Abstract
Mitochondria serve a pivotal role in the regulation of apoptosis or programmed cell death. Recent studies have demonstrated that reactive electrophiles induce mitochondrion-dependent apoptosis. We hypothesize that covalent modification of specific mitochondrial proteins by reactive electrophiles serves as a trigger leading to the initiation of apoptosis. In this study, we identified protein targets of the model biotin-tagged electrophile probes N-iodoacetyl-N-biotinylhexylene-diamine (IAB) and 1-biotinamido-4-(4′-[maleimidoethylcyclohexane]carboxamido)butane (BMCC) in HEK293 cell mitochondrial fractions by liquid chromatography-tandem mass spectrometry (LC-MS-MS). These electrophiles reproducibly adducted a total of 1693 cysteine residues that mapped to 809 proteins. Protein modifications were selective in that only 438 cysteine sites in 1255 cysteinyl peptide adducts (35%) and 362 of the 809 identified protein targets (45%) were adducted by both electrophiles. Of these, approximately one-third were annotated to the mitochondria following protein database analysis. IAB initiated apoptotic events including cytochrome c release, caspase-3 activation, and poly(ADP-ribose)polymerase (PARP) cleavage, whereas BMCC did not. Of the identified targets of IAB and BMCC, 44 were apoptosis-related proteins, and adduction site specificity on these targets differed between the two probes. Differences in sites of modification between these two electrophiles may reveal alkylation sites whose modification triggers apoptosis.
Introduction
Mitochondria govern the balance between life and death in eukaryotic cells. Through respiration, electron transport, and oxidative phosphorylation, mitochondria supply most of the bioenergetic requirements for life. Mitochondria also serve as integrators for apoptosis, in which cellular stresses and receptor-mediated death-signaling pathways cause the release of mitochondrial apoptogenic factors, including cytochrome c, apoptosis-inducing factor (AIF),1 and secondary mitochondrial activator of caspases/direct IAP binding protein with low pI (SMAC/DIABLO) into the cytosol. These factors trigger downstream events such as caspase activation, DNA fragmentation, inhibition of DNA repair proteins, membrane blebbing, and cell fragmentation (1). It is believed that mitochondria release these factors through the opening of the permeability transition pore (PTP) channel, although other pathways may also contribute (2, 3).
Apoptosis can be triggered by electrophilic xenobiotic metabolites and electrophilic products of oxidative stress that covalently modify nucleophilic sites in DNA and proteins to form adducts (4–6). Although the initiation of apoptosis by DNA damage has been extensively studied (7), proteins also are major targets of electrophiles. The characteristics of protein damage associated with apoptosis have been explored only to a limited extent. Cysteine thiols are thought to be particularly important targets for pro-apoptotic damage, as these nucleophilic residues are easily modified by oxidants and electrophiles and are critical in regulating certain aspects of cellular signaling (8–10). Protein covalent binding can trigger apoptosis, at least in part through modification of critical cellular protein thiols (11–14). Other studies indicate that thiol-modifying reagents can induce the mitochondrial permeability transition (15–20), thus providing a possible mechanistic basis for the induction of apoptosis by reactive electrophiles.
Mitochondria generate reactive oxygen species, especially under conditions of cellular stress, and this enhanced production of reactive oxygen species may lead to oxidative stress (21). Oxidative stress also leads to the generation of reactive electrophilic products of lipid oxidation. 4-Hydroxy-2-nonenal (HNE) and related aldehydic products of lipid peroxidation induced apoptosis in human colorectal carcinoma and endothelial cells (5, 22, 23). The electrophilic lipid 15-deoxy-Δ12,14-prostaglandin J2 induced apoptosis in a human neuroblastoma cell line (24) and in cultured hepatocytes harvested from Sprague—Dawley rat livers (25). Apoptosis was induced in human cell lines by the anticarcinogen isothiocyanates sulforaphane and phenethyl isothiocyanate (26, 27) and in animal models by electrophilic metabolites of the carcinogens acrylonitrile and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (28, 29). Although it is clear that electrophiles induce apoptosis, the mechanism is poorly understood. Given that pro-apoptitic electrophiles can covalently modify proteins and that the mitochondrial permeability transition plays a key role in apoptosis, it seems probable that alkylation of mitochondrial protein targets contributes to apoptosis. However, any test of this hypothesis will require identification of mitochondrial protein targets of pro-apoptotic electrophiles.
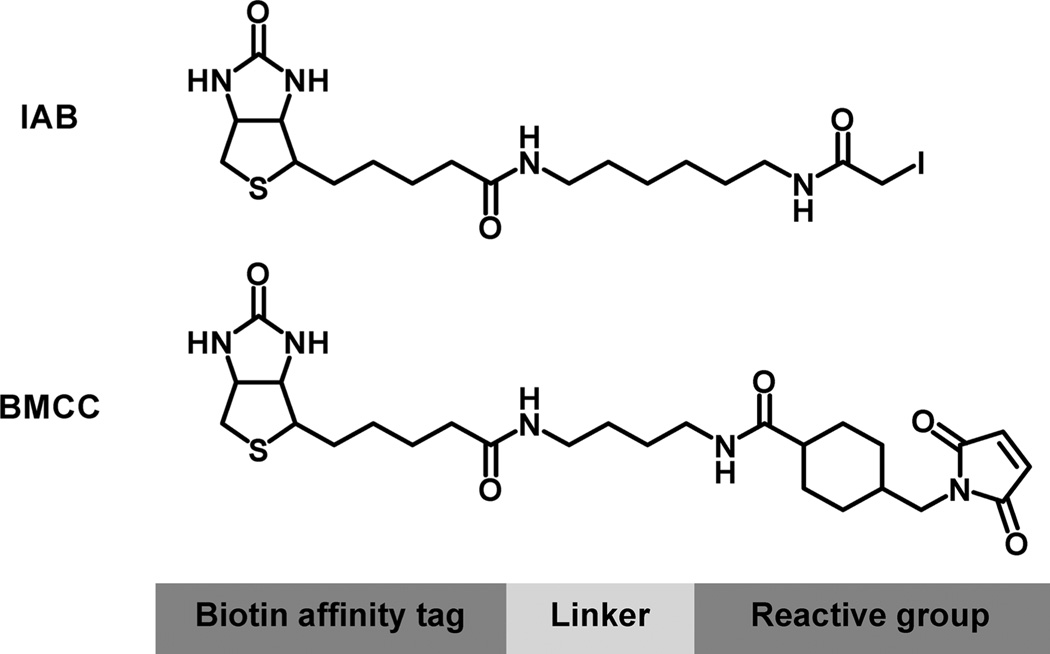
We have recently used biotinylated electrophile probes to investigate the scope and characteristics of protein covalent binding to subcellular proteomes (30, 31). The iodoacetamido probe N-iodoacetyl-N-biotinylhexylenediamine (IAB) and N-alkylmaleimido probe 1-biotinamido-4-(4′-[maleimidoethylcy-clo-hexane]carboxamido)butane (BMCC) (Figure 1) display reaction chemistries analogous to many electrophilic metabolites and endogenous electrophiles. Adducts formed with these probes have been mapped to over a thousand specific cysteines in cytoplasmic, nuclear, and microsomal proteins (30, 31). Moreover, adduction appears to be associated with different biological effects, as IAB, but not BMCC-induced endoplasmic reticulum (ER) stress in cells exposed to these probes (31).
Figure 1.
Structures of reactive electrophile probes used in this study.
Here, we have extended this approach to characterize alkylation-induced apoptosis in human embryonic kidney 293 (HEK293) cells. Although both compounds form protein adducts, IAB induces apoptosis, whereas BMCC does not. We identified IAB and BMCC adducts formed in isolated HEK293 mitochondrial fractions that were treated with both probes. These analyses mapped over 1200 cysteine thiol targets of IAB and BMCC, a significant fraction of which are on mitochondrial proteins. IAB and BMCC displayed distinct differences in the alkylation of mitochondrial protein targets. These differences in site-specific modification between the two electrophiles may govern the relationship between protein damage and apoptosis.
Experimental Procedures
Caution
IAB and BMCC are hazardous alkylating agents and should be handled with care.
Chemicals and Reagents
IAB, BMCC, and the bicinchoninic assay kit were purchased from Pierce Biochemicals (Rockford, IL). Streptavidin Sepharose high-performance beads were obtained from GE Healthcare (Uppsala, Sweden). Trypsin Gold was purchased from Promega (Madison, WI). Complete Mini protease inhibitor cocktail was obtained from Roche Diagnostics (Mannheim, Germany). Dulbecco’s modified Eagle’s medium (DMEM), phosphate-buffered saline (PBS), 0.5% trypsin in PBS, Novex Colloidal blue stain kit, and trypan blue were purchased from Invitrogen (Carlsbad, CA). Sources of primary antibodies were as follows: antibodies against cytochrome c (raised in rabbit, catalog no. 630105, BD Clontech, Palo Alto, CA), cytochrome oxidase 4 (Cyto Ox 4) (mouse, catalog no. 630105, BD Clontech), caspase-3, (mouse, catalog no. ab7850, Abcam, Cambridge, MA), poly(ADP-ribose)-polymerase (PARP) (rabbit, catalog no. 9542, Cell Signaling, Beverly, MA), actin (mouse, catalog no. 8226, Abcam), glutathione S-transferase (rabbit, catalog no. ab14502, Abcam), histone 2A (rabbit, catalog no. 4271, Abcam), Grp78/Bip (rabbit, catalog no. 610979, Abcam), and biotin (mouse, catalog no. 1297597, Roche, Indianapolis, IN). IRDye 700 conjugated affinity goat antimouse and antirabbit secondary antibodies were purchased from Rockland Immunochemicals (catalog no. A21076 and A21058, respectively, Gilbertsville, PA). The corresponding IRDye 800 sary antibodies were also obtained from Rockland (catalog no. 610-432-020 and 611-132-122, respectively). Formic acid (98–100%) was acquired from EM Science (Darmstadt, Germany). All other chemicals were obtained from Sigma (St. Louis, MO) and Fisher Chemical Co. (Fair Lawn, NJ).
Cell Culture
HEK293 cells were purchased from American Type Culture Corporation (ATCC, Manassas, VA). Cells were seeded on plates in DMEM supplemented with 10% fetal bovine serum and antibiotic/antimycotic at 10 mL/L and grown to 80% confluency at 37 °C and 95% air/5% CO2.
Lactate Dehydrogenase (LDH) Leakage Assay
Cells were washed once with PBS and then treated with IAB (50–200 µM), BMCC (100–400 µM), staurosporine (2 µM), or DMSO vehicle delivered in DMEM without phenol red for 6–24 h. The maximum DMSO concentration in the medium was 1.2% (v/v). LDH leakage was determined as described in the LDH-based in vitro toxicology assay kit (Sigma).
Preparation and Immunoblot Analyses of Subcellular Fractions
Cells were treated for 24 h with IAB, BMCC, staurosporine, or vehicle at the concentrations indicated above for the LDH leakage assay. Three different types of preparations were carried out for immunoblot analysis: cytosolic fractions to detect cytochrome c, whole cell lysates to detect PARP cleavage, and whole cell lysates to monitor caspase-3 activation. Cytosolic fractions were isolated by digitonin treatment as described previously (32) with the following modifications. Harvested cells were resuspended in ice-cold 70 mM Tris buffer, pH 7.5, containing 250 mM sucrose and protease inhibitors (Sigma, catalog no. P4830). For detection of PARP cleavage, whole cell lysates were prepared as described previously (5). For caspase-3 activation, cells were scraped into the media, washed twice with ice-cold PBS, and then resuspended in 312.5 mM HEPES, pH 7.5, containing 31.25% sucrose (w/v) and 0.3125% CHAPS (w/v) at a concentration of 108 cells/mL. Cells were lysed by freeze–thawing and then incubation on ice for 15 min. Following centrifugation at 15000g for 20 min, supernatant (whole cell lysate) was collected.
Mitochondria of untreated HEK293 cells were purified by Percoll gradient centrifugation as described previously (33) with the following modifications. After gradient centrifugation, the protein band (approximately 30% down the Percoll gradient) was collected with a Pasteur pipet, diluted with 0.25 M mannitol, 25 mM HEPES-NaOH, pH 7.5, 0.5 mM EGTA, and mammalian protease inhibitor cocktail (Sigma) (buffer A), and then centrifuged at 10000g for 15 min to remove the Percoll. The pellet was washed with buffer A, and the purified mitochondrial pellet was resuspended in 10 mM HEPES buffer, pH 7.5. Protein concentrations for cellular fractions were determined with the bicinchoninic acid assay kit (Pierce).
Proteins (20–50 µg per lane) were resolved by sodium dodecyl sulfate—polyacrylamide gel electrophoresis (SDS-PAGE) using 10% NuPAGE Novex Bis-Tris precast gels (Invitrogen) in either NuPAGE MOPS SDS or MES SDS running buffer (Invitrogen). They were transferred to polyvinylidene difluoride membranes (Invitrogen), blocked with 1:1 TBS: blocking buffer for near-infrared fluorescence Western blotting (Rockland, Gilbertsville, PA), and then probed with antibodies against cytochrome c, Cyto Ox 4, biotin, caspase-3, PARP, actin, glutathione S-transferase, histone 2A, or Grp78/Bip. Membranes were incubated with IRDye700 or IRDye800 conjugated affinity goat antimouse or antirabbit secondary antibodies and then analyzed on a LI-COR Odyssey Imaging system (Lincoln, NE) with a preset setting “membrane” and resolution of 169 µm.
Caspase Activation Colorimetric Assay
Cells were treated for 24 h with IAB, BMCC, staurosporine, or vehicle at the concentrations indicated above for the LDH leakage assay. Cell treatment was done for four replicate experiments. For each experiment, whole cell lysates were prepared as described above for immunoblot detection of caspase-3. Caspase-3 activities in lysates (45 µg protein per reaction) were measured with the CaspACE Colorimetic Assay System kit (Promega, Madison, WI). The assay reaction mixtures were incubated for 24 h at 37 °C. Reactions were carried out in triplicate and were measured spectroscopically at 405 nm using a 96 well plate reader (Dynex Technologies, Chantilly, VA).
Incubation of HEK293 Cell Mitochondria with Electrophile Probes and Analysis of Adducted Proteins
Mitochondria (4 mg mL−1 protein) were incubated with 100 µM IAB or BMCC in 10 mM HEPES buffer, pH 7.5, at 37 °C for 30 min. The reaction was quenched with 10 mM dithiothreitol and placed on ice. NuPAGE LDS sample buffer (Invitrogen) was added to quenched protein mixtures in a ratio of 1:3 (v:v) and incubated at 70 °C for 10 min. SDS-PAGE separation of proteins (100 µg protein per gel lane; four lanes total), in-gel digestion, and peptide extraction were carried out as described previously (31) with the following modifications. SDS-PAGE gel lanes were cut into 20 slices (2–6 mm lengthwise per slice) comprising the entire molecular weight range prior to cutting of individual slices into 1 mm cubes. Following the drying of gel cubes in vacuo, they were rehydrated with trypsin (1:25 approximate trypsin/protein mass ratio) in 25 mM ammonium bicarbonate for 10 min at room temperature and then incubated for 12–16 h at 37 °C.
Streptavidin Capture of Biotinylated (Adducted) Peptides
Streptavidin sepharose beads were prewashed three times with 100 mM ammonium bicarbonate and then diluted with ammonium bicarbonate to achieve a 50:50 (v/v) bead/buffer slurry. The peptide digests in 200 µL of ammonium bicarbonate were mixed with 450 µL of the slurry, and the suspension was mixed with rotation at room temperature for 2 h. The mixture was centrifuged at 8000g for 2 min, and the liquid supernatant was removed. The beads containing the biotinylated peptides were washed in the following sequence: twice with 1 mL of 100 mM ammonium bicarbonate, three times with 1 mL of 1 M NaCl, twice with 1 mL of 100 mM ammonium bicarbonate, and twice with 1 mL of H2O. For each wash, beads were resuspended in the wash solution and then centrifuged at 8000g for 2 min, and the supernatant was discarded. Biotinylated peptides were then eluted by washing the beads with 70:25:5 acetonitrile/H2O/formic acid (300 µL per elution). The first elution was carried out at 4 °C overnight with mixing by rotation. The second and third elutions were at 70 °C for 30 min. Following each elution, beads were centrifuged at 8000g for 2 min, and the supernatant was collected. The three elution supernatants were combined. Prior to liquid chromatography-tandem mass spectrometry (LC-MS-MS) analysis, samples were concentrated in vacuo in a SpeedVac vacuum centrifuge (Thermo-Fisher Scientific, Waltham, MA) to 10 µL and then diluted with 60 µL of 0.1% formic acid.
LC-MS-MS Analyses and Identification of Electrophile-Adducted Peptides
Peptide digests were analyzed by reverse-phase LC-MS-MS as described previously (31). IAB- and BMCC-adducted peptide sequences were determined by search of MS-MS spectra against the UniRef 100 database (downloaded June 2005) with TurboSequest (ThermoElectron, San Jose, CA) (34). Sequence-spectrum matches were filtered through a custom-designed software and database system as described previously (31) with the following modifications: S-Carboxamidomethylation of Cys (+57 amu), oxidation of Met (+16 amu), IAB adduction at Cys [+382.2 and +398.2 amu (oxidized adduct)], and BMCC adduction at Cys [+533.7 and +549.7 amu (oxidized adduct)] were specified as dynamic modifications. Criteria for acceptance of MS-MS spectrum to database sequence matches for adducted peptides have been described previously (30). Sequest outputs were analyzed with a custom-designed software and database system called CHIPS (Complete Hierarchical Integration of Protein Searches), which enables filtering of Sequest output files based on Sequest output parameters for sequence-spectrum matches and other criteria. Sequence-spectrum assignments were accepted based on the following filtering criteria: (i) All peptide sequence assignments were required to result from fully tryptic cleavages; (ii) all peptides possessed the appropriate reactive electrophile adduct mass; (iii) adducted peptides needed to be present in at least three out of five sample sets in the IAB and BMCC experiments to be accepted; (iv) singly, doubly, and triply charged ions were accepted if their XCorr scores were greater than 2, 2.5, and 3, respectively; and (v) all putative matches were confirmed by visual inspection of the spectra. Peptide identifications from all 20 SDS-PAGE gel slice fractions were combined in the database system. Identifications were accepted when the same adducted peptide was found in at least three out of five replicate experiments.
Results and Discussion
Toxicity and Induction of Apoptosis End Points by IAB and BMCC
We chose IAB and BMCC as model electrophile probes because these two compounds display chemistries that are commonly observed in biologically relevant electrophiles generated by metabolism of xenobiotics and by endogenous oxidation of lipids. IAB undergoes SN2 nucleophilic displacement reactions with cellular nucleophiles and mimics aliphatic epoxides, alkyl halides, and episulfonium ions. BMCC is a Michael acceptor whose chemistry mimics that of α,β-unsaturated carbonyl metabolites and lipid oxidation products, as well as quinones and quinone methides. Both compounds are highly reactive toward protein cysteine thiols but also may modify other nucleophiles.
Our previous studies with IAB [or its congener (+)-biotinyl-iodoacetamidyl-3,6-dioxaoctanediamine, (PEO-IAB)] and BMCC indicated that both the iodoacetamido and the N-alkylmaleimido probes alkylate comparable numbers of protein targets, as determined by LC-MS-MS analyses (30, 31). These studies also indicated that the overlap in targets alkylated by the two electrophiles is about 20%, which indicates different selectivities. These probes display notable differences in toxicity and induction of stress responses. IAB induced ER stress in HEK293 cells, whereas BMCC did not (31). IAB also induced activation of the stress-activated transcription factor Nrf2 in HepG2 cells and HEK293 cells, whereas BMCC did not (35).
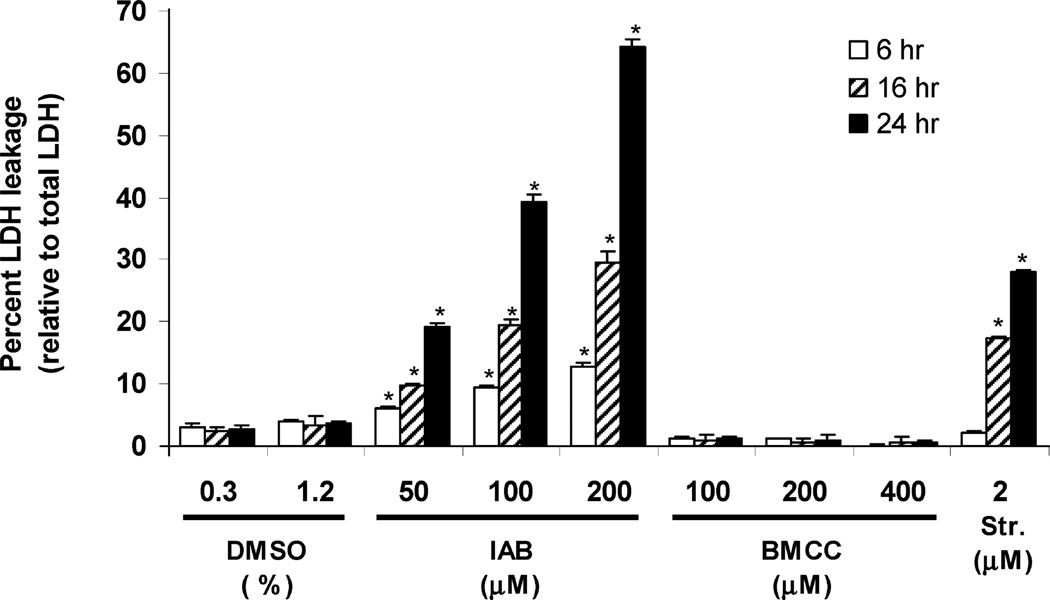
We began these studies with a focus on the toxicity differences between these compounds. We first measured the leakage of LDH from HEK293 cells, which is due to lysis of the cell membrane due either to primary necrosis or to necrosis secondary to apoptosis, following electrophile treatment for 6, 16, or 24 h. (Figure 2). Cytotoxicity of IAB increased with treatment time, and treatments with 50 and 100 µM IAB increased LDH leakage up to 19 and 39%, respectively, at 24 h. The cytotoxic response was similar to that of staurosporine, a potent inducer of apoptosis (36). In contrast, little cytotoxicity was caused by BMCC, which produced only 1% LDH leakage (100 µM, 24 h).
Figure 2.
Cytotoxicity of IAB and BMCC in HEK293 cells. Cells were treated with indicated concentrations of IAB, BMCC, and staurosporine (positive control) for 6 (empty bars), 12 (hatched bars), or 24 h (filled bars). Cytotoxicity was measured as LDH leakage from the cells and is reported as a percent of total LDH in the cells. Results were determined from three independent experiments (n = 3). *p < 0.001 as compared to respective DMSO controls.
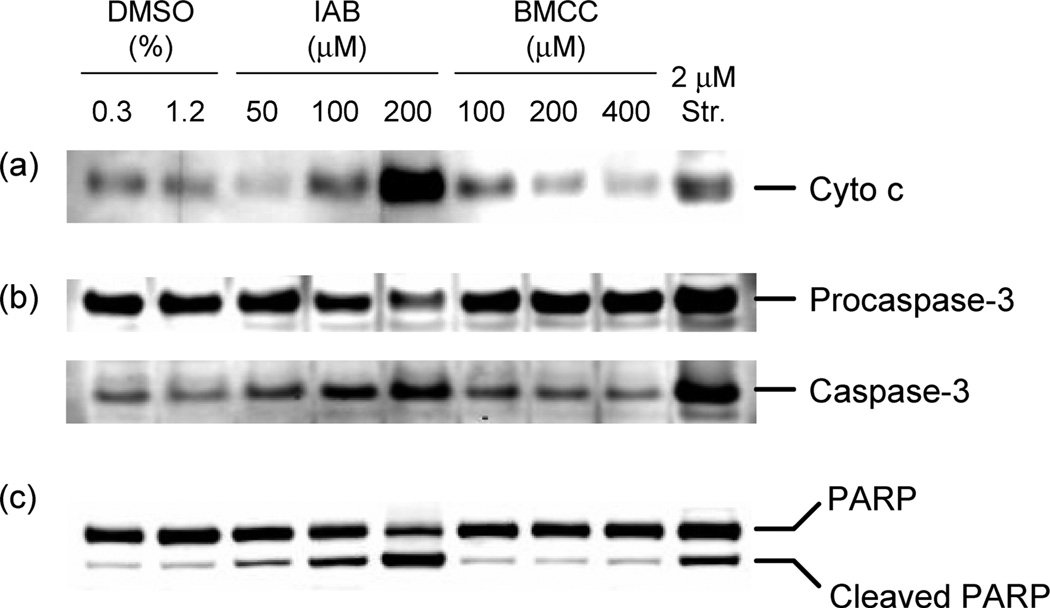
We next tested the ability of IAB and BMCC to induce apoptosis in HEK293 cells. IAB (50–200 µM) caused the release of cytochrome c from the mitochondria to the cytosol after 24 h of treatment (Figure 3A). This event is characteristic of mitochondrion-dependent apoptosis (1). In contrast, BMCC (100–400 µM) did not induce cytochrome c release at concentrations up to 400 µM (Figure 3A). PARP cleavage by caspase 3 is a hallmark biochemical characteristic of apoptosis (37), and caspase-3 activation was induced by IAB to a similar extent to that caused by staurosporine (Figure 3B). IAB (50–200 µM) induced cleavage of the DNA repair protein PARP from a full length 116 kDa form to a cleaved (89 kDa) form, whereas BMCC (up to 400 µM) did not induce PARP cleavage after 24 h of treatment (Figure 3C). Overall, these results demonstrate that IAB induces mitochondrion-dependent apoptosis, but BMCC does not, even at concentrations up to 400 µM, which is the limit of BMCC aqueous solubility.
Figure 3.
Apoptosis was induced by IAB but not by BMCC in HEK293 cells. Cells were treated with indicated concentrations of IAB or BMCC for 24 h. (a) Mitochondrial cytochrome c release was measured in cytosolic fractions. (b) Caspase-3 activation and (c) PARP cleavage were measured in whole cell lysates.
Identification of Protein Cysteinyl Adducts of IAB and BMCC
The major objective of the analyses described below was to identify mitochondrial proteins with a high reactivity toward the electrophile probes. As in our previous studies (30, 31), we exposed isolated mitochondrial fractions rather than intact cells to IAB and BMCC. This makes intrinsic reactivity toward electrophiles the predominant factor determining adduction, rather than the ability of the electrophiles to penetrate cells and reach mitochondrial targets. Indeed, physiologically relevant electrophiles may arise from both inside and outside cells or within mitochondria. The major analytical challenge that we faced in the analysis of mitochondrial protein targets is the high proportion of transmembrane, membrane-associated, and other hydrophobic proteins in mitochondrial preparations exposed to the biotin electrophile probes. We employed an analytical approach similar to what we described previously (31). The major elements of the approach are a 1D SDS-PAGE separation to resolve mitochondrial proteins, including hydrophobic proteins by molecular weight, in-gel digestion to generate tryptic peptides, streptavidin capture of biotinylated (adducted) peptides from the digests, and LC-MS-MS analysis of captured peptides.
Mitochondrial fractions were purified by Percoll gradient centrifugation of the crude mitochondrial fraction (27000g pellet). The purity of these fractions was determined by Western blot detection of representative organelle markers as shown in the Supporting Information. Percoll gradient centrifugation enriched the mitochondria relative to the crude mitochondria and the whole cell lysate based on band intensities of cytochrome oxidase IV band (mitochondrial membrane-bound protein). Glutathione-S-transferase was not present in the Percoll-purified fractions, which suggested that the purified mitochondrial fraction was minimally contaminated by cytosolic proteins. Detectable amounts of actin (cytoskeleton marker), Grp78/Bip (ER), and PARP (nucleus) were present in the purifed fractions. Our protein adduct inventories suggested that these traditionally employed markers provide an inadequate index of mitochondrial purity (see below).
Mitochondria were incubated with IAB and BMCC in 10 mM HEPES buffer at pH 7.4. This hypotonic buffer maintained near-physiological pH for mitochondrial proteins but would cause swelling and rupture of inner mitochondrial membranes, which enabled comparable exposure of mitochondrial outer membrane, intermembrane, and matrix proteins to the electrophile probes.
Incubations of mitochondrial preparations with IAB and BMCC were terminated by the addition of excess dithiothreitol, which readily reacts with and neutralizes the excess electrophiles. The proteins then were resolved by SDS-PAGE, reduced with dithiothreitol, and then alkylated with iodoacetamide. Following in-gel tryptic digestion, the adducted peptides were captured with immobilized streptavidin. Following high-salt washes to remove nonbiotinylated peptides, the biotinylated peptide adducts were eluted with 70:25:5 acetonitrile/H2O/formic acid at temperatures up to 70 °C. This elution method was found to elute a greater number of adducted peptides from streptavidin than than the room temperature elution that we described previously (30, 31). To compare the two methods, tryptic digests of seven different gel fractions from IAB-treated purified mitochondria were loaded onto streptavidin sepharose beads, washed, and then eluted by each of the methods. The high-temperature method consistently identified at least 2.7-fold more protein targets as shown in Supporting Information Figure S2. One complication of the high-temperature elution is the elution of some streptavidin protein monomer with the adducted peptides. Consequently, the enriched adduct mixture was refrigerated at 4 °C overnight to precipitate the contaminating streptavidin. The supernatant was isolated from the precipitate by centrifugation and was then analyzed by LC-MS-MS.
Peptide adduct sequence matches were assigned based on Universal Protein (UniProt) human database matches by the Sequest algorithm (34); this program accounts for the expected mass change from cysteine modifications by IAB or BMCC. All identified peptide adducts contained cysteine modifications of +382.2 Da for IAB, +533.7 Da for BMCC, or the corresponding S-oxidized products (+398.2 and +549.7 Da, respectively). These adducts are the SN2 reaction product of the cysteine thiol and the iodoacetamido group of IAB or Michael addition product of the cysteine thiol and the N-ethylmaleimido group of BMCC. Sequence-spectrum matches that met the minimum requirements as outlined in the Experimental Procedures were accepted. Several representative MS-MS spectra-sequence assignments are shown in Supporting Information Figures S5–S14.
Mitochondrial Protein Targets
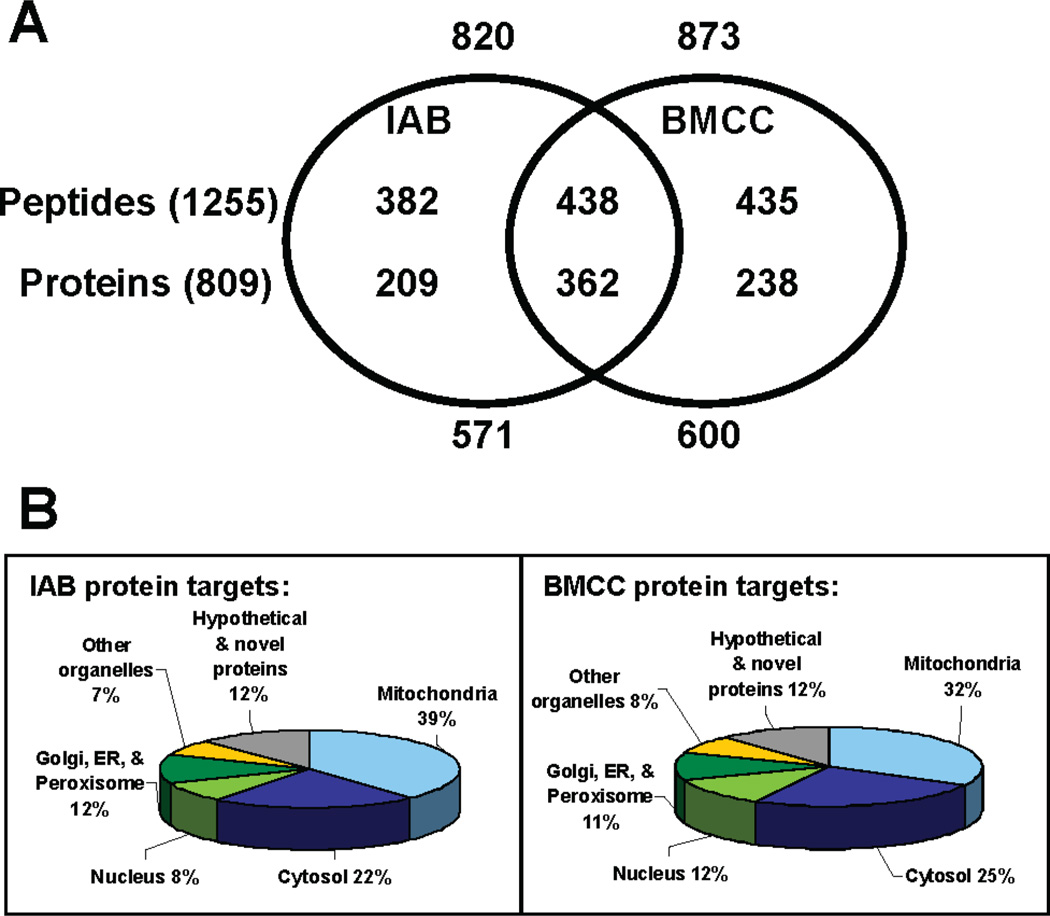
The complete list of the protein targets from the in vitro reaction of purified mitochondria with the two biotin-tagged electrophiles is given in Supporting Information Table 1. A Venn diagram summarizing the overlap of cysteine adducts of IAB and BMCC is shown in Figure 4A. There were a total of 1693 IAB or BMCC adducts identified at 1255 cysteine sites. IAB modified 820 of these cysteine adducts, which were mapped to 571 proteins. Similarly, BMCC adducted 873 cysteines, which mapped to 600 proteins. One striking feature of the data set is the difference in cysteine thiol reactivity between the two electrophiles. Only a modest fraction of the identified targets (438 peptides or 35% of the total identified adduct sites; 362 proteins or 45% of the total identified protein targets) was adducted by both IAB and BMCC. This result is similar to our observations with adduction of cytosolic and nuclear proteins (30) and of microsomal proteins (31). However, 209 proteins were selectively adducted by IAB, whereas 238 proteins were selectively adducted by BMCC. Of the proteins adducted by both electrophiles, different cysteine residues were targeted in some cases. An example is adenine nucleotide translocase 3 (ANT3), a mitochondrial inner membrane protein reportedly involved in apoptosis. BMCC covalently modifies Cys159 and Cys256, whereas IAB only reacts with Cys256 (Table 1).
Figure 4.
(a) Venn diagram of the overlap of protein and cysteine adduct targets of IAB and BMCC. (b) Distribution of primary subcellular location for IAB and BMCC protein targets annotated from the UniProt database (http://www.pir.uniprot.org/).
Table 1.
Protein Targets of IAB and BMCC Related to Apoptosis
| position of adducted
Cys |
|||||
|---|---|---|---|---|---|
| protein type | protein name | UniProt KB prot ID | IAB | BMCC | refs |
| permeability transition pore complex | ADP/ATP translocase 2 | P05141 | 256 | 256 | 53 |
| ADP/ATP translocase 3 | P12236 | 256 | 159, 256 | 54 | |
| creatine kinase,
sarcomeric mitochondrial precursor |
P17540 | 317 | 317 | 55 | |
| hexokinase, type I | P19367 | 628 | 628 | 38 | |
| splice isoform 3 of VDAC2 | P45880-3 | 13 | 13 | 56 | |
| VDAC1 | P21796 | 126, 231 | 126, 231 | 57 | |
| VDAC2 | P45880 | 62, 91 | 62, 91, 242 | 56 | |
| VDAC3 | Q9Y277 | 36, 65, 229 | 36, 65 | 58 | |
| preprotein import translocation | import inner membrane | O43615 | 343 | 59 | |
| translocase subunit
TIM44, mitochondrial precursor |
|||||
| import inner
membrane translocase subunit TIM50, mitochondrial precursor |
Q3ZCQ8 | 236 | 60 | ||
| probable
mitochondrial import receptor subunit TOM40 homologue |
O96008 | 74 | 74, 76, 86 | 61 | |
| thioredoxin | peroxiredoxin 1 | Q06830 | 83 | 173 | 62 |
| peroxiredoxin 4 | Q13162 | 51, 245 | 51, 245 | 63 | |
| peroxiredoxin 5,
mitochondrial precursor |
P30044 | 100 | 64 | ||
| thioredoxin | P10599 | 72 | 65 | ||
| thioredoxin
domain containing protein 5 precursor |
Q8NBS9 | 121, 217, 247, 350 | 121, 128, 247, 254, 381 | 66 | |
| thioredoxin reductase
2, mitochondrial precursor |
Q9NNW7 | 168 | 67, 68 | ||
| thioredoxin,
mitochondrial precursor |
Q99757 | 90 | 32, 69 | ||
| thioredoxin-dependent peroxide reductase, mitochondrial precursor |
P30048 | 229 | 229 | 70 | |
| thioredoxin-like 5 | Q9BRA2 | 43 | 71 | ||
| heat shock proteins | 60 kDa heat shock
protein, mitochondrial precursor |
P10809 | 237, 442, 447 | 237, 442, 447 | 72 |
| heat shock 70 kDa protein 1 | P08107 | 306 | 17, 306 | 73 | |
| heat shock 70 kDa protein 4 | P34932 | 34 | 73 | ||
| heat shock cognate 71 kDa protein | P11142 | 17 | 17, 63 | 73 | |
| heat shock protein 75
kDa, mitochondrial precursor |
Q12931 | 573 | 501, 573 | 74, 75 | |
| STIP1 homology and
U box-containing protein 1 |
Q9UNE7 | 199 | 48, 83 | 50 | |
| stress-70
protein, mitochondrial precursor |
P38646 | 317, 366, 608 | 317, 608 | 76, 77 | |
| assorted | 38 kDa FK-506
binding protein homologue |
Q14318 | 170, 217, 238 | 138, 170, 217, 238 | 78 |
| aconitate
hydratase, mitochondrial precursor |
Q99798 | 385 | 79 | ||
| caspase-3 precursor | P42574 | 163 | 80 | ||
| caspase-6 precursor | P55212 | 163 | 80 | ||
| catalase | P04040 | 376 | 376 | 81 | |
| cofilin 1 | P23528 | 138 | 138 | 39 | |
| ETHE1 protein,
mitochondrial precursor |
O95571 | 170 | 82 | ||
| FEM-1-like death
receptor binding protein |
Q9UK73 | 186 | 83 | ||
| FK506-binding protein
8 variant |
Q53GU3 | 137 | 78 | ||
| macrophage
migration inhibitory factor |
P14174 | 80 | 84 | ||
| oxidoreductase HTATIP2 | Q9BUP3 | 172 | 172 | 85 | |
| peptidylprolyl isomerase
A, isoform 2 |
P62937 | 101 | 101 | 86 | |
| programmed cell
death 6-interacting protein |
Q8WUM4 | 250 | 87 | ||
| programmed cell death protein
8, mitochondrial precursor |
O95831 | 256 | 88 | ||
| sphingosine-1-phosphate lyase 1 | O95470 | 285 | 285 | 89 | |
| tax1-binding protein 1 | Q86VP1 | 675 | 90 | ||
| transmembrane GTPase MFN2 | O95140 | 348 | 91 | ||
Approximately one-third of all of the identified protein targets were annotated as having mitochondrial localizations in the (http://www.pir.uniprot.org) or Bioinfomatic Harvester (http://harvester.embl.de) protein databases. Our finding that about two-thirds of the identified protein targets were not annotated as having mitochondrial localization is perhaps surprising. Two factors may contribute to this finding. First, despite our use of a widely accepted method for isolation of mitochondria, our mitochondrial preparations were not rigorously purified and were inevitably contaminated with proteins considered to be markers for other subcellular organelles (Supporting Information Figure S1). Second, some of the proteins that we identified may have multiple subcellular localizations, some of which are not represented in current databases. Indeed, it has been demonstrated previously that proteins can translocate from one organelle to another. Two protein targets, hexokinase (38) and cofilin (39), have been shown to translocate from the cytoplasm and nucleus, respectively, to the mitochondria. Thus, cross-contamination and multiorganelle distribution of some proteins presumably account for our observation. Similar findings have been reported by others who have done shotgun proteomic analyses on subcellular fractions (40, 41).
The human genome encodes an estimated 1000–2000 proteins in mitochondria (42, 43). In this study, IAB or BMCC covalently modified a total of 809 proteins in purified mitochondrial fractions (Figure 4). Previously, the two largest proteomic studies of human mitochondria identified 615 proteins from purified heart mitochondria and 680 proteins from mitochondria of T leukemia cells (44, 45). The relatively higher number of identifications in our study may reflect selective enrichment of cysteine-adducted peptides by streptavidin capture. This enables a broader sampling of proteins, although only electrophile targets are captured.
The total of over 800 protein targets identified in this study is dramatically higher than results from previous studies identifying protein targets of reactive electrophiles and oxidants. Marley and co-workers identified 51 mitochondrial protein targets of thiol-reactive (4-iodobutyl)triphenylphosphonium from rat heart mitochondria by LC-MS-MS analysis (46). In a separate study, 29 protein targets were identified from reaction of HEK293 whole cell lysate and the electrophilic lipid 15-deoxy-Δ12,14-prostaglandin J2 by two-dimensional isoelectric focusing, SDS-PAGE, and silver staining (47). Suh and co-workers employed a differential labeling approach, a thiol-reactive biotin-maleimide (structurally similar to BMCC) and 2D SDS-PAGE, to identify 10 mitochondrial proteins that underewent thiol oxidation in alcohol-exposed human hepatoma cells (48). The identified targets included heat shock protein 60, protein disulfide isomerase, protein disulfide isomerase A3, and mitochondrial aldehyde dehydrogenase, all of which were identified in our present study. Our data confirm previous work but provide a greatly expanded list of mitochondrial proteins that are targeted by reactive electrophiles.
We hypothesize that protein modifications by these electrophiles could act as sensors to trigger events of apoptosis. Although our identification of mitochondrial protein targets per se does not provide a test of this hypothesis, consideration of the identities of the identified targets and their known involvement in apoptotic mechanisms is worth examining and could provide the basis for new mechanistic studies. The differential induction of apoptosis by our probes (IAB induces apoptosis, and BMCC does not) and differential labeling of protein targets provide an interesting framework for consideration. A listing of IAB and BMCC protein targets associated with apoptosis is compiled in Table 1. The PubMed references offer representative evidence for the role of protein targets in apoptosis. (The literature citations can be accessed by clicking the weblinks.)
The three major classes that emerged from this compilation were pore channel proteins (PTP channel, preprotein translocation), thioredoxin family proteins, and heat shock proteins. Adduction of proteins in these classes may offer new insights on mechanisms of electrophile-induced apoptosis.
One proposed mechanism of cytochrome c release is the opening of the PTP channel resulting in the mitochondrial permeability transition, loss of transmembrane potential (Δψm), mitochondrial swelling, and rupture of the outer membrane (3). Covalent modification of cysteine thiols of ANT, a putative member of the PTP channel, coincided with membrane permeabilization (15). Modification of the ANT and other PTP channel members identified in this study may induce pore opening and cytochrome c release.
The adduction of several apoptosis-related proteins differed for the two electrophiles that differed in inducing apoptosis (Table 1 and Figure 3). IAB reproducibly adducted mitochondrial thioredoxin at Cys 90 whereas BMCC adducts were not detected. This protein is vital in redox regulation, defense against oxidative stress, refolding of disulfide-containing proteins, and regulation of transcription factors (49). Another protein target of note is the C terminus of Hsp70-interacting protein (CHIP), which is also known as STIP1 homology and U box-containing protein 1. It regulates the activation of the stress response of heat shock proteins and protects against stress-induced apoptosis in mice (50). IAB only modified Cys199, whereas BMCC did not modify this residue but instead modified Cys48 and Cys83.
IAB and BMCC adducted several apoptosis-related protein targets at the same cysteine residue. For example, both electrophiles modified voltage-dependent anion-selective channel protein 1 at Cys126 and Cys231 (Table 1). However, this does not mean that adduction by the two probes was equivalent. The adduct data were qualitative (i.e., whether cysteine adducts were detected or not). A quantitative analysis of adduction by IAB or BMCC could reveal differences in adduction kinetics and degrees of modification that correlate with biological effect.
Cysteine residues that are specifically modified by IAB could be triggers for IAB-induced apoptosis, although we have no direct evidence to support this hypothesis. It is nevertheless reasonable to postulate that our IAB target list contains one or more triggers for apoptosis. We note in this context that we have previously demonstrated that adduction selectivity of these probes accounts for their selectivity in stimulating activation of the transcription factor Nrf2 (35) and in mediating inhibition of protein phosphatase 2A (51). Similarly, previous reports have demonstrated that iodoacetamide and N-ethylmaleimide, respective analogues of IAB and BMCC, differentially inactivated bovine ferrochelatase (52), probably due to the modification of different cysteine residues. As we have noted previously, IAB and BMCC also contain slightly different linker structures that may contribute to the alkylation differences that we observed (30, 31), although we cannot estimate the magnitude of these effects. We note also that our electrophile incubations were done in the absence of glutathione, a critical protectant against protein alkylation. Although the presence of glutatione could possibly alter specific protein—electrophile interactions (due perhaps to allosteric effects), the major effect of glutathione would be to increase the electrophile concentration required to alkylate the same targets. Thus, the protein targets identified here would be expected to be most reactive in intact cells.
In summary, our work demonstrates that two different thiol-reactive electrophile probes alkylate mitochondrial proteins with different selectivities. Furthermore, among the identified protein targets involved in apoptosis, these probes have distinct adduction profiles that may explain their differing pro-apoptotic activities. These observations demonstrate that adduction specificity reflects different biological effects. Accordingly, analyses of site-specific protein alkylation by electrophilic xenobiotic metabolites and endogenous electrophiles generated from oxidative stress and inflammation could predict mechanisms of toxicity, including apoptosis. Further work is necessary to test this hypothesis, but our observations reduce the problem to evaluation of the roles of specific protein targets.
Supplementary Material
Acknowledgment
We thank Drs. Simona G. Codreanu, Chuan Ji, Nah-Young Shin, and Ying Xiong for helpful discussions. We are also grateful for financial support from the National Institutes of Health (Grants T32 ES010056, ES000267, and ES007028).
Footnotes
Abbreviations: AIF, apoptosis-inducing factor; ANT, adenine nucleotide translocase; IAB, N-iodoacetyl-N-biotinylhexylenediamine; BMCC, 1-biotinamido-4-(4′-[maleimidoethylcyclo-hexane]-carboxamido)-butane; Cyto Ox 4, cytochrome oxidase 4; DMEM, Dulbecco’s modified Eagle’s medium; ER, endoplasmic reticulum; HEK293, human embryonic kidney 293 cells; HNE, 4-hydroxy-2-nonenal; HPLC, high-performance liquid chromatography; LC-MS-MS, liquid chromatography-tandem mass spectrometry; LDH, lactate dehydrogenase; PARP, poly(ADP-ribose)polymerase; PBS, phosphate-buffered saline; PEO-IAB, (+)-biotinyl-iodoacetamidyl-3,6-dioxaoctanediamine; PTP, permeability transition pore; SDS-PAGE, sodium dodecyl sulfate—polyacrylamide gel electrophoresis; SMAC/DIABLO, secondary mitochondrial activator of caspases/direct IAP binding protein with low pI; TIM, translocase of inner membrane; TOM, translocase of outer membrane; UniProt, Universal Protein Resource human database; VDAC, voltage-dependent anion channel protein.
Supporting Information Available: Western blots comparing the purity of the whole cell homogenate and crude and purified mitochondrial fractions (Figure S1); comparison between streptavidin elution methods from a previous study by Dennehy and co-workers (30) and the present study on a number of protein targets identified by LC-MS-MS (Figure S2); distribution of the number of cysteine adducts on identified protein targets of BMCC and IAB (Figure S3); induction of caspase-3 activity by IAB but not BMCC by the colorimetric CaspACE assay system (Figure S4); several representative MS-MS spectra of protein targets of IAB and BMCC (Figures S5–S14); and tables showing the protein targets (Table S1) and protein motifs and features (Table S2) of IAB and BMCC. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Green DR, Kroemer G. Pharmacological manipulation of cell death: clinical applications in sight? J. Clin. Invest. 2005;115:2610–2617. doi: 10.1172/JCI26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zamzami N, Kroemer G. The mitochondrion in apoptosis: how Pandora’s box opens. Nat. Rev. Mol. Cell Biol. 2001;2:67–71. doi: 10.1038/35048073. [DOI] [PubMed] [Google Scholar]
- 3.Von Ahsen O, Waterhouse NJ, Kuwana T, Newmeyer DD, Green DR. The ‘harmless’ release of cytochrome c. Cell Death Differ. 2000;7:1192–1199. doi: 10.1038/sj.cdd.4400782. [DOI] [PubMed] [Google Scholar]
- 4.Marnett LJ, Riggins JN, West JD. Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J. Clin. Invest. 2003;111:583–593. doi: 10.1172/JCI18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji C, Amarnath V, Pietenpol JA, Marnett LJ. 4-hydroxynonenal induces apoptosis via caspase-3 activation and cytochrome c release. Chem. Res. Toxicol. 2001;14:1090–1096. doi: 10.1021/tx000186f. [DOI] [PubMed] [Google Scholar]
- 6.West JD, Marnett LJ. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem. Res. Toxicol. 2006;19:173–194. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- 7.Rich T, Allen RL, Wyllie AH. Defying death after DNA damage. Nature. 2000;407:777–783. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 8.Davis W, Ronai Z, Tew KD. Cellular thiols and reactive oxygen species in drug-induced apoptosis. J. Pharmacol. Exp. Ther. 2001;296:1–6. [PubMed] [Google Scholar]
- 9.Finkel T. Reactive oxygen species and signal transduction. IUBMB Life. 2001;52:3–6. doi: 10.1080/15216540252774694. [DOI] [PubMed] [Google Scholar]
- 10.Rhee SG, Bae YS, Lee SR, Kwon J. Hydrogen peroxide: A key messenger that modulates protein phosphorylation through cysteine oxidation. Sci. STKE. 2000:E1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- 11.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Water B, Wang Y, Asmellash S, Liu H, Zhan Y, Miller E, Stevens JL. Distinct endoplasmic reticulum signaling pathways regulate apoptotic and necrotic cell death following iodoacetamide treatment. Chem. Res. Toxicol. 1999;12:943–951. doi: 10.1021/tx990054q. [DOI] [PubMed] [Google Scholar]
- 13.Rao RV, Peel A, Logvinova A, del RG, Hermel E, Yokota T, Goldsmith PC, Ellerby LM, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program: Role of the ER chaperone GRP78. FEBS Lett. 2002;514:122–128. doi: 10.1016/s0014-5793(02)02289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powis G, Briehl M, Oblong J. Redox signalling and the control of cell growth and death. Pharmacol. Ther. 1995;68:149–173. doi: 10.1016/0163-7258(95)02004-7. [DOI] [PubMed] [Google Scholar]
- 15.Costantini P, Belzacq AS, Vieira HL, Larochette N, de Pablo MA, Zamzami N, Susin SA, Brenner C, Kroemer G. Oxidation of a critical thiol residue of the adenine nucleotide translocator enforces Bcl-2-independent permeability transition pore opening and apoptosis. Oncogene. 2000;19:307–314. doi: 10.1038/sj.onc.1203299. [DOI] [PubMed] [Google Scholar]
- 16.Vieira HL, Belzacq AS, Haouzi D, Bernassola F, Cohen I, Jacotot E, Ferri KF, El Hamel C, Bartle LM, Melino G, Brenner C, Goldmacher V, Kroemer G. The adenine nucleotide translocator: A target of nitric oxide, peroxynitrite, and 4-hydroxynonenal. Oncogene. 2001;20:4305–4316. doi: 10.1038/sj.onc.1204575. [DOI] [PubMed] [Google Scholar]
- 17.Majima E, Koike H, Hong YM, Shinohara Y, Terada H. Characterization of cysteine residues of mitochondrial ADP/ATP carrier with the SH-reagents eosin 5-maleimide and N-ethyl-maleimide. J. Biol. Chem. 1993;268:22181–22187. [PubMed] [Google Scholar]
- 18.Jones JG, Otieno S, Barnard EA, Bhargava AK. Essential and nonessential thiols of yeast hexokinase. Reactions with iodoacetate and iodoacetamide. Biochemistry. 1975;14:2396–2403. doi: 10.1021/bi00682a020. [DOI] [PubMed] [Google Scholar]
- 19.Otieno S, Bhargava AK, Barnard EA, Ramel AH. Essential thiols of yeast hexokinase: Alkylation by a substrate-like reagent. Biochemistry. 1975;14:2403–2410. doi: 10.1021/bi00682a021. [DOI] [PubMed] [Google Scholar]
- 20.Subbarao B, Kenkare UW. Reaction of brain hexokinase with tetranitromethane: Oxidation of essential thiol groups. Arch. Biochem. Biophys. 1977;181:8–18. doi: 10.1016/0003-9861(77)90477-5. [DOI] [PubMed] [Google Scholar]
- 21.Chakraborti T, Das S, Mondal M, Roychoudhury S, Chakraborti S. Oxidant, mitochondria and calcium: An overview. Cell. Signaling. 1999;11:77–85. doi: 10.1016/s0898-6568(98)00025-4. [DOI] [PubMed] [Google Scholar]
- 22.West JD, Ji C, Duncan ST, Amarnath V, Schneider C, Rizzo CJ, Brash AR, Marnett LJ. Induction of apoptosis in colorectal carcinoma cells treated with 4-hydroxy-2-nonenal and structurally related aldehydic products of lipid peroxidation. Chem. Res. Toxicol. 2004;17:453–462. doi: 10.1021/tx034248o. [DOI] [PubMed] [Google Scholar]
- 23.Jian W, Arora JS, Oe T, Shuvaev VV, Blair IA. Induction of endothelial cell apoptosis by lipid hydroperoxide-derived bifunctional electrophiles. Free Radical Biol. Med. 2005;39:1162–1176. doi: 10.1016/j.freeradbiomed.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Kondo M, Shibata T, Kumagai T, Osawa T, Shibata N, Kobayashi M, Sasaki S, Iwata M, Noguchi N, Uchida K. 15-Deoxy-Delta(12,14)-prostaglandin J(2): the endogenous electrophile that induces neuronal apoptosis. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7367–7372. doi: 10.1073/pnas.112212599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landar A, Shiva S, Levonen AL, Oh JY, Zaragoza C, Johnson MS, Darley-Usmar VM. Induction of the permeability transition and cytochrome c release by 15-deoxy-Delta12,14-prostaglandin J2 in mitochondria. Biochem. J. 2006;394:185–195. doi: 10.1042/BJ20051259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomson SJ, Brown KK, Pullar JM, Hampton MB. Phenethyl isothiocyanate triggers apoptosis in Jurkat cells made resistant by the overexpression of Bcl-2. Cancer Res. 2006;66:6772–6777. doi: 10.1158/0008-5472.CAN-05-3809. [DOI] [PubMed] [Google Scholar]
- 27.Singh AV, Xiao D, Lew KL, Dhir R, Singh SV. Sulforaphane induces caspase-mediated apoptosis in cultured PC-3 human prostate cancer cells and retards growth of PC-3 xenografts in vivo. Carcinogenesis. 2004;25:83–90. doi: 10.1093/carcin/bgg178. [DOI] [PubMed] [Google Scholar]
- 28.Ghanayem BI, Elwell MR, Eldridge SR. Effects of the carcinogen, acrylonitrile, on forestomach cell proliferation and apoptosis in the rat: Comparison with methacrylonitrile. Carcinogenesis. 1997;18:675–680. doi: 10.1093/carcin/18.4.675. [DOI] [PubMed] [Google Scholar]
- 29.Tithof PK, Elgayyar M, Schuller HM, Barnhill M, Andrews R. 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a nicotine derivative, induces apoptosis of endothelial cells. Am. J. Physiol. Heart Circ. Physiol. 2001;281:H1946–H1954. doi: 10.1152/ajpheart.2001.281.5.H1946. [DOI] [PubMed] [Google Scholar]
- 30.Dennehy MK, Richards KAM, Wernke GW, Shyr Y, Liebler DC. Cytosolic and nuclear protein targets of thiol-reactive electrophiles. Chem. Res. Toxicol. 2006;19:20–29. doi: 10.1021/tx050312l. [DOI] [PubMed] [Google Scholar]
- 31.Shin NY, Liu Q, Stamer SL, Liebler DC. Protein targets of reactive electrophiles in human liver microsomes. Chem. Res. Toxicol. 2007;20:859–867. doi: 10.1021/tx700031r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Cai J, Murphy TJ, Jones DP. Overexpressed human mitochondrial thioredoxin confers resistance to oxidant-induced apoptosis in human osteosarcoma cells. J. Biol. Chem. 2002;277:33242–33248. doi: 10.1074/jbc.M202026200. [DOI] [PubMed] [Google Scholar]
- 33.Davidson CE, Reese BE, Billingsley ML, Yun JK. Stannin, a protein that localizes to the mitochondria and sensitizes NIH-3T3 cells to trimethyltin and dimethyltin toxicity. Mol. Pharmacol. 2004;66:855–863. doi: 10.1124/mol.104.001719. [DOI] [PubMed] [Google Scholar]
- 34.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 35.Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J. Biol. Chem. 2005;280:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 36.Belmokhtar CA, Hillion J, Segal-Bendirdjian E. Staurosporine induces apoptosis through both caspase-dependent and caspase-independent mechanisms. Oncogene. 2001;20:3354–3362. doi: 10.1038/sj.onc.1204436. [DOI] [PubMed] [Google Scholar]
- 37.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 38.Majewski N, Nogueira V, Robey RB, Hay N. Akt inhibits apoptosis downstream of BID cleavage via a glucose-dependent mechanism involving mitochondrial hexokinases. Mol. Cell. Biol. 2004;24:730–740. doi: 10.1128/MCB.24.2.730-740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chua BT, Volbracht C, Tan KO, Li R, Yu VC, Li P. Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat. Cell Biol. 2003;5:1083–1089. doi: 10.1038/ncb1070. [DOI] [PubMed] [Google Scholar]
- 40.Yi EC, Marelli M, Lee H, Purvine SO, Aebersold R, Aitchison JD, Goodlett DR. Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis. 2002;23:3205–3216. doi: 10.1002/1522-2683(200209)23:18<3205::AID-ELPS3205>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 41.Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 42.McDonald TG, Van Eyk JE. Mitochondrial proteomics. Undercover in the lipid bilayer. Basic Res. Cardiol. 2003;98:219–227. doi: 10.1007/s00395-003-0417-8. [DOI] [PubMed] [Google Scholar]
- 43.DiMauro S, Schon EA. Nuclear power and mitochondrial disease. Nat. Genet. 1998;19:214–215. doi: 10.1038/883. [DOI] [PubMed] [Google Scholar]
- 44.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 45.Rezaul K, Wu L, Mayya V, Hwang SI, Han D. A systematic characterization of mitochondrial proteome from human T leukemia cells. Mol Cell Proteomics. 2005;4:169–181. doi: 10.1074/mcp.M400115-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marley K, Mooney DT, Clark-Scannell G, Tong TT, Watson J, Hagen TM, Stevens JF, Maier CS. Mass tagging approach for mitochondrial thiol proteins. J. Proteome Res. 2005;4:1403–1412. doi: 10.1021/pr050078k. [DOI] [PubMed] [Google Scholar]
- 47.Levonen AL, Landar A, Ramachandran A, Ceasar EK, Dickinson DA, Zanon G, Morrow JD, Darley-Usmar VM. Cellular mechanisms of redox cell signaling: Role of cysteine modification in controlling antioxidant defenses in response to electrophilic lipid oxidation products. Biochem. J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suh SK, Hood BL, Kim BJ, Conrads TP, Veenstra TD, Song BJ. Identification of oxidized mitochondrial proteins in alcohol-exposed human hepatoma cells and mouse liver. Proteomics. 2004;4:3401–3412. doi: 10.1002/pmic.200400971. [DOI] [PubMed] [Google Scholar]
- 49.Powis G, Montfort WR. Properties and biological activities of thioredoxins. Annu. Rev. Biophys. Biomol. Struct. 2001;30:421–455. doi: 10.1146/annurev.biophys.30.1.421. [DOI] [PubMed] [Google Scholar]
- 50.Dai Q, Zhang C, Wu Y, McDonough H, Whaley RA, Godfrey V, Li HH, Madamanchi N, Xu W, Neckers L, Cyr D, Patterson C. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Codreanu SG, Adams DG, Dawson ES, Wadzinski BE, Liebler DC. Inhibition of protein phosphatase 2A activity by selective electrophile alkylation damage. Biochemistry. 2006;45:10020–10029. doi: 10.1021/bi060551n. [DOI] [PubMed] [Google Scholar]
- 52.Dailey HA. Effect of sulfhydryl group modification on the activity of bovine ferrochelatase. J. Biol. Chem. 1984;259:2711–2715. [PubMed] [Google Scholar]
- 53.Le Bras M, Borgne-Sanchez A, Touat Z, El Dein OS, Deniaud A, Maillier E, Lecellier G, Rebouillat D, Lemaire C, Kroemer G, Jacotot E, Brenner C. Chemosensitization by knockdown of adenine nucleotide translocase-2. Cancer Res. 2006;66:9143–9152. doi: 10.1158/0008-5472.CAN-05-4407. [DOI] [PubMed] [Google Scholar]
- 54.Zamora M, Granell M, Mampel T, Vinas O. Adenine nucleotide translocase 3 (ANT3) overexpression induces apoptosis in cultured cells. FEBS Lett. 2004;563:155–160. doi: 10.1016/S0014-5793(04)00293-5. [DOI] [PubMed] [Google Scholar]
- 55.Schlattner U, Tokarska-Schlattner M, Wallimann T. Mitochondrial creatine kinase in human health and disease. Biochim. Biophys. Acta. 2006;1762:164–180. doi: 10.1016/j.bbadis.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 56.Cheng EH, Sheiko TV, Fisher JK, Craigen WJ, Korsmeyer SJ. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science. 2003;301:513–517. doi: 10.1126/science.1083995. [DOI] [PubMed] [Google Scholar]
- 57.Abu-Hamad S, Sivan S, Shoshan-Barmatz V. The expression level of the voltage-dependent anion channel controls life and death of the cell. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5787–5792. doi: 10.1073/pnas.0600103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rahmani Z, Huh KW, Lasher R, Siddiqui A. Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. J. Virol. 2000;74:2840–2846. doi: 10.1128/jvi.74.6.2840-2846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Wada J, Hashimoto I, Eguchi J, Yasuhara A, Kanwar YS, Shikata K, Makino H. Therapeutic approach for diabetic nephropathy using gene delivery of translocase of inner mitochondrial membrane 44 by reducing mitochondrial superoxide production. J. Am. Soc. Nephrol. 2006;17:1090–1101. doi: 10.1681/ASN.2005111148. [DOI] [PubMed] [Google Scholar]
- 60.Guo Y, Cheong N, Zhang Z, De Rose R, Deng Y, Farber SA, Fernandes-Alnemri T, Alnemri ES. Tim50, a component of the mitochondrial translocator, regulates mitochondrial integrity and cell death. J. Biol. Chem. 2004;279:24813–24825. doi: 10.1074/jbc.M402049200. [DOI] [PubMed] [Google Scholar]
- 61.Ott M, Norberg E, Walter KM, Schreiner P, Kemper C, Rapaport D, Zhivotovsky B, Orrenius S. The mitochondrial TOM complex is required for tBid/Bax-induced cytochrome c release. J. Biol. Chem. 2007;282:27633–27639. doi: 10.1074/jbc.M703155200. [DOI] [PubMed] [Google Scholar]
- 62.Berggren MI, Husbeck B, Samulitis B, Baker AF, Gallegos A, Powis G. Thioredoxin peroxidase-1 (peroxiredoxin-1) is increased in thioredoxin-1 transfected cells and results in enhanced protection against apoptosis caused by hydrogen peroxide but not by other agents including dexamethasone, etoposide, and doxorubicin. Arch. Biochem. Biophys. 2001;392:103–109. doi: 10.1006/abbi.2001.2435. [DOI] [PubMed] [Google Scholar]
- 63.Crowley-Weber CL, Payne CM, Gleason-Guzman M, Watts GS, Futscher B, Waltmire CN, Crowley C, Dvorakova K, Bernstein C, Craven M, Garewal H, Bernstein H. Development and molecular characterization of HCT-116 cell lines resistant to the tumor promoter and multiple stress-inducer, deoxy-cholate. Carcinogenesis. 2002;23:2063–2080. doi: 10.1093/carcin/23.12.2063. [DOI] [PubMed] [Google Scholar]
- 64.Yuan J, Murrell GA, Trickett A, Landtmeters M, Knoops B, Wang MX. Overexpression of antioxidant enzyme peroxiredoxin 5 protects human tendon cells against apoptosis and loss of cellular function during oxidative stress. Biochim. Biophys. Acta. 2004;1693:37–45. doi: 10.1016/j.bbamcr.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 65.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sullivan DC, Huminiecki L, Moore JW, Boyle JJ, Poulsom R, Creamer D, Barker J, Bicknell R. EndoPDI, a novel protein-disulfide isomerase-like protein that is preferentially expressed in endothelial cells acts as a stress survival factor. J. Biol. Chem. 2003;278:47079–47088. doi: 10.1074/jbc.M308124200. [DOI] [PubMed] [Google Scholar]
- 67.Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, Schmahl W, Chien KR, Wurst W, Bornkamm GW, Brielmeier M. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol. Cell. Biol. 2004;24:9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cenas N, Prast S, Nivinskas H, Sarlauskas J, Arner ES. Interactions of nitroaromatic compounds with the mammalian selenoprotein thioredoxin reductase and the relation to induction of apoptosis in human cancer cells. J. Biol. Chem. 2006;281:5593–5603. doi: 10.1074/jbc.M511972200. [DOI] [PubMed] [Google Scholar]
- 69.Nonn L, Williams RR, Erickson RP, Powis G. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol. Cell. Biol. 2003;23:916–922. doi: 10.1128/MCB.23.3.916-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nonn L, Berggren M, Powis G. Increased expression of mitochondrial peroxiredoxin-3 (thioredoxin peroxidase-2) protects cancer cells against hypoxia and drug-induced hydrogen peroxide-dependent apoptosis. Mol. Cancer Res. 2003;1:682–689. [PubMed] [Google Scholar]
- 71.Jeong W, Chang TS, Boja ES, Fales HM, Rhee SG. Roles of TRP14, a thioredoxin-related protein in tumor necrosis factor-alpha signaling pathways. J. Biol. Chem. 2004;279:3151–3159. doi: 10.1074/jbc.M307959200. [DOI] [PubMed] [Google Scholar]
- 72.Beere HM. “The stress of dying”: The role of heat shock proteins in the regulation of apoptosis. J. Cell Sci. 2004;117:2641–2651. doi: 10.1242/jcs.01284. [DOI] [PubMed] [Google Scholar]
- 73.Beere HM, Wolf BB, Cain K, Mosser DD, Mahboubi A, Kuwana T, Tailor P, Morimoto RI, Cohen GM, Green DR. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2000;2:469–475. doi: 10.1038/35019501. [DOI] [PubMed] [Google Scholar]
- 74.Pridgeon JW, Olzmann JA, Chin LS, Li L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Masuda Y, Shima G, Aiuchi T, Horie M, Hori K, Nakajo S, Kajimoto S, Shibayama-Imazu T, Nakaya K. Involvement of tumor necrosis factor receptor-associated protein 1 (TRAP1) in apoptosis induced by beta-hydroxyisovalerylshikonin. J. Biol. Chem. 2004;279:42503–42515. doi: 10.1074/jbc.M404256200. [DOI] [PubMed] [Google Scholar]
- 76.Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 77.Jin J, Hulette C, Wang Y, Zhang T, Pan C, Wadhwa R, Zhang J. Proteomic identification of a stress protein, mortalin/mthsp70/GRP75: relevance to Parkinson disease. Mol. Cell. Proteomics. 2006;5:1193–1204. doi: 10.1074/mcp.M500382-MCP200. [DOI] [PubMed] [Google Scholar]
- 78.Wang HQ, Nakaya Y, Du Z, Yamane T, Shirane M, Kudo T, Takeda M, Takebayashi K, Noda Y, Nakayama KI, Nishimura M. Interaction of presenilins with FKBP38 promotes apoptosis by reducing mitochondrial Bcl-2. Hum. Mol. Genet. 2005;14:1889–1902. doi: 10.1093/hmg/ddi195. [DOI] [PubMed] [Google Scholar]
- 79.Murakami K, Ishida K, Watakabe K, Tsubouchi R, Naruse M, Yoshino M. Maltol/iron-mediated apoptosis in HL60 cells: Participation of reactive oxygen species. Toxicol. Lett. 2006;161:102–107. doi: 10.1016/j.toxlet.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 80.Fernandes-Alnemri T, Litwack G, Alnemri ES. Mch2, a new member of the apoptotic Ced-3/Ice cysteine protease gene family. Cancer Res. 1995;55:2737–2742. [PubMed] [Google Scholar]
- 81.Bai J, Cederbaum AI. Catalase protects HepG2 cells from apoptosis induced by DNA-damaging agents by accelerating the degradation of p53. J. Biol. Chem. 2003;278:4660–4667. doi: 10.1074/jbc.M206273200. [DOI] [PubMed] [Google Scholar]
- 82.Higashitsuji H, Higashitsuji H, Nagao T, Nonoguchi K, Fujii S, Itoh K, Fujita J. A novel protein overexpressed in hepatoma accelerates export of NF-kappa B from the nucleus and inhibits p53-dependent apoptosis. Cancer Cell. 2002;2:335–346. doi: 10.1016/s1535-6108(02)00152-6. [DOI] [PubMed] [Google Scholar]
- 83.Chan SL, Tan KO, Zhang L, Yee KS, Ronca F, Chan MY, Yu VC. F1Aalpha, a death receptor-binding protein homologous to the Caenorhabditis elegans sex-determining protein, FEM-1, is a caspase substrate that mediates apoptosis. J. Biol. Chem. 1999;274:32461–32468. doi: 10.1074/jbc.274.45.32461. [DOI] [PubMed] [Google Scholar]
- 84.Shen L, Hu J, Lu H, Wu M, Qin W, Wan D, Li YY, Gu J. The apoptosis-associated protein BNIPL interacts with two cell proliferation-related proteins, MIF and GFER. FEBS Lett. 2003;540:86–90. doi: 10.1016/s0014-5793(03)00229-1. [DOI] [PubMed] [Google Scholar]
- 85.King FW, Shtivelman E. Inhibition of nuclear import by the proapoptotic protein CC3. Mol. Cell. Biol. 2004;24:7091–7101. doi: 10.1128/MCB.24.16.7091-7101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cande C, Vahsen N, Kouranti I, Schmitt E, Daugas E, Spahr C, Luban J, Kroemer RT, Giordanetto F, Garrido C, Penninger JM, Kroemer G. AIF and cyclophilin A cooperate in apoptosis-associated chromatinolysis. Oncogene. 2004;23:1514–1521. doi: 10.1038/sj.onc.1207279. [DOI] [PubMed] [Google Scholar]
- 87.Chatellard-Causse C, Blot B, Cristina N, Torch S, Missotten M, Sadoul R. Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J. Biol. Chem. 2002;277:29108–29115. doi: 10.1074/jbc.M204019200. [DOI] [PubMed] [Google Scholar]
- 88.Cande C, Cohen I, Daugas E, Ravagnan L, Larochette N, Zamzami N, Kroemer G. Apoptosis-inducing factor (AIF): A novel caspase-independent death effector released from mitochondria. Biochimie. 2002;84:215–222. doi: 10.1016/s0300-9084(02)01374-3. [DOI] [PubMed] [Google Scholar]
- 89.Oskouian B, Sooriyakumaran P, Borowsky AD, Crans A, Dillard-Telm L, Tam YY, Bandhuvula P, Saba JD. Sphingosine-1-phosphate lyase potentiates apoptosis via p53- and p38-dependent pathways and is down-regulated in colon cancer. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17384–17389. doi: 10.1073/pnas.0600050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.De Valck D, Jin DY, Heyninck K, Van de Craen M, Contreras R, Fiers W, Jeang KT, Beyaert R. The zinc finger protein A20 interacts with a novel anti-apoptotic protein which is cleaved by specific caspases. Oncogene. 1999;18:4182–4190. doi: 10.1038/sj.onc.1202787. [DOI] [PubMed] [Google Scholar]
- 91.Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, Santel A, Fuller M, Smith CL, Youle RJ. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J. Cell Biol. 2002;159:931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.