 1d).
Mll2 mediates H3K4me3 at bivalent gene promoters
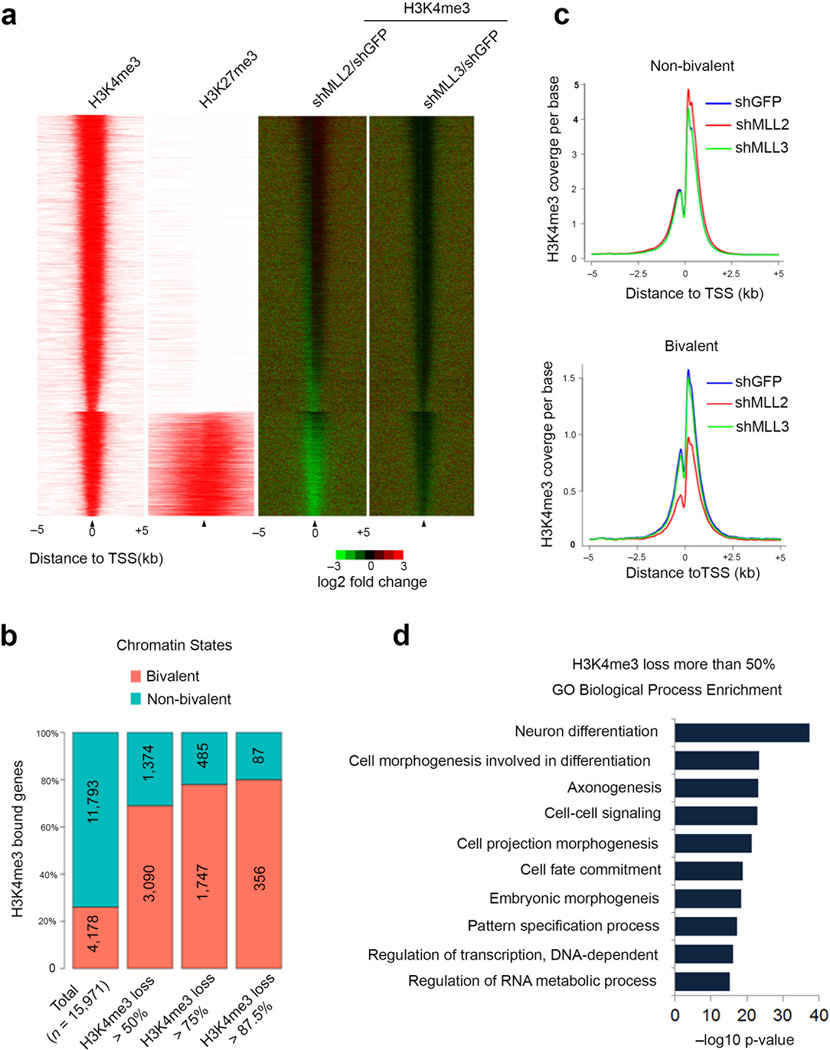
In order to test the generality of the requirement of Mll2 for marking the promoters of bivalent genes, we generated heat maps of fold change for H3K4me3 in Mll2 and Mll3 knockdown cells. By separating H3K4me3 enriched promoters into bivalent or non-bivalent classes based on H3K27me3 occupancy, it was reproducibly observed that bivalently-marked promoters have reduced H3K4me3 levels in Mll2-depleted, but not Mll3-depleted embryonic stem cells (Fig. 2a and Supplementary Fig. 2). Of the 4464 Ensemble gene promoters that had two fold or greater reduction in H3K4me3 in the Mll2 knockdown embryonic stem cells, 70% were bivalently marked, with a tendency for the most affected genes to be in the bivalent class (Fig. 2b).
Figure 2.
Mll2 is required for the implementation of bivalency genome-wide. (a) The H3K4me3 occupancy change in the Mll2- and Mll3-depleted mouse embryonic stem cells for all H3K4me3 enriched genes. Left: ChIP-seq enrichment profiles for +/− 5kb around the TSS of all H3K4me3 enriched genes. Bivalent genes are shown as a separate group from the H3K4me3-only modified genes. Right: H3K4me3 occupancy log2 fold-change after depletion of Mll2 (shMLL2/shGFP) or Mll3 (shMLL3/shGFP) measured +/− 5kb around TSS. (b) Percentage of bivalent and non-bivalent genes with H3K4me3 occupancy loss. The percentages are shown at four levels: total H3K4me3-enriched genes, genes with more than 50% decrease of H3K4me3, genes with more than 75% decrease, and genes with more than 87.5% decrease. Numbers shown are the total number of genes for each level. H3K27me3 data in a,b are from Wamstad et al.20 (c) Average-gene occupancy plots of H3K4me3 in wild-type (shGFP) (blue line), shMll2 (red line) and shMll3 (green line) knockdown embryonic stem cells. Top and bottom panels show non-bivalent and bivalent genes, respectively. Plots are averaged from both H3K4me3 ChIP-seq biological replicates. (d) Gene Ontology analysis of genes with H3K4me3 loss after Mll2 knockdown. Benjamini-corrected p-values are shown.
Average gene analysis of H3K4me3 around the TSS (+/− 5kb) revealed that the knock-down of MLL2 significantly reduced the H3K4me3 coverage for bivalent genes (p < 2.2e-16), while there was not sufficient evidence (p =1) that MLL2 knock-down reduces H3K4me3 coverage at non-bivalent genes (Welch Two-Sample t-Test; one-sided), further indicating that Mll2 is the major enzyme responsible for trimethylation of H3K4 on bivalent gene promoters in embryonic stem cells. Gene Ontology (GO) analysis of genes losing H3K4me3 in Mll2 knockdown cells showed enrichment for genes involved in differentiation (Fig. 2d).
Bivalency is not required for rapid induction of genes
In order to understand Mll2’s role in gene expression in embryonic stem cells, we performed RNA-seq in control and Mll2-knockdown cells. We could detect some changes in the expression pattern of bivalently-marked genes in the embryonic stem state (Supplementary Fig. 3a,b), however, since these genes are lowly expressed, analysis of their expression due to loss of Mll2 needs to be further evaluated. A previous study has reported that Mll2−/− embryonic stem cells are viable and retain pluripotency18. In agreement with this result, we found that Mll2 depletion in mouse embryonic stem cells by RNAi has no detectable effect on self-renewal of embryonic stem cells as demonstrated by alkaline phosphatase staining (Supplementary Fig. 3c). This was further supported by RNA-seq data for control and Mll2-depleted mouse embryonic stem cells that showed pluripotent genes such as Nanog and Oct4 were unaltered in expression (Supplementary Fig. 3d). Our RNA-seq studies also demonstrated that Mll2, but not any other H3K4 methylase, had reduced expression in the Mll2 shRNA-treated cells (Supplementary Fig. 3e), further supporting our conclusion that the H3K4me3 mark at bivalent genes is deposited by Mll2 and not by other COMPASS family members.
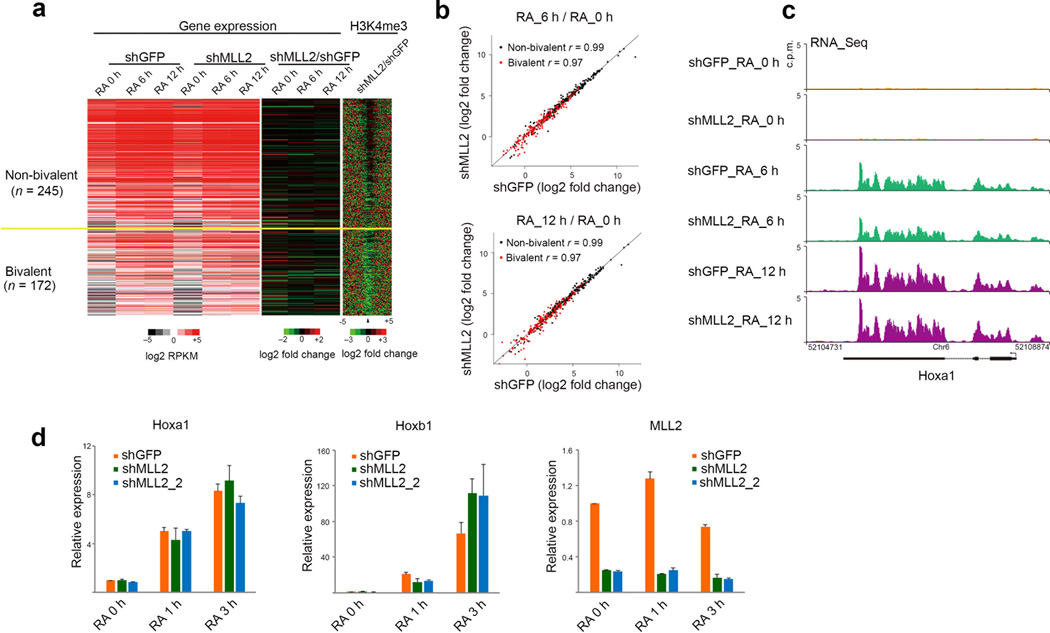
We next asked what was the consequence of Mll2’s role in setting up bivalently marked promoters for future transcriptional activation of these genes. Therefore, we treated control and Mll2-depleted embryonic stem cells with retinoic acid (RA) for 6 and 12 hours and analyzed gene expression by RNA-seq. About 400 genes were significantly induced by 6h or ~ 500 genes by 12h of RA treatment (Fig. 3a). Our analyses suggest that there was no major defect in the induction kinetics for either non-bivalent or bivalent genes in the Mll2 knockdown cells (Fig 3b,c). To determine whether induction kinetics were altered at earlier time-points, we analyzed the response to RA induction for Hoxa1 and Hoxb1, which were previously demonstrated to be early targets of RA induction5 (Fig. 3d). These analyses demonstrate that the loss of Mll2 (using two independent shRNA constructs) had no significant effect on their induction (Fig. 3d).
Figure 3.
Mll2 depletion has little effect on transcriptional induction kinetics in response to retinoic acid. (a) Expression analysis of mouse V6.5 embryonic stem cells infected with GFP shRNA (shGFP) or Mll2 shRNA (shMll2) lentivirus and induced to differentiate with 2µM retinoic acid for 6 (RA 6) or 12 hours (RA 12). Heatmaps of expression and H3K4me3 occupancy are shown. Genes with expression increased by 6 and 12 hours of RA treatment are sorted by the wild-type occupancy of H3K4me3 and separated as bivalent or not bivalent. Left: Expression heatmap in control and shMLL2 treated cells. Middle: log2 fold changes for expression after Mll2 RNAi. Induction defects would appear as an increase in green in the 6 and 12 hour timepoints. Right: log2 fold change of H3K4me3 occupancy. (b) Scatter plots of log2 fold changes in expression after RA induction at 6 hours (top panel) or 12 hours (bottom panel). Each dot represents a gene that was induced by 6 hours. Correlation coefficients are calculated in the log2 scale. (c) Gene expression track examples before and after RA induction. (d) Induction kinetics at one and three hours RA treatment. Results are shown as means and s.d., (n=2 technical replicates, representative of 3 biological replicate experiments).
DISCUSSION
Here, we have presented evidence that Mll2 is required broadly and specifically for the trimethylation of H3K4 at most bivalently-marked promoters in mouse embryonic stem cells. We demonstrated that (i) depletion of Mll2 via lentivirus-mediated RNAi had no effect on the bulk levels of the mono-, di-, and trimethylation of H3K4 and other histone modifications; (ii) Mll2 was required for the establishment of H3K4me3 at all four Hox gene clusters and other bivalently-marked promoters; (iii) the reduced H3K4me3 levels at bivalent promoters had no major effect on the rapid induction of gene expression in response to retinoic acid. These findings are particularly interesting in light of the Mll1 and Mll2 knockout mouse phenotypes. Mll1 is required for mid-to-late developmental processes including hematopoietic development19, whereas Mll2 expression is detected at very early stages and a loss-of-function mutation results in severe developmental retardation and lethality14,15. Mll2’s role in forming the bivalent state in embryonic stem cells could help explain its demonstrated restricted role during early mammalian development14.
Our findings that loss of H3K4me3 at bivalent promoters had little effect on early induction kinetics in response to differentiation cues suggest that bivalency may have a subtler role in differentiation. Similarly, although we did find plenty of bivalent genes with increased H3K27me3 upon Mll2 knockdown, the Hox clusters did not show an obvious increase in this modification. Interestingly, reduction of PRC2 components in embryonic stem cells also had relatively subtle effects compared to the prevalence of the bivalent state8,9. This could reflect a limitation of the embryonic stem cell culture model, as opposed to the precise timing requirements of cell signaling and the acquisition of the proper cell identity during development of an organism. It might also be worth testing the expression and induction capability of bivalently marked promoters under both PRC2 null and Mll2 RNAi conditions.
In addition to the loss of H3K4me3 at bivalent promoters we also observed loss of H3K4me3 at other sites in the genome with low H3K4me3 and low transcription, without enrichment for H3K27me3. Both bivalent and other low H3K4me3 sites could have low levels of methylation because they recruit low levels of Mll2. The difference between Mll2-dependent bivalent promoters and non-bivalent promoters could be that the bivalent promoters also recruit PRC2 and H3K27me3. Thus the permissive environment of embryonic stem cell chromatin could allow a low level of Mll2-dependent H3K4me3 and a concomitant low level of transcription. Nonetheless, our identification of Mll2 as the regulator of H3K4 trimethylation at bivalent promoters establishes a basis for molecular investigation of the function of bivalency in early differentiation9 and perhaps the role this modification may play in cancer cell plasticity and growth.
ONLINE METHODS
ES cell culture and lentiviral based knockdown
Mouse V6.5 embryonic stem cells were cultured in 0.1% gelatin-coated tissue culture flasks with irradiated mouse embryonic fibroblast (MEF) feeder cells. Cells were grown in DMEM supplemented with 15% fetal bovine serum (Hyclone), 2 mM L-glutamine, 0.1mM nonessential amino acids (Stemcell Technologies) and 1000 U/ml recombinant LIF (Millipore). For Mll2 and Mll3 knockdown, mouse V6.5 ES cells were infected with lentiviruses expressing short-hairpin RNA (shRNA) for Mll2 (shMLL2: GGAGAACTCTGATTGAGAAAG; shMLL2_2: GCTATGAAGACAATGACTATG) or Mll3 (shMLL3: GGAGAGGACTACAAATGAAGT) for 24 hours and then were subjected to selection with 2ug/ml puromycin for 5 days. For ChIP-seq and RNA-seq experiments, cells were grown for one passage without feeder cells for 30 min before harvesting. For all trans retinoic acid (RA, Sigma-Aldrich) treatment, cells were induced with 2 µm RA for indicated time points and subjected to RNA extraction following removal of feeder cells.
Antibodies
Antibodies against H3K4me1, H3K4me2 and H3K4me3 are generated in our lab. H3K27me3 antibody is purchased from Active Motif (#39155). UbH2B antibody is from Cell Signaling (#5546).
ChIP-seq
ChIP assays were performed as previously described21. Briefly, 5×107 cells were crosslinked with 1% paraformaldehyde at room temperature for 10 min and sonicated to generate chromatin fragments of 200–600bp. ChIP-seq data for H3K27me3 for track examples are from GSE122414 and H3K27me3 data for enrichment profiles and bivalent gene definition are from GNomEx accession 44R20.
ChIP-Seq data Analysis
Reads were aligned to the mouse genome (UCSC mm9) using the Bowtie aligner v0.12.9 allowing uniquely mapping reads only and allowing up to three mismatches22. Reads were extended to 150 bases toward the interior of the sequenced fragment and normalized to total reads aligned (reads per million; RPM).
Peak detection was done using MACS v1.4.223 for all samples. Associated control samples were used to determine statistical enrichment at a FDR < 0.05. Additional enrichment was called for H3K27me3 using the broad domain peak detector SICER v1.124 at the FDR < 1e-8, window size of 200, and gap size of 600.
The high-confidence enriched regions were used to calculate ChIP-Seq occupancy and depict enrichment profiles. Gene annotations and transcript start site information were from Ensembl 67 excluding pseudogenes and polymorphic_pseudogenes. Genes with H3K4me3 enriched within 1kb upstream of the TSS of any isoforms were defined as H3K4me3 bound genes, and H3K4me3 bound genes were associated with the nearest H3K4me3 enriched region. Each bound gene’s H3K4me3 occupancy was calculated as the averaged coverage of its associated enriched region. The bivalent genes were defined as the H3K4me3 bound genes that are also H3K27me3 enriched in the gene body.
The enrichment profiles in Fig. 2c are shown as a binary value of enriched or not enriched. The rows were H3K4me3 bound genes and separated into two groups of bivalent or not bivalent genes and then sorted by H3K4me3 occupancy. The isoform’s start site with the maximum H3K4me3 occupancy was selected as the TSS of that gene. The longest isoform was selected from the isoforms of the same gene with the same H3K4me3 occupancy. The profiles of coverage fold change in Fig. 2c are shown as the log2 ratios of knockdown over wild-type coverage. The rows were sorted the same as the enrichment profiles. Gene regions spanning 5 kb on either side of TSS were binned into 25 bp windows and are shown from 5’ to 3’.
RNA-Seq data Analysis
Reads for each sample were aligned to the mouse genome UCSC mm9 and to gene annotations from Ensembl 67 using TopHat v2.0.825 with bowtie1 v0.12.9, allowing uniquely mapping reads only and allowing up to two mismatches. Cuffdiff v1.3.0 was used to quantify RPKM values, to perform differential expression analysis at FDR < 0.05, and to assess statistically sufficient read coverage for each gene26.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Egidy, A. Peak, A. Perera and the rest of the Stowers Molecular Biology core for help with Illumina sequencing. We are grateful to the Stowers Tissue Culture core for assistance with the generation and maintenance of cell lines. We thank L. Shilatifard for editorial assistance. These studies were supported in part by the National Cancer Institute grant R01CA150265 to A.S.
Footnotes
ACCESSION CODE
ChIP-seq and expression data have been deposited at GEO under the accession number GSE48172.
AUTHOR CONTRIBUTIONS
D.H. and M.A.M. performed the experiments, A.S.G., X.G. and M.C. performed bioinformatics analysis, and all authors were involved in preparation of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 3.Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin C, et al. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC) Genes Dev. 2011;25:1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasini D, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 7.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu M, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herz HM, et al. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 14.Glaser S, et al. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin. 2009;2:5. doi: 10.1186/1756-8935-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser S, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 16.Milne TA, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 17.Wang P, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol Biol Cell. 2007;18:2356–2366. doi: 10.1091/mbc.E06-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004;14:2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Wamstad JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES FOR ONLINE METHODS
- 21.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.