Abstract
Telomeres, the ends of linear eukaryotic chromosomes, are characterized by the presence of multiple repeats of a short DNA sequence. This telomeric DNA is protected from illicit repair by telomere-associated proteins, which in mammals form the shelterin complex. Replicative polymerases are unable to synthesize DNA at the extreme ends of chromosomes, but in unicellular eukaryotes such as yeast and in mammalian germ cells and stem cells, telomere length is maintained by a ribonucleoprotein enzyme known as telomerase. Recent work has provided insights into the mechanisms of telomerase recruitment to telomeres, highlighting the contribution of telomere-associated proteins including TPP1 in humans, Ccq1 in S. pombe, and Cdc13 and Ku in S. cerevisiae.
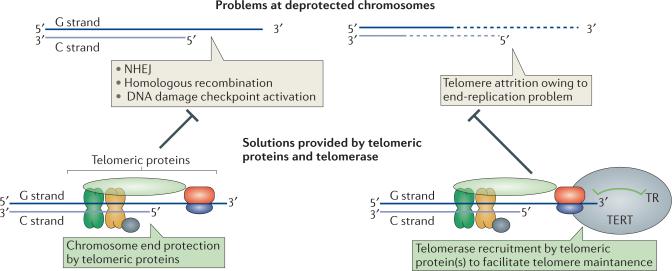
The proper maintenance of linear chromosomes requires countering two biological problems: the chromosome end-protection problem and the chromosome end-replication problem. The ends of linear chromosomes, such as those observed in eukaryotes, must be distinguished from broken DNA ends that require repair. In the absence of such a distinction, linear chromosomes are prone to illicit DNA end-joining and recombinational events that result in deleterious consequences such as end-to-end chromosomal fusions. Such reactions define the chromosome end-protection problem1. The gradual loss of sequence information at the extreme end of chromosomes owing to incomplete replication by DNA polymerases (which only synthesize DNA in the 5’ to 3’ direction and are unable to fill in the gap left behind by the 5’ most RNA primer) defines the chromosome end-replication problem2.
Telomerase, a unique enzyme that contains a protein reverse transcriptase (TERT) and a template-containing RNA component (TR), facilitates the solution of both chromosome end-related problems. By synthesizing multiple tandem repeats of DNA (called telomeric DNA) encoded by its RNA template, telomerase compensates for the erosion of DNA ends during replication (Fig. 1) and provides the docking sites for telomeric proteins that bind specifically to the ends of chromosome to distinguish them from broken DNA ends (Fig. 1). Although most protists, fungi, plants and animals employ a telomerase, Drosophila species use a retrotransposon mechanism to overcome chromosome end-protection and end-replication3.
Figure 1.
Chromosome end-protection versus telomerase recruitment and action. Deprotected, protein-free telomeric DNA (G/C strand duplex at top) could be a substrate for illicit DNA end-joining and cell cycle checkpoint activation events at the telomere. By specifically binding telomeric DNA, telomeric proteins protect chromosome ends from such deleterious processes. DNA sequences are lost at the ends of chromosomes due to incomplete replication by DNA polymerases. Telomerase is recruited to telomeres via interaction with specific telomeric proteins to extend chromosome ends and thereby counter DNA lost from incomplete replication. In the absence of telomerase recruitment or action, telomeric DNA shortens, ultimately leading to cell senescence. Furthermore, shorter telomeric DNA results in a loss of bound telomeric proteins, which could result in deprotected chromosome ends.
The action of telomerase is required for the survival of continuously dividing cells such as those of unicellular eukaryotes. In mammals, telomerase is active in the germ line and in stem cells, but its expression in somatic cells may lead to or predispose to cancer. In the absence of a telomere-maintenance mechanism, the telomeres of such proliferating cells shrink to the point when the cells stop dividing (replicative senescence). Telomerase has also been linked to ageing, as telomere loss may result in tissue atrophy, stem cell depletion and deficient tissue regeneration4. In humans, loss-of-function mutations in either TERT or TR have been associated with dyskeratosis congenita and cases of aplastic anaemia and pulmonary fibrosis5.
Since its discovery by Greider and Blackburn, telomerase has been studied at the level of structure, function, biology and medicine6-9. In this Review we focus on telomerase recruitment to telomeres, addressing the conceptual dilemma of how a chromosome end bound tightly by telomeric end-capping proteins also allows telomerase action. We describe, and compare and contrast the mechanisms of telomerase recruitment in three systems: human, budding yeast and fission yeast.
Telomeres
Telomeric DNA
Telomeric DNA is typically composed of multiple repeats of a short sequence, often G+T-rich in the strand that extends 5’-to-3’ towards the chromosome end. The length and sequence of each DNA repeat are encoded by the telomerase RNA template (see below). The number of repeats per telomere varies widely between species, from a fixed 4.5 repeats of G4T4 in the ciliate Oxytricha nova, to ~350-500 bp in S. cerevisiae, variable numbers of GGTTAG repeats encompassing 10 – 15 kb in humans and 20 – 50 kb in certain mouse and rat species1. Telomeric DNA is mostly double-stranded, but ends with a single-stranded G-rich 3’ overhang or tail that serves two key functions: providing primers for extension by telomerase, and binding specific telomeric proteins to cap chromosome ends and ensure genome stability1 (see below). Alternative structures formed by telomeric ssDNAs include G quadruplexes10, 11 and t-loops12 (Box1).
Proteins that cap ds and ss telomeric DNA
The mammalian chromosome end is capped by a protein complex known as shelterin that is composed of six proteins: TRF1, TRF2, RAP1, TIN2, POT1 and TPP1. S. pombe telomeres contain a similar complex that contains the Taz1 (a TRF homologue), Rap1, Pot1, Tpz1, Poz1 and Ccq1. By contrast, S. cerevisiae does not contain a shelterin-like complex, but instead contains chromosome end-binding proteins such as Rap1, Cdc13, Stn1 and Ten19 (see below).
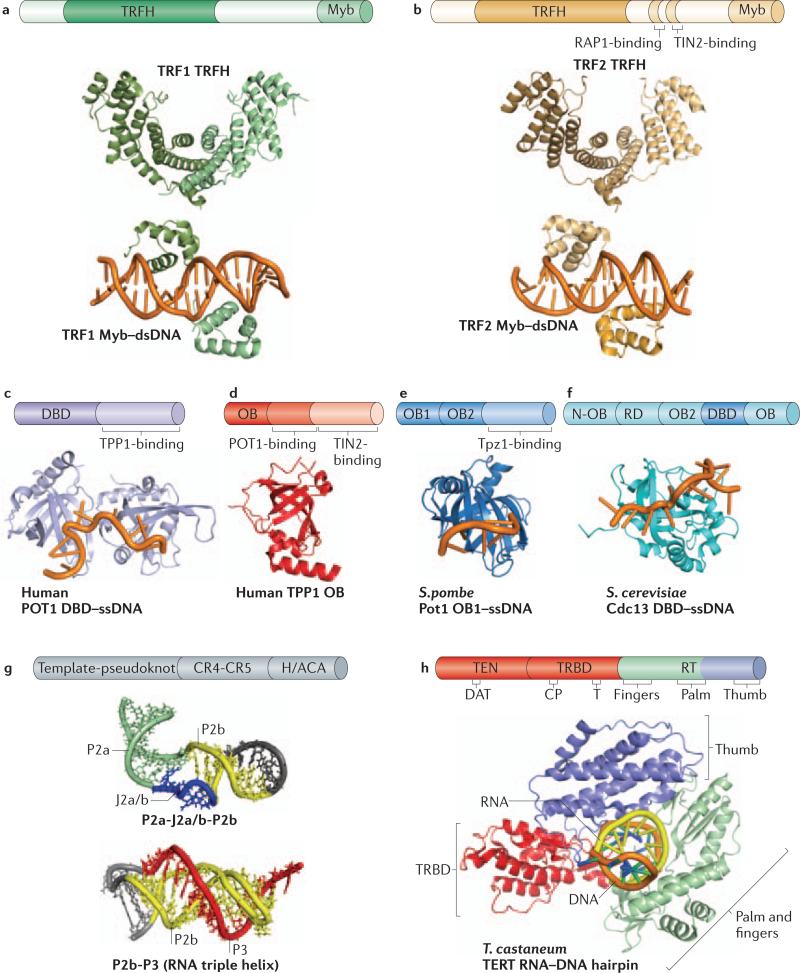
A dedicated set of proteins binds the ds segment of telomeric DNA in mammals and in yeast. The mammalian proteins are TRF1 and TRF2, each of which contains a TRF homology (TRFH) domain that allows homodimerization13 (Fig. 2a,b) and a DNA-binding Myb domain that provides high-affinity binding to ds telomeric DNA14-16 (Fig. 2a,b). Taz117 binds ds telomeric DNA in S. pombe. Although no obvious structural homologues of TRF or Taz1 exist in S. cerevisiae, Rap1 binds ds telomeric DNA as well as certain internal sequences in this organism18. The C-terminal portion of Rap1 binds Rif119 and Rif220 to form the ds telomeric DNA end-protection complex. Rap1 has homologues in humans and S. pombe that associate with telomeres, but these proteins lack the ability to bind DNA directly and are retained at telomeres via their interaction with TRF2 and Taz1, respectively21, 22.
Figure 2.
Structures of telomeric proteins and telomerase components. a, Crystal structure of TRFH domain (PDB: 1H6O) and dsDNA-bound Myb domain of human TRF1 (PDB: 1W0T). b, Crystal structure of TRFH domain (1H6P) and dsDNA-bound Myb domain of human TRF2 (PDB: 1W0U). c, Crystal structure of ssDNA-bound DNA-binding domain (DBD) of human POT1, comprised of two OB folds (PDB: 1XJV). d, Crystal structure of the N-terminal OB domain of human TPP1 (PDB: 2I46). e, NMR structure of ssDNA-bound DBD of S. cerevisiae Cdc13 (PDB: 1S40). f, Crystal structure of ssDNA-bound structure of the first OB domain (OB1) of S. pomble Pot1 (PDB: 1QZG). g, NMR structures of template-pseudoknot (PK) fragments of human TR containing the indicated secondary structural elements; top structure (PDB: 2L3E); bottom structure (PDB: 2K96). The ‘P’ and ‘J’ elements are RNA double-helical paired regions and joining segments, respectively. h, Crystal structure of T. castaneum TERT with a hybrid RNADNA hairpin representing a putative telomerase-primer-template ternary complex (PDB: 3KYL).
The G-rich single-stranded tail of telomeres is protected from illicit DNA end-joining events by telomere-specific DNA-binding proteins in ciliated protozoa, yeast and mammals. The first discovered protein was the TEBPα-TEBPβ heterodimer in the ciliate O. nova, and the crystal structure of its complex with telomeric ssDNA revealed the chromosome tail essentially buried by the protein23. The OB (oligonucleotide/oligosaccharide-binding) domains used for DNA recognition in TEBPα-TEBPβ turned out to be a general feature of telomere end-capping proteins.
POT1 and TPP1 are evolutionarily related to TEBPα and TEBPβ, respectively. Indeed, mammalian POT1 binds with high affinity and specificity to the ss G-rich 3’ tail using two OB fold elements that collectively form the DNA binding domain (DBD) of POT1(Fig. 2c), and this interaction is enhanced by ternary complex formation with TPP124-30. A centrally positioned domain of TPP1 binds POT1 (Fig. 2d), and this interaction is essential for recruitment of POT1 to telomeres31-34. A recent EM study has shown that POT1 TPP1 can completely coat long stretches of ss telomeric DNA to form compact, ordered structures, suggesting that such structures may exist in vivo35.
S. pombe Pot1-Tpz1 (Pot1-Tpz1) (Fig. 2e) is functionally homologous to the mammalian POT1-TPP1 complex in that it binds ss telomeric DNA with high affinity and specificity24, 36-38. However, whereas human POT1 binds ss telomeric DNA as a monomer using two N-terminal tandem OB domains27, S. pombe Pot1 can bind 2.5 repeats of S. pombe telomeric DNA (GGTTAC)2GGT as a monomer39 or two repeats (GGTTAC)2 as a dimer38.
Budding yeast also cap their telomeres with OB-fold proteins, but they are evolutionarily distinct from POT1-TPP1. The G-rich ss tail in S. cerevisiae is bound by the CST (Cdc13-Stn1-Ten1) complex40-42. Cdc13 binds the ss telomeric DNA with high affinity and specificity using a centrally positioned OB domain40, 43-45 (Fig. 2f) and homodimerizes using an N-terminal OB domain46. Thus, the CST complex may be dimeric. Because all CST members are composed mainly of OB fold domains analogous to the replication protein A (RPA) heterotrimer, CST has been proposed to be a telomere-specific RPA47.
The shelterin complex
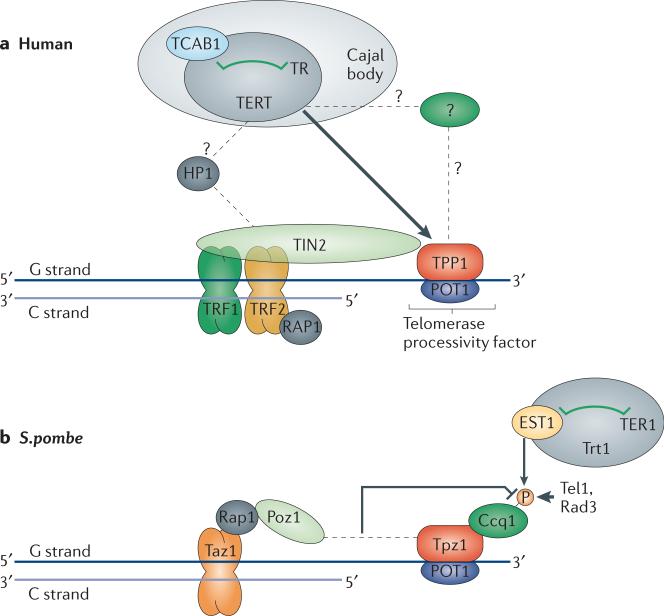
The mammalian shelterin complex (Table 1; yeast homologues are also indicated) brings together ss and ds telomeric DNA-binding proteins by bridging protein partners to protect the natural ends from being recognized as sites of DNA damage. TIN2 is a crucial bridging component that not only links TRF1 and TRF2, but also connects TRF1 and TRF2 to POT1-TPP148-52 (Fig. 3a). TIN2 uses an N-terminal region to bind TRF2 , and a FxLxP amino acid motif at the C-terminus of TIN2 facilitates TRF1 binding50, 53. Bridging of TRF1 and TRF2 by TIN2 is crucial for the existence of a complete shelterin complex because TRF1 does not heterodimerize or interact otherwise with TRF213. TIN2 uses a region in its N-terminal domain to bind to the C-terminal portion of TPP1 to recruit it to telomeres33, 51, 52 (Fig. 2d). Because the TRF2 and TPP1 binding sites reside on the N-terminal domain of TIN2, TIN2 binding to TRF2 and TPP1 could potentially occur in a mutually exclusive manner, but data showing stabilization of TRF1–TIN2–TRF2 in the presence of TPP1 support a model whereby the four shelterin components coexist in a single complex54.
Table 1.
Telomere and telomerase components in humans, S. cerevisiae, and S. pombe
| Human | Fission yeast | Budding yeast | Function |
|---|---|---|---|
| Telomeric proteins | |||
| TRF1 | Taz1 | NA | Bind dsDNA |
| TRF2 | Taz1 | NA | Bind dsDNA |
| Rap1 | Rap1 | Rap1 | Fission yeast protein binds dsDNA |
| TIN2 | Poz1 | NA | Bridge telomeric proteins via protein-protein interactions |
| POT1 | Pot1 | NA | Bind ssDNA |
| TPP1 | Tpz1 | NA | Enhance ssDNA binding of POT1; telomerase recruitment (for human TPP1) |
| NA | Ccq1 | NA | Telomerase recruitment |
| CTC1 | NA | NA | Part of a telomeric RPA-like complex |
| STN1 | Stn1 | Stn1 | Part of a telomeric RPA-like complex |
| TEN1 | Ten1 | Ten1 | Part of a telomeric RPA-like complex |
| NA | NA | Cdc13 | Binds ssDNA; part of a telomeric RPA-like complex |
| Telomere-associated proteins or complexes | |||
| Ku70-Ku80 | Ku70-Ku80 | Ku70-Ku80 | Contributes to telomeric heterochromatin |
| XPF-ERCC1 | NA | NA | 3′ overhang processing |
| EXO1 | NA | Exo1 | 5′ end resection |
| Apollo | NA | NA | 5′ end resection |
| MRE11 complex | NA | NA | TRF2 binding |
| RAD51D [Au: is the D meant to be there?] | NA | NA | Telomere protection against attrition and fusion |
| Tankyrase | NA | NA | PARsylation of TRF1 and resolution of sister telomere cohesion prior to mitosis |
| ORC | -NA | NA | Prevention of telomere circles |
| 9-1-1 complex | NA | NA | Telomere maintenance |
| RecQ helicases | NA | NA | TRF2 binding |
| HP1 | NA | NA | Sister telomere cohesion |
| Telomerase core components | |||
| TERT | Trt1 | Est2 | Telomerase reverse transcriptase protein component |
| TR | Ter1 | Tlc1 | Telomerase RNA component |
| Telomerase accessory factors | |||
| Dyskerin | NA | NA | Binds and stabilizes TR |
| EST1 | Est1 | Est1 | Telomerase recruitment (for) |
| NA | NA | Est3 | Associates with Est2; is essential in vivo |
| NA | NA | Ku70-80 | Nuclear localization of telomerase |
| NA | Sm, Lsm | Sm | Telomerase biogenesis, nuclear localization |
| TCAB1 | NA | NA | Localizes telomerase in Cajal bodies |
| Reptin | NA | NA | Associates with TERT in a cell cycle-dependent manner |
| Pontin | NA | NA | Associates with TERT in a cell cycle-dependent manner |
| PINX1 | NA | NA | Inhibitor of telomerase |
| HSP90 | NA | Hsp82 | Associates with telomerase |
| p23 | NA | Sba1 | Associates with telomerase |
Figure 3.
Models for telomerase recruitment in humans and fission yeast. a, Model for telomerase recruitment in humans. The first step of human telomerase recruitment involves the redistribution of telomerase from Cajal bodies (site for accumulation of recruitment-competent telomerase RNP) to telomeres driven by the telomerase-TPP1 interaction105, 107, 108. This recruitment is TIN2-dependent because TIN2 is required for recruiting TPP1 to telomeres107. It is possible that the TPP1-telomerase interaction is mediated by a yet-to-be-identified Ccq1 homologue in humans. Once at telomeres, telomerase action during S-phase is coordinated with sister telomere cohesion, which is facilitated by the recruitment of HP1, mediated by the PTVML binding-motif of TIN2122, 123. Telomerase extension then proceeds in a processive manner facilitated by bound POT1-TPP1, which reduces the primer dissociation rate and increases the translocation efficiency of telomerase30, 106, 115. Putative interactions are indicated by dashed arrows. b, Model for telomerase recruitment in fission yeast. The Pot1-Tpz1-Ccq1 complex (telomerase stimulatory) associates with the Taz1-Rap1-Poz1 complex (telomerase inhibitory) at long telomeres37. Such an association is facilitated by the presumably high local concentration of Taz1-Rap1 proteins at such long telomeres and results in the inhibition of Ccq1 phosphorylation at T93 by Tel1 and Rad337, 127, 128. At short telomeres however, the Pot1-Tpz1-Ccq1 connection to Taz1-Rap1 is lost owing to lower protein concentration of Taz1-Rap1, facilitating phosphorylation of Ccq1, which results in Est1-Ccq1 binding via the interaction between the 14-3-3-like domain of Est1 and the T93-phosphorylated face of Ccq137, 127, 128. Although telomerase is recruited to telomeres via interaction of Trt1 to Ccq1-bound Est1, this state would be in a ‘closed’ configuration with regards to telomerase action, possibly because telomerase tethered to shelterin is somehow unable to extend telomeres. A conformational switch that replaces phosphorylated Ccq1 with TER1 at the phosphoSer/RNA binding surface of the 14-3-3-like domain of Est1 would relieve the telomerase-shelterin tether and lead to an ‘open’ configuration of telomerase that allows it to carry out telomere elongation134.
Although the various binary interactions that join the six shelterin components to one another are known, the number and identity of shelterin sub-complexes are not established. Removal of mammalian TRF1 led to a depletion of TRF2 from telomeres, an observation that is unanticipated if TRF1 and TRF2 were present in mutually exclusive complexes50, 55. Moreover, co-immunoprecipitation and gel-filtration analysis have shown that TRF1 and TRF2 are part of the same complex that also contains RAP1, TIN2, TPP1 and POT155. However, similar studies have also shown the presence of stable TRF2-RAP1 and TRF2-RAP1-TIN2-POT1 complexes in cell extracts, suggesting sub-complexes of shelterin might co-exist in cells along with the complete complex50. Furthermore, a study that used quantitative immunoblotting to determine the stoichiometry of shelterin components in human cells showed that TRF1, TRF2, RAP1 and TIN2 are present at comparable molar equivalents, but POT1 and TPP1 are present at lower levels56. Thus, much of TRF1-TRF2-RAP1-TIN2 may not be associated with POT1-TPP1.
Regardless of whether shelterin functions as one complex or multiple subcomplexes, its importance in protecting chromosome ends cannot be understated. Knockdown of individual components of shelterin leads to deprotection of chromosome ends, activating DNA damage response pathways at telomeres34. A recent study of a conditional TRF1-TRF2 double knockout mouse in which shelterin was completely removed from telomeres revealed six independent DNA damage response pathways that shelterin normally prevents from occurring at chromosome ends57.
In addition to the telomeric proteins discussed above, other proteins that are not telomere specific have also been shown to be present at telomeres and to carry out important functions there (Table 1).
Telomerase structure and function
The RNA subunits: a conserved core with elaborations
TR is a core subunit of telomerase that includes, among other important elements, the template for DNA synthesis58. TRs from different organisms vary substantially in length. Ciliate TRs are short (Tetrahymena thermophila TR is 159 nt), mammalian TRs are intermediate in length (human TR is 451 nt and mouse TR is 397 nt59), and TR subunits in yeast such as S. cerevisiae (TLC1; 1,157 nt)60 and S. pombe (TER1; 1,213 nt)61, 62 are much longer.
TR is involved at multiple stages of telomerase biogenesis and function. For example, TR provides a template boundary element that limits the extent of reverse transcription63-65. In the absence of such an element, telomerase would ‘read-through’ past the template to incorporate extra non-telomeric nucleotides. The pseudoknot and triple helix66, 67 (Fig. 2g) contribute to catalysis, perhaps by orienting the primer-template duplex in the enzyme active site68. TR has also been shown to contribute to the enzyme processivity69, 70, which allows multiple rounds of telomeric DNA additions after a single primer-binding step. Using biochemical experiments and single-molecule Förster resonance energy transfer (FRET) measurements, T. thermophila TR has been proposed to position the template within the active site and to aid template translocation during repeat addition processivity71. An alternative, more passive model proposes that the strand separation and template realignment processes of T. thermophila TR translocation occur outside the telomerase active site, followed by binding of the realigned hybrid to the active site72.
Another essential property of TR is to bind TERT. Vertebrate TRs bind their cognate TERT subunit using the template-pseudoknot domain and an additional domain known as the CR4-CR5 domain73-76 (Fig. 2g).
In addition to binding TERT, TRs have distinct RNA structural or sequence elements that bind various telomerase accessory proteins. The 3’ ends of mammalian TRs contain an H/ACA box that is associated with RNA-binding proteins dyskerin, NHP2, NOP10 and GAR1, which ensures TR stability and telomerase RNP biogenesis8, 77. Also included in the 3’ domain of mammalian TRs is a Cajal box (CAB) motif, which allows binding to the WD repeat-containing protein known as TCAB1 (also known as WDR79) and consequent localization of TR in Cajal bodies, where the telomerase RNP is packaged for delivery to telomeres78, 79. Similarly, yeast TRs bind a variety of protein cofactors. S. cerevisiae TLC1 binds the Est1 regulatory subunit80 and the dsDNA-binding Ku heterodimer81, 82 (see below), as well as Sm proteins83. Sm is required for S. cerevisiae telomerase function, probably because it is necesarry for nuclear import of TLC183. In S. pombe the Sm complex first binds to the Sm-binding site on TER1 to facilitate spliceosomal cleavage and Tgs1-mediated 5’-capping of the RNA84, 85. Next, the Lsm (Like-Sm) complex Lsm2-8 replaces the Sm complex on TER1. This allows binding of the catalytic Trt1 subunit to TER1 and protects the mature TER1 RNA from exonucleolytic degradation85. Complexes containing human telomerase and Sm proteins have also been observed but seem dispensable for telomerase function86.
Yeast TRs contain long protein-binding ‘arms’ protruding from the catalytic core of the RNA. These arms are ’flexible’ with respect to their sequence, length and positional requirements87,88. It has therefore been hypothesized that TLC1 binds to its protein partners in a ‘beads’ (protein partners) on a ‘string’ (RNA linkers) manner such that the main function of the RNA arms is to recruit the protein partners and retain them in the same complex to facilitate telomerase function. To evaluate the importance of conformational flexibility of the RNA arms, a recent study replaced the natural bulges and internal loops in TLC1 with perfect double-helical RNA. The resulting mutant, TSAT, fully reconstituted telomerase activity in vitro and, although deficient in RNA accumulation, it could carry out telomere length maintenance in vivo89. When the RNA expression levels were equalized, the telomeres of cells expressing the TSA-T mutant were longer than those of wild-type cells, suggesting that the flexibility of wild-type TLC1 comes at some cost of telomerase activity89.
The TERT subunit provides the active site for catalysis
In addition to the RNA subunit, the telomerase core enzyme contains the reverse transcriptase subunit TERT90, 91. TERT contains roughly 1,000 (± ~150) amino acids and is conserved among organisms as disparate as humans92, S. cerevisiae (in which it is known as Est2)90, S. pombe (in which it is known as Trt1)92 and T. thermophila. Based on structure and function, the TERT polypeptide can be subdivided into three major domains: the telomerase essential N-terminal (TEN) domain, the TERT RNA-binding domain (TRBD) and the reverse transcriptase (RT) domain, which contains the fingers, palm and thumb (also known as the C-terminal extension) subdomains and the active site for reverse transcription90 (Fig. 2h).
The crystal structure of a putative TERT from the flour beetle Tribolium castaneum93, 94 has provided more detailed structural information. Although definitive identification of this protein as an authentic telomerase subunit awaits the discovery of its RNA subunit, the presence of a T-motif provides confidence that it is either TERT or a closely related protein. The structure revealed close contacts between the TRBD and the thumb of the RT that result in a closed ring-like tertiary structure with a large cavity at its center, which is sufficiently large to bind the primer-template duplex93. A co-crystal structure of the T. castaneum protein bound to a RNA–DNA hairpin mimicking a template-primer pair confirmed this idea and provided further insights into the various molecular contacts at and near the active site94 (Fig. 2h). The structure showed that the fingers and palm of the RT interact with the backbone of the RNA arm of the hairpin to place the template in the active site, whereas two motifs of RT -- the T-pocket (which contains the T-motif residues) and the CP-pocket -- bind a region upstream of the templating region on the hairpin that presumably mimics the template boundary element of TR. Although the fingers and palm hold the RNA template, the thumb of the RT binds and secures the DNA primer part of the hairpin.
The TEN domain of TERT has been implicated in providing the ‘anchor site’ that binds telomeric DNA upstream from the primer-template interaction95, 96, although other domains contribute as well97. The crystal structure of the isolated TEN domain of T. thermophila TERT revealed a novel protein fold with a groove on the surface that is important for DNA-primer binding and telomerase activity. A photocrosslink between a DNA primer maps to Trp187 within the groove, which suggests that the surface encompassing this residue is involved in DNA binding98. A similar anchor site resides within the TEN domain of human TERT99, 100. In addition, the human TEN domain contains the DAT motif101, which has been implicated in telomerase recruitment, and the yeast TEN domain contributes to binding the Est3 accessory subunit102.
In addition to the catalytic core of telomerase composed of TERT and TR, several telomerase accessory factors (Table 1) assist in mammalian and yeast telomerase assembly, maturation, recruitment and activation. Additional components of T. thermophila telomerase have been recently reviewed8.
Human telomere-telomerase interactions
Telomerase recruitment
One predictable outcome of loss of chromosome end-protection would be telomere elongation, resulting from unregulated access of telomerase to telomeres. Indeed, expression of shelterin mutants defective in end-protection or siRNA-mediated knockdown of shelterin components leads to over-elongation of telomeres26, 48, 51, 103-105. In contrast, cells carrying a mutant TPP1 that lacks the OB domain (TPP1ΔOB) do not show enhanced telomere elongation; instead, TPP1ΔOB seems to protect telomeres as efficiently as the wild-type protein105. Furthermore, TERT-TPP1 binding has been observed in pull-down experiments, and the binding is lost with TPP1ΔOB105. Consistent with a direct telomerase-TPP1 interaction, telomerase has been shown to act preferentially on telomeric primers coated with POT1-TPP1 when such substrates are present together with protein-free primers in vitro106. Thus, the OB fold of TPP1 (Fig. 2f) is proposed to be the telomerase-binding component of the shelterin complex that facilitates telomerase association with chromosome ends that are otherwise protected by this complex (Fig. 3a).
Furthermore, cell biological studies have confirmed that TPP1 recruits telomerase to chromosome ends. Immunofluorescence-fluorescence and in situ hybridization (IF-FISH) to telomeres, as well as telomere ChIP using antibodies against TERT, was used to assess and quantify telomerase recruitment107. RNAi-mediated knockdown of individual human shelterin components revealed that TPP1 and TIN2, but not POT1, were essential for recruiting telomerase to telomeres in HeLa cells. The loss of telomerase recruitment owing to TPP1 silencing resulted in telomerase accumulation in Cajal bodies, and recruitment was rescued by the expression of a wild-type construct that was resistant to the silencing RNA. This recruitment role is conserved in other mammals, as conditional deletion of TPP1 prevented telomerase recruitment to mouse telomeres108. In agreement with the telomerase-binding data presented above, expression of the TPP1ΔOB construct in a TPP1 knockdown background failed to rescue telomerase recruitment to telomeres107. More recently, the TPP1 OB domain was shown to be sufficient to recruit telomerase to an artificially constructed non-telomeric locus in the human genome109, and particular amino acids on the surface of the TPP1 OB were required for both telomerase binding110, 111 and recruitment to telomeres109, 111. Thus, the TPP1 OB domain facilitates binding of telomerase to TPP1 and recruitment of telomerase to telomeres, highlighting this domain as a key shelterin component involved in facilitating telomerase action at telomeres (Fig. 3a).
In vitro, TPP1 may be bound to ss telomeric DNA through its interaction with POT1. However, recruitment via POT1 is not rate-limiting in vivo, suggesting that TPP1 is recruited to telomeres primarily through its interaction with TIN2. Indeed, knockdown of TIN2 results in reduction of telomerase recruitment to telomeres to levels similar to what is observed upon TPP1 knockdown107.
Although an interaction with TPP1 is central to the recruitment of telomerase to mammalian telomeres, this event must be cell-cycle regulated because telomerase (TERT and TR) only accumulates at telomeres at detectible levels during S-phase112, 113. Hence, akin to the situation in fission yeast and budding yeast (see below), it seems highly probable that cell cycle-dependent phosphorylation of telomeric components provides a switch for triggering recruitment of telomerase in mammals. However, the telomeric protein substrates and the involved cell cycle-related kinases remain unidentified in mammals.
Telomerase activation and processivity
The assignment of POT1-TPP1 as the mammalian counterpart of the ciliate ssDNA-binding complex TEBPα-TEBPβ led to the simplistic prediction that POT1-TPP1 would be an inhibitor of telomerase; after all, POT1-TPP1 and telomerase bind to the same G-rich tail of the chromosome. This prediction turned out to be partly true, as telomerase failed to efficiently extend a primer that was bound by POT1 at its 3’ end (the site of action of telomerase) in vitro30, 114. However, addition of both POT1 and TPP1 to telomerase preparations resulted in a 2-3 fold increase in telomerase processivity, and addition of TPP1 alone resulted in a 2-fold increase in total telomerase activity30, 106, 115. Although it has not yet been possible to measure the importance of processivity-activation by POT1-TPP1 on telomerase function in vivo, we note that only with such enhanced processivity would one or two rounds of telomerase binding and extension be sufficient to give the 60 nucleotides of telomeric DNA synthesis per end seen in a single cell cycle116. Are the processivity-stimulation and telomerase-recruitment activities of TPP1 a manifestation of the same TPP1-telomerase interaction? Recent studies have revealed that the same patch of amino acids on the surface of the TPP1 OB domain that is crucial for telomerase binding and recruitment also mediates telomerase processivity stimulation111 and telomere elongation in vivo109, 111.
What is the mechanism by which POT1-TPP1 increases the processivity of telomerase? A step-by-step analysis of the telomerase catalytic cycle using a combination of kinetic and equilibrium approaches106 revealed that POT1-TPP1 increases telomerase processivity by decreasing primer dissociation and improving template translocation. Furthermore, a single POT1-TPP1-DNA interaction is necessary and sufficient for stimulating telomerase processivity, suggesting that once the POT1-TPP1-telomerase tether is formed on ss telomeric DNA, it is maintained throughout telomerase action on that DNA, independently of additional POT1-DNA binding events. Both the decrease of primer dissociation and increase of translocation by POT1-TPP1 are most easily explained by a physical TPP1-telomerase interaction on the primer being extended. An alternative model in which POT1-TPP1 stimulation is solely due to its DNA-binding activity (for example, preventing G-quadruplex formation) would predict that the stimulation would occur independent of the species from which the telomerase was obtained. However, each organism's POT1-TPP1 is most efficient in stimulating its cognate telomerase115, arguing against the processivity stimulation arising exclusively from primer DNA-protein interactions.
TIN2 also has a role in promoting telomerase action that is independent of its interaction with TPP1. Indeed, mutations in TIN2117-119 outside its TPP1-binding site are associated with dyskeratosis congenita (DC), a disease of compromised telomerase function120. The TIN2 DC mutations were shown to cause telomere shortening without any signs of chromosome end-deprotection, suggesting that the mutations interfere with telomerase function121. Furthermore, TR and TPP1 immunoprecipitated with TIN2, but the levels of TR, but not TPP1, associated with TIN2 decreased in the presence of the TIN2 DC mutant121. This further confirmed that the telomerase defect in the TIN2 DC mutants is not due to disruption of the TIN2-TPP1 interaction.
The involvement in DC of TIN2 mutations that do not affect its interaction with other shelterin components suggested that either TIN2 directly binds telomerase (using the protein region that harbours the DC mutations) or that TIN2 binds another factor that regulates telomerase at telomeres. Evidence for the latter possibility came from studies that showed binding of heterochromatin protein 1 (HP1) to a HP1-binding motif (PTVML) in the region of TIN2 that also harbours the DC mutations122 (Fig. 3a). Indeed, TIN2 from patient cells containing the TIN2 DC mutations failed to bind HP1, suggesting that disruption of the HP1-TIN2 interaction contributes to the DC phenotype122. The HP1-binding site on TIN2 was shown to be required for sister chromatid cohesion at telomeres, as mutation of the PTVML motif resulted in an increased separation of sister telomeres122. HP1 was also shown to localize with TIN2 and to participate in cohesion at telomeres in the S phase of the cell cycle122. Importantly, disruption of the HP1-TIN2 interaction led to a moderate decrease in telomere length, suggesting that failure to undergo telomeric cohesion leads to some telomere shortening. Strikingly, an N-terminal deletion mutant of TIN2 that shows drastic telomere elongation in a telomerase-dependent manner failed to show telomere lengthening in the background of mutated PTVML122. It is possible that cohered sister telomeres are better substrates for telomerase122, 123, which has been proposed to function as a dimer124-126, but more work is required to understand how these activities are related.
Fission yeast telomere-telomerase interactions
Chromosome end-protection in S. pombe is similar to what occurs in mammals (see above). Recent studies have identified three proteins in S. pombe —Tpz1, Ccq1 and Poz137 — that, together with Pot1, Taz1 and Rap1, form the shelterin-like complex in this species (Fig. 3b). As mentioned above, Pot1-Tpz1 is functionally homologous to POT1-TPP1 from mammals and can thereby mediate chromosome end-protection24, 37, 38. In addition, deletion of either Pot1 or Tpz1 leads to depletion of telomeric DNA and circularization of chromosomes (a common mode of survival in S. pombe when chromosome end-protection is compromised). However, telomerase recruitment to telomeres in S. pombe seems to occur through a distinct mechanism that involves the S. pombe-specific shelterin protein Ccq137 (Fig. 3b). Interestingly, unlike TPP1, Ccq1 does not seem to affect S. pombe telomerase activity, as Trt1 or Tpz1 immunoprecipitates from ccq1Δ cells showed telomerase activity that was comparable to that of wild-type cells37.
Ccq1 is the telomerase recruitment factor in fission yeast
The first indication that Ccq1 may have a role in facilitating telomerase action at telomeres came from the observation that Ccq1 deletion led to a ~200 bp reduction in telomere length37. By contrast, Poz1 deletion resulted in a drastic increase (up to 2 kb) in telomere length, suggesting that Poz1 is a negative regulator of telomerase37. In addition, the telomerase activity co-immunoprecipitated with Tpz1 (which is similar to the telomerase activity of Trt1 immunoprecipitates) was lost when Ccq1 was deleted, but not when Poz1 was deleted. The effects of Ccq1 on telomere length occurred through the telomerase pathway, because cells lacking both Ccq1 and Trt1 showed similar reduction in telomere length as cells lacking just Trt137. Finally, epistasis experiments examining Poz1-Taz1 double mutants or Poz1-Rap1 double mutants confirmed that Taz1, Rap1 and Poz1 function along a single telomerase-inhibition pathway, because none of the double deletions exacerbated the telomere shortening phenotype of the respective single deletions37.
However, Ccq1 was important for telomerase recruitment to telomeres when assayed by Trt1-telomere ChIP127, 128. Hence, Ccq1 seems to be a recruitment factor for S. pombe telomerase (Fig. 3b). Surprisingly, Ccq1-Poz1 double mutant cells lost telomeres more rapidly than Trt1-Poz1 double mutants, suggesting that Ccq1 also has a telomerase-independent contribution to telomere maintenance37. It is possible that Ccq1 homologues might exist in higher eukaryotes, leading to recruitment of telomerase to telomeres in those organisms37. However, such a protein would likely have to bridge TPP1 and telomerase, because TPP1 has been shown to pull down telomerase from cell lysates105.
Regulation of telomerase recruitment via phosphorylation
The roles of ATM and ATR kinases in mammalian and fission yeast telomere biology are complex. Although they are normally inhibited by shelterin from triggering a DNA-damage response at telomeres, they are also recruited to telomeres during S phase and G2 phase for telomere maintenance1, 34, 129-131. Deletion of both Tel1 (ATM) and Rad3 (ATR) in S. pombe led to a failure to recruit telomerase to telomeres and a complete loss of telomeres, leading to chromosome circularization132, 133. The same double deletion also resulted in a decrease in Ccq1 recruitment to telomeres, suggesting that Ccq1 might be a substrate for the kinases Tel1 and Rad3 and that phosphorylation of Ccq1 could be crucial for telomerase recruitment to telomeres.
Further work revealed that Ccq1 interacts with the telomerase accessory subunit Est1127 via the N-terminal half of Ccq1 and three basic amino acids at the N-terminal 14-3-3-like domain of Est1127. Phosphorylation of Ccq1 amino acid T93 was crucial for the Ccq1-Est1 interaction, for telomerase recruitment to telomeres and for telomere length maintenance127, 128. T93 was shown to be phosphorylated by Tel1 and Rad3, resolving the role of these kinases in S. pombe telomerase regulation127, 128. This phosphorylation event is inhibited by the telomerase-inhibitory complex Taz1-Rap1-Poz1 127 (Fig. 3b). Consistent with this, shorter telomeres exhibited Ccq1 hyperphosphorylation, probably because they have a lower concentration of the Taz1-Rap1-Poz1 complex127.
How does Ccq1 binding to Est1, a telomerase regulatory subunit of S. pombe telomerase, lead to telomerase recruitment to telomeres? A fragment of TER1 (nucleotides 415–507) has been shown to bind specifically to Est1 in a yeast three-hybrid screen. Through an unbiased random mutagenesis screen of Est1, the authors of this study found that three amino acids (L48, R194 and K252) that reside in the T93-binding 14-3-3-like domain of Est1 are crucial for the interaction of Est1 with TER1134. This raised the intriguing possibility that Est1 binding to TER1 and to Ccq1 may be mutually exclusive, with important ramifications for telomerase recruitment to telomeres in S. pombe: substitution of phosphorylated Ccq1 by TER1 at the 14-3-3-like domain of Est1 may convert telomerase to its active or open state134 (Fig. 3b).
Budding yeast telomere-telomerase interactions
The protein components of budding yeast telomerase were discovered through pioneering genetic screens that identified four ever-shorter telomeres (est) genes, Est1-4135, 136. Est2 (the TERT subunit) and regulatory subunits Est1 and Est3 together with the TR subunit, TLC160, make up the telomerase holoenzyme of S. cerevisiae. Est4 was found to be Cdc13, a telomeric DNA-binding protein identified previously in a cell cycle regulation screen135, 136. Cdc13 is not considered a telomerase subunit because it does not co-immunoprecipitate with TLC1 RNA or with active telomerase137, but instead associates with Stn1 and Ten1 to form the trimeric CST complex41 (see above).
Cdc13 and Est1 mediate telomerase recruitment
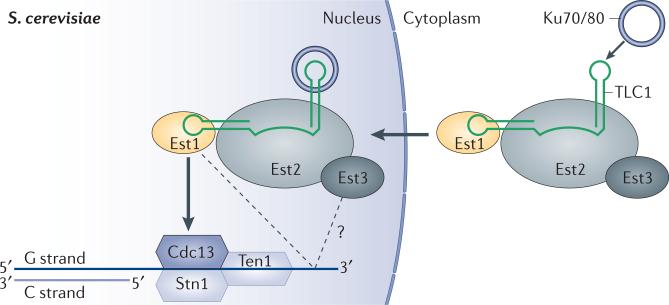
How is budding yeast telomerase recruited to telomeric DNA? Cdc13 and Est1 were initially implicated in this event because of the following observations. First, Cdc13 and Est1 bound ss DNA of telomeric sequence 40, 138 (Fig. 4). Next, Cdc13 and Est1 were shown to weakly interact with each other in yeast two-hybrid experiments40, 139. Finally, Est1 was shown to bind TLC1140, thereby establishing a possible Est2-TLC1-Est1-Cdc13-DNA connectivity for telomerase recruitment in S. cerevisiae (Fig. 4). One prediction of this model is that est mutations of Cdc13 that disrupt its connection to telomerase could be rescued if the mutant were fused covalently to Est1. Indeed, a covalent fusion of Est1 to a telomerase-defective Cdc13est mutant rescued the est phenotype (that is, short telomeres)141. Furthermore, covalently tethering Est2 to Cdc13 eliminated the requirement of Est1 for telomerase recruitment, suggesting that the primary function of Est1 in telomerase recruitment is to bridge Cdc13 to telomerase141. Specifically, genetic data indicated that E252 of Cdc13 forms a salt-bridge with K444 of Est1 such that single-charge reversals at either of these positions lead to disruption of the Est1-Cdc13 connection (leading to telomere shortening), whereas double-charge reversals at these positions restore the salt-bridge and rescue Est1-Cdc13 binding142. This salt bridge may perhaps not be required for binding per se but for a subsequent step143.
Figure 4.
Model for telomerase recruitment in budding yeast. The Ku heterodimer binds TLC1 (the telomerase RNA) and promotes nuclear import and retention of the RNP. The interaction between ssDNA-bound Cdc13 and Est1 is responsible for recruiting telomerase to budding yeast telomeres. It is not known whether Cdc13 is part of the CST complex when it recruits telomerase to telomeres. The Est1-ssDNA interaction may further strengthen the telomerase-telomere tether. A putative Est2-dependent Est3-ssDNA interaction proposed for certain Candida species could serve as yet another mechanism for telomerase recruitment.
Additional results support Cdc13-Est1 functioning in telomerase recruitment in S. cerevisiae. Using an engineered system, tethering either Cdc13 or Est1 near a double-strand DNA break promoted telomerase recruitment and de novo telomere formation144. Furthermore, although Est2 is first recruited to telomeres in G1/early S phase by the Ku-dependent pathway (see below), those levels are insufficient to prevent an est phenotype; wild-type Est2 levels at telomeres require Est1 to be both telomere-associated and bound to TLC1 RNA145. The interaction between Est1 and Cdc13 is facilitated by Cdk1-mediated phosphorylation of Cdc13146, 147.
In addition to its interaction with Cdc13, Est1 has been reported to promote telomerase activation148, possibly by recruiting the Est3 subunit149 (see below). Because Est1 can bind ss DNA directly, it has been proposed that Est1 may present the 3’ end of the chromosome in a conformation conducive for telomerase action141 and/or stabilize telomerase on telomeres150, 151.
The Est3 subunit
The telomerase regulatory Est3 subunit of budding yeast is predicted to fold into an OB domain that resembles the OB domain of the mammalian telomere protein TPP1152, 153, suggesting that Est3 might have some function in the recruitment of telomerase to telomeres. A group of amino acids in Est3 has been found to bind to the TEN domain of Est2, which helps to explain how Est3 is incorporated into the telomerase holoenzyme without binding TLC1, and the inclusion of Est3 in the telomerase preparation stimulates telomerase activity in vitro 102. These observations alone do not implicate Est3 in recruitment. However, the Est3 of another yeast species, Candida, which is a larger protein than that of S. cerevisiae with additional structural elements, has been proposed to be functionally analogous to the OB domain of TPP1154; these authors proposed that binding of Est3 to Est2 causes a conformational change that reveals a DNA-binding site that may be involved in telomerase recruitment (Fig. 4). Additional studies are needed to test this model.
Ku promotes telomerase nuclear localization
The Ku70-80 heterodimer, best known for its DNA-binding function in the nonhomologous end-joining (NHEJ) pathway, binds a 48 nucleotide stem-loop of TLC1 to facilitate telomerase accumulation at telomeres during the G1 phase of the budding yeast cell cycle82, 145, 155. It was initially proposed that Ku could bind the end of a chromosome (dsDNA) and telomerase (TLC1) simultaneously, suggesting an elegant model for telomerase recruitment81, 155, 156. However, purified S. cerevisiae Ku is unable to form a ternary complex with a DNA substrate (that mimics the end of a chromosome) and its binding site on TLC1 RNA; the binding of these two nucleic acids is mutually exclusive157. Thus, it seems unlikely that Ku directly bridges telomerase with telomeric DNA.
It is now apparent that Ku's role in facilitating telomerase recruitment is first and foremost at the level of nuclear localization (Fig. 4). TLC1 biogenesis involves export of the nuclear RNA to the cytoplasm followed by import back into the nucleus158. Using FISH to visualize TLC1 localization, it was shown that the nuclear retention of TLC1 was lost in the absence of Ku70, suggesting that Ku70 is important for importing TLC1 back into the nucleus158. Moreover, Ku80 deletion mutants that are deficient in TLC1-binding (and DNA binding) failed to accumulate telomerase in the nucleus157. Although nuclear localization of telomerase by Ku is necessary for telomerase recruitment in G1, it is presumably not sufficient; other factors such as Mps3 (a membrane-spanning SUN domain protein found at the nuclear periphery) are involved159, 160.
Although the telomerase-associated functions of Ku have only been described in certain budding yeast, Ku has additional functions in forming telomeric heterochromatin and repressing gene expression that require its dsDNA-binding activity and may be broadly conserved161-163. In budding yeast these telomeric functions have been genetically separated from Ku's NHEJ function164. Ku also contributes to the localization of telomeres at the nuclear envelope, and this in turn affects telomerase extension160.
Conclusion
Recent biochemical and functional studies have revealed salient features of telomerase recruitment in humans, budding yeast and fission yeast. The recruitment mechanisms in humans and S. pombe are similar but also exhibit organism-specific nuances. Homologous proteins bind the ssDNA tails of chromosomes in humans and S. pombe (POT1-TPP1 and Pot1-Tpz1, respectively) and are involved in telomerase recruitment. However, an S. pombe-specific protein, Ccq1, bridges telomerase to Pot1-Tpz1 in this organism. Although TPP1 seems to directly connect human telomerase with telomeres, it is also possible that humans have a Ccq1-like protein that has not been identified. If a functional Ccq1 homologue were discovered in humans, it would provide a unifying principle for telomerase recruitment in these two organisms.
Another important aspect of telomerase recruitment in S. pombe is the phosphorylation and activation of Ccq1 by the ATM and ATR-like kinases, Tel1 and Rad3. It remains to be seen how protein modifications may regulate the recruitment of human telomerase.
In S. cerevisiae the ssDNA-binding protein Cdc13 binds the Est1 subunit of telomerase to facilitate telomerase recruitment. In addition, the Ku heterodimer binds the telomerase RNA subunit to mediate nuclear retention of telomerase. The different architecture of the telomeric protein complex and the unique roles of Est1 and the Ku heterodimer separate the budding yeast telomerase recruitment mechanism from that of fission yeast and humans. The recently discovered structural homology between budding yeast Est3 and the OB domain of human TPP1 has provoked the reexamination of possible mechanistic conservation between some aspects human and budding yeast telomerase recruitment, a subject of current research.
Box 1. Alternative telomeric DNA structures.
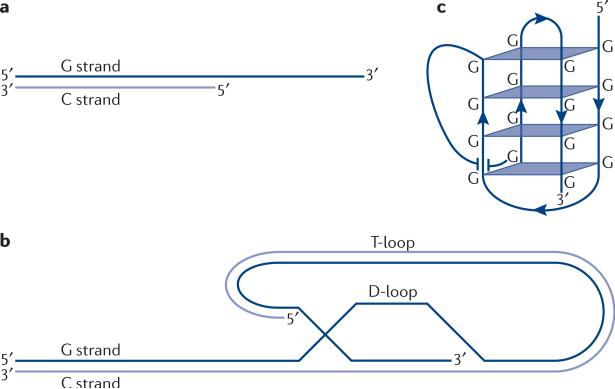
Telomeric DNA ends with a single-stranded G-ring 3’ overhang that is a ligand for telomeric proteins and a substrate for telomerase (see the figure, part a). However, alternative structures can be adopted. Telomere loops (t-loops) are one such example. They are lariat-like configurations that arise by strand invasion of the ssDNA tail into the upstream ds telomeric DNA (see the figure, part b). Electron microscopy (EM) of photocrosslinked telomeric DNA first showed the existence of t-loops12, and EM studies on native (un-crosslinked) telomeric chromatin released from chicken erythrocytes and mouse lymphocytes revealed similar structures. T-loop formation is not thermodynamically favorable, so it is not surprising that protein factors have been implicated in t-loop assembly12, 165. Because the t-loop would be a highly protected, inert structure, future work will be needed to address the question of how t-loops are opened up to allow telomerase to access the chromosome end. Another common structure is G-quadruplex DNA (see the figure, part c). This configuration is built from G-quartets, which are square-planar arrays of four Gs hydrogen-bonded by Hoogsteen base-pairing; atomic structures of both parallel and antiparallel G-quadruplexes have been solved. Intramolecular G-quadruplexes form spontaneously in vitro, as long as the DNA sequence contains at least four blocks of Gs and the ionic conditions are appropriate. Given the thermodynamic imperative for G-quadruplex formation, it is perhaps not surprising that telomeric proteins can catalyze their folding and unfolding166-168. All of the known chromosome end-capping proteins bind unfolded telomeric DNA and ignore the G-quadruplex169, 170, providing a strong argument that most telomeric DNA in cells is not folded into quadruplexes. However, there is evidence that these structures occur in vivo171. They may serve to limit telomerase extension in vitro166, 172, 173, and this could certainly be useful in vivo. Another possibility is that they provide an emergency solution to the end-protection problem when capping proteins have been displaced. A big challenge to interrogating the function of G-quadruplex and t-loop DNAs is that it's difficult to ‘knock out’ a DNA structure using genetics and determine the phenotype. Although G-quadruplexes can be knocked out by changing the telomerase template sequence, the resultant novel repeats do not bind telomeric proteins, so the phenotypes are indirect.
Online summary.
The ends of linear chromosomes pose two problems to eukaryotic cells: the end-protection problem and the end-replication problem. End protection is necessary to prevent natural ends of chromosomes from being recognized as DNA breaks that require DNA repair, and end-replication is required to compensate for the inability of replicative polymerases to copy the extreme ends of chromosomes.
The telomerase ribonucleoprotein enzyme utilizes its internal RNA template for replicating the ends of chromosomes to furnish telomeric DNA, which is bound by telomeric proteins that facilitate chromosome end protection.
How does telomerase get recruited to chromosome? The telomerase-telomere connection is mediated by TPP1 (in humans), Ccq1 (in fission yeast) and Cdc13 (in budding yeast).
The amino-terminal OB domain of human TPP1 binds telomerase, recruits telomerase to telomeres, and increases the processivity of the telomerase enzyme.
Ccq1, an S. pombe-specific telomeric protein, recruits telomerase to telomeres in this organism. Phosphorylation of Ccq1 by the Tel1 and Rad3 kinases regulates its binding to the Est1 subunit of S. pombe telomerase.
Binding of the heterodimeric Ku protein to its binding site on TLC1, the RNA subunit of S. cerevisiae telomerase, facilitates localization of telomerase in the nucleus of this organism. The interaction between the single-stranded telomeric DNA-binding protein Cdc13 and the telomerase subunit Est1 tethers budding yeast telomerase to telomeres.
Acknowledgements
T.R.C. is an investigator of the Howard Hughes Medical Institute. Our telomerase research is funded in part by NIH grants R01 GM099705 to T.R.C. and K99 CA167644 to J.N.
Glossary terms
- Dyskeratosis congenita
A rare genetic disease, characterized by abnormal skin pigmentation, nail dystrophy, and mucosal leukoplakia, caused by telomere shortening owing to mutations in telomerase core components (TERT and TR), telomerase accessory factors (such as dyskerin, TCAB1, Nhp2, and Nop10), or the shelterin protein TIN2.
- Cajal bodies
Conserved nuclear organelles that are involved in the biogenesis of small nuclear ribonucleoprotein (snRNP) particles and maturation of the telomerase RNP.
- Sm proteins
Sm proteins form a ring that binds to a specific U-rich sequence near the 3’ ends of small nuclear RNAs involved in mRNA splicing, allowing nuclear importation of the RNP.
- Photocrosslink
The formation of covalent adducts between nucleotides and/or amino acids that are adjacent in a complex using light energy, generally in the ultraviolet range.
- T-motif
An amino acid sequence preceding the RT motifs that is conserved among TERT proteins but not apparent in other reverse transcriptases.
- Immunofluorescence
Localizationof a protein (or other macromolecule) within cells or tissues using a specific antibody that is derivatized with a fluorescent probe (direct immunofluorescence) or using a fluorescently labelled secondary antibody (indirect immunofluorescence).
- Fluorescence in situ hybridization
Localization of specific nucleic acid sequences in the cell by specific annealing of fluorescently labelled antisense oligonucleotide probes.
- ChIP
(Chromatin immunoprecipitation). A technique that allows isolation of DNA sequences bound to a protein of interest using specific antibodies.
- Processivity
Repeat addition processivity is the addition of multiple telomeric repeats following a single primer-binding event.
- Nonhomologous end-joining
A pathway for repair of DNA double-strand breaks by directly ligating the broken ends, without the need for a homologous template.
- Hoogsteen base-pairing
Non-Watson-Crick pairing that involves the N7 atom of a purine and is important in stabilizing triplex and quadruplex nucleic acid structures.
Biography
Thomas R. Cech is Distinguished Professor of Chemistry & Biochemistry and Director of the BioFrontiers Institute at the University of Colorado-Boulder. He received the Nobel Prize (Chemistry, 1989) for the discovery of RNA catalysis and served as president of HHMI from 2000-2009. In 2009 he returned to Boulder to study telomerase and long noncoding RNAs and to teach undergraduates.
Jayakrishnan Nandakumar is a research associate in T. Cech's laboratory at the University of Colorado-Boulder. His postdoctoral work has focused on the mechanisms by which telomeric proteins facilitate chromosome end protection and end replication. He conducted his thesis work at the Sloan-Kettering institute in New York.
References
- 1.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB. Telomere end-replication problem and cell aging. J Mol Biol. 1992;225:951–960. doi: 10.1016/0022-2836(92)90096-3. [DOI] [PubMed] [Google Scholar]
- 3.Pardue ML, DeBaryshe PG. Drosophila telomeres: A variation on the telomerase theme. Fly (Austin) 2008;2:101–110. doi: 10.4161/fly.6393. [DOI] [PubMed] [Google Scholar]
- 4.Jaskelioff M, et al. Telomerase reactivation reverses tissue degeneration in aged telomerasedeficient mice. Nature. 2010;469:102–106. doi: 10.1038/nature09603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dokal I. Dyskeratosis congenita. Hematology Am Soc Hematol Educ Program. 2011;2011:480–486. doi: 10.1182/asheducation-2011.1.480. [DOI] [PubMed] [Google Scholar]
- 6.Cifuentes-Rojas C, Shippen DE. Telomerase regulation. Mutat Res. 2012;730:20–27. doi: 10.1016/j.mrfmmm.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis KA, Wuttke DS. Telomerase and telomere-associated proteins: structural insights into mechanism and evolution. Structure. 2012;20:28–39. doi: 10.1016/j.str.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. RNA. 2012;18:1747–1759. doi: 10.1261/rna.034629.112. [This excellent review also includes the Tetrahymena telomerase holoenzyme, which is not covered in detail in the current article.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zakian VA. Telomeres: the beginnings and ends of eukaryotic chromosomes. Exp Cell Res. 2012;318:1456–1460. doi: 10.1016/j.yexcr.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 11.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 12.Griffith JD, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 13.Fairall L, Chapman L, Moss H, de Lange T, Rhodes D. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol Cell. 2001;8:351–361. doi: 10.1016/s1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- 14.Court R, Chapman L, Fairall L, Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6:39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishikawa T, et al. Solution structure of a telomeric DNA complex of human TRF1. Structure. 2001;9:1237–1251. doi: 10.1016/s0969-2126(01)00688-8. [DOI] [PubMed] [Google Scholar]
- 16.Hanaoka S, Nagadoi A, Nishimura Y. Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities. Protein Sci. 2005;14:119–130. doi: 10.1110/ps.04983705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 18.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 19.Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 20.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 21.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 22.Chikashige Y, Hiraoka Y. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol. 2001;11:1618–1623. doi: 10.1016/s0960-9822(01)00457-2. [DOI] [PubMed] [Google Scholar]
- 23.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 24.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 25.Baumann P, Podell E, Cech TR. Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol Cell Biol. 2002;22:8079–8087. doi: 10.1128/MCB.22.22.8079-8087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 27.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 28.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 29.Wu L, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 30.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [This study reported the stimulation of telomerase in vitro by the POT1-TPP1 complex and also solved the high-resolution crystal structure of the N-terminal OB domain of TPP1.] [DOI] [PubMed] [Google Scholar]
- 31.Hockemeyer D, et al. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat Struct Mol Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 32.Guo X, et al. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu D, et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 34.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 35.Taylor DJ, Podell ER, Taatjes DJ, Cech TR. Multiple POT1-TPP1 proteins coat and compact long telomeric single-stranded DNA. J Mol Biol. 2011;410:10–17. doi: 10.1016/j.jmb.2011.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 37.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [This study reported the identification of S. pombe telomeric proteins -- Ccq1, Poz1, and Tpz1 -- and demonstrated that Ccq1 is a positive regulator of telomere length.] [DOI] [PubMed] [Google Scholar]
- 38.Nandakumar J, Cech TR. DNA-induced dimerization of the single-stranded DNA binding telomeric protein Pot1 from Schizosaccharomyces pombe. Nucleic Acids Res. 2011;40:235–244. doi: 10.1093/nar/gkr721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschuler SE, Dickey TH, Wuttke DS. Schizosaccharomyces pombe protection of telomeres 1 utilizes alternate binding modes to accommodate different telomeric sequences. Biochemistry. 2011;50:7503–7513. doi: 10.1021/bi200826a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNAbinding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 41.Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 42.Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson EM, Halsey WA, Wuttke DS. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res. 2002;30:4305–4313. doi: 10.1093/nar/gkf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 45.Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, Wuttke DS. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J Mol Biol. 2004;338:241–255. doi: 10.1016/j.jmb.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 46.Sun J, et al. Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha. Cell Res. 2011;21:258–274. doi: 10.1038/cr.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 48.Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SH, et al. TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem. 2004;279:43799–43804. doi: 10.1074/jbc.M408650200. [DOI] [PubMed] [Google Scholar]
- 50.Ye JZ, et al. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem. 2004;279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 51.Ye JZ, et al. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y, et al. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 54.O'Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci U S A. 2006;103:11874–11879. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 56.Takai KK, Hooper SM, Blackwood SL, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2009 doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–597. doi: 10.1126/science.1218498. [By conditional deletion of the entire mouse shelterin complex, the authors revealed six DNA damage signalling pathways that are normally repressed at telomeres.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 59.Blasco MA, Funk W, Villeponteau B, Greider CW. Functional characterization and developmental regulation of mouse telomerase RNA. Science. 1995;269:1267–1270. doi: 10.1126/science.7544492. [DOI] [PubMed] [Google Scholar]
- 60.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 61.Webb CJ, Zakian VA. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol. 2008;15:34–42. doi: 10.1038/nsmb1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leonardi J, Box JA, Bunch JT, Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol. 2008;15:26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- 63.Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17:2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tzfati Y, Fulton TB, Roy J, Blackburn EH. Template boundary in a yeast telomerase specified by RNA structure. Science. 2000;288:863–867. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]
- 65.Box JA, Bunch JT, Zappulla DC, Glynn EF, Baumann P. A flexible template boundary element in the RNA subunit of fission yeast telomerase. J Biol Chem. 2008;283:24224–24233. doi: 10.1074/jbc.M802043200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 67.Shefer K, et al. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol Cell Biol. 2007;27:2130–2143. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol. 2008;15:634–640. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen JL, Greider CW. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. Embo J. 2003;22:304–314. doi: 10.1093/emboj/cdg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lai CK, Miller MC, Collins K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol Cell. 2003;11:1673–1683. doi: 10.1016/s1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berman AJ, Akiyama BM, Stone MD, Cech TR. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat Struct Mol Biol. 2011;18:1371–1375. doi: 10.1038/nsmb.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qi X, et al. RNA/DNA hybrid binding affinity determines telomerase template-translocation efficiency. EMBO J. 2011;31:150–161. doi: 10.1038/emboj.2011.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bley CJ, et al. RNA-protein binding interface in the telomerase ribonucleoprotein. Proc Natl Acad Sci U S A. 2011;108:20333–20338. doi: 10.1073/pnas.1100270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 75.Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 76.Chen JL, Opperman KK, Greider CW. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 2002;30:592–597. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 78.Venteicher AS, et al. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peterson SE, et al. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 82.Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 84.Box JA, Bunch JT, Tang W, Baumann P. Spliceosomal cleavage generates the 3' end of telomerase RNA. Nature. 2008;456:910–914. doi: 10.1038/nature07584. [DOI] [PubMed] [Google Scholar]
- 85.Tang W, Kannan R, Blanchette M, Baumann P. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature. 2012;484:260–264. doi: 10.1038/nature10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007;28:773–785. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci U S A. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zappulla DC, Goodrich K, Cech TR. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol. 2005;12:1072–1077. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]
- 89.Lebo KJ, Zappulla DC. Stiffened yeast telomerase RNA supports RNP function in vitro and in vivo. RNA. 2012;18:1666–1678. doi: 10.1261/rna.033555.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lingner J, et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 91.Meyerson M, et al. hEST2, the putative human telomerase catalytic subunit gene, is upregulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 92.Nakamura TM, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 93.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 94.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 95.Lue NF. A physical and functional constituent of telomerase anchor site. J Biol Chem. 2005;280:26586–26591. doi: 10.1074/jbc.M503028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moriarty TJ, Ward RJ, Taboski MA, Autexier C. An anchor site-type defect in human telomerase that disrupts telomere length maintenance and cellular immortalization. Mol Biol Cell. 2005;16:3152–3161. doi: 10.1091/mbc.E05-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Finger SN, Bryan TM. Multiple DNA-binding sites in Tetrahymena telomerase. Nucleic Acids Res. 2008;36:1260–1272. doi: 10.1093/nar/gkm866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Romi E, et al. High-resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. Proc Natl Acad Sci U S A. 2007;104:8791–8796. doi: 10.1073/pnas.0703157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wyatt HD, Tsang AR, Lobb DA, Beattie TL. Human telomerase reverse transcriptase (hTERT) Q169 is essential for telomerase function in vitro and in vivo. PLoS One. 2009;4:e7176. doi: 10.1371/journal.pone.0007176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jurczyluk J, et al. Direct involvement of the TEN domain at the active site of human telomerase. Nucleic Acids Res. 2011;39:1774–1788. doi: 10.1093/nar/gkq1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Armbruster BN, Banik SS, Guo C, Smith AC, Counter CM. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol Cell Biol. 2001;21:7775–7786. doi: 10.1128/MCB.21.22.7775-7786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Talley JM, DeZwaan DC, Maness LD, Freeman BC, Friedman KL. Stimulation of yeast telomerase activity by the ever shorter telomere 3 (Est3) subunit is dependent on direct interaction with the catalytic protein Est2. J Biol Chem. 2011;286:26431–26439. doi: 10.1074/jbc.M111.228635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 104.Ye JZ, de Lange T. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat Genet. 2004;36:618–623. doi: 10.1038/ng1360. [DOI] [PubMed] [Google Scholar]
- 105.Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [This study demonstrated that the N-terminal OB domain of TPP1 binds telomerase.] [DOI] [PubMed] [Google Scholar]
- 106.Latrick CM, Cech TR. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Abreu E, et al. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol. 2010;30:2971–2982. doi: 10.1128/MCB.00240-10. [This study reported the requirement of the N-terminal OB domain of TPP1 in telomerase recruitment to human telomeres.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tejera AM, et al. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell. 2010;18:775–789. doi: 10.1016/j.devcel.2010.03.011. [This study reported the role of TPP1 in telomerase recruitment to mouse telomeres.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhong FL, et al. TPP1 OB-Fold Domain Controls Telomere Maintenance by Recruiting Telomerase to Chromosome Ends. Cell. 2012;150:481–494. doi: 10.1016/j.cell.2012.07.012. [This study reported the sufficiency of the N-terminal OB domain of TPP1 in telomerase recruitment to human telomeres and identified key amino acids on TPP1 and TERT required for telomerase recruitment.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sexton AN, Youmans DT, Collins K. Specificity Requirements for Human Telomere Protein Interaction with Telomerase Holoenzyme. J Biol Chem. 2012 doi: 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nandakumar J, et al. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature (Advanced Online Publication) 2012 doi: 10.1038/nature11648. [This study identified a patch of amino acids on the surface of the N-terminal OB domain of TPP1 that mediates telomerase binding, telomerase recruitment, telomerase processivity stimulation, and telomere lengthening.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jady BE, Richard P, Bertrand E, Kiss T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell. 2006;17:944–954. doi: 10.1091/mbc.E05-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 115.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhao Y, et al. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol Cell. 2011;42:297–307. doi: 10.1016/j.molcel.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Savage SA, et al. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Hum Genet. 2008;82:501–509. doi: 10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood. 2008;112:3594–3600. doi: 10.1182/blood-2008-05-153445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sasa GS, Ribes-Zamora A, Nelson ND, Bertuch AA. Three novel truncating TINF2 mutations causing severe dyskeratosis congenita in early childhood. Clin Genet. 2012;81:470–478. doi: 10.1111/j.1399-0004.2011.01658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bessler M, Wilson DB, Mason PJ. Dyskeratosis congenita. FEBS Lett. 2010;584:3831–3838. doi: 10.1016/j.febslet.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang D, He Q, Kim H, Ma W, Songyang Z. TIN2 protein dyskeratosis congenita missense mutants are defective in association with telomerase. J Biol Chem. 2011;286:23022–23030. doi: 10.1074/jbc.M111.225870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Canudas S, et al. A role for heterochromatin protein 1gamma at human telomeres. Genes Dev. 2011;25:1807–1819. doi: 10.1101/gad.17325211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Houghtaling BR, Canudas S, Smith S. A role for sister telomere cohesion in telomere elongation by telomerase. Cell Cycle. 2012;11:19–25. doi: 10.4161/cc.11.1.18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wenz C, et al. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 2001;20:3526–3534. doi: 10.1093/emboj/20.13.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Prescott J, Blackburn EH. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moser BA, Chang YT, Kosti J, Nakamura TM. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat Struct Mol Biol. 2011;18:1408–1413. doi: 10.1038/nsmb.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yamazaki H, Tarumoto Y, Ishikawa F. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 2012;26:241–246. doi: 10.1101/gad.177873.111. [References 127 and 128 demonstrated how the ATM-ATR kinases of S. pombe, Tel1 and Rad3, phosphorylate Ccq1 to facilitate Est1 binding and telomerase recruitment.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 130.Sabourin M, Zakian VA. ATM-like kinases and regulation of telomerase: lessons from yeast and mammals. Trends Cell Biol. 2008;18:337–346. doi: 10.1016/j.tcb.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Moser BA, et al. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009;28:810–820. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moser BA, Subramanian L, Khair L, Chang YT, Nakamura TM. Fission yeast Tel1(ATM) and Rad3(ATR) promote telomere protection and telomerase recruitment. PLoS Genet. 2009;5:e1000622. doi: 10.1371/journal.pgen.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Naito T, Matsuura A, Ishikawa F. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet. 1998;20:203–206. doi: 10.1038/2517. [DOI] [PubMed] [Google Scholar]
- 134.Webb CJ, Zakian VA. Schizosaccharomyces pombe Ccq1 and TER1 bind the 14-3-3-like domain of Est1, which promotes and stabilizes telomerase-telomere association. Genes Dev. 2012;26:82–91. doi: 10.1101/gad.181826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 136.Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- 138.Virta-Pearlman V, Morris DK, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 139.Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 140.Zhou J, Hidaka K, Futcher B. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol Cell Biol. 2000;20:1947–1955. doi: 10.1128/mcb.20.6.1947-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [This study showed that Est1 can bridge Est2-TLC1 and Cdc13 at the telomere by demonstrating that the requirement of Est1 for telomerase requirement can be waived if the Est2 polypeptide is fused with the Cdc13 protein.] [DOI] [PubMed] [Google Scholar]
- 142.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 143.Wu Y, Zakian VA. The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc Natl Acad Sci U S A. 2011;108:20362–20369. doi: 10.1073/pnas.1100281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bianchi A, Negrini S, Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 145.Chan A, Boule JB, Zakian VA. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 2008;4:e1000236. doi: 10.1371/journal.pgen.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tseng SF, Shen ZJ, Tsai HJ, Lin YH, Teng SC. Rapid Cdc13 turnover and telomere length homeostasis are controlled by Cdk1-mediated phosphorylation of Cdc13. Nucleic Acids Res. 2009;37:3602–3611. doi: 10.1093/nar/gkp235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li S, et al. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Taggart AK, Teng SC, Zakian VA. Est1p as a cell cycle-regulated activator of telomerebound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [This study demonstrated that Est1, Est2, and Cdc13 are associated with telomeres during telomere replication in the S phase of the cell cycle, and suggested a telomerase-activating function for Est1 upon its binding to Cdc13.] [DOI] [PubMed] [Google Scholar]
- 149.Tuzon CT, Wu Y, Chan A, Zakian VA. The Saccharomyces cerevisiae telomerase subunit Est3 binds telomeres in a cell cycle- and Est1-dependent manner and interacts directly with Est1 in vitro. PLoS Genet. 2011;7:e1002060. doi: 10.1371/journal.pgen.1002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.DeZwaan DC, Freeman BC. The conserved Est1 protein stimulates telomerase DNA extension activity. Proc Natl Acad Sci U S A. 2009;106:17337–17342. doi: 10.1073/pnas.0905703106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.DeZwaan DC, Freeman BC. Is there a telomere-bound ‘EST’ telomerase holoenzyme? Cell Cycle. 2010;9:1913–1917. doi: 10.4161/cc.9.10.11536. [DOI] [PubMed] [Google Scholar]
- 152.Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol. 2008;15:990–997. doi: 10.1038/nsmb.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yu EY, Wang F, Lei M, Lue NF. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol. 2008;15:985–989. doi: 10.1038/nsmb.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yen WF, Chico L, Lei M, Lue NF. Telomerase regulatory subunit Est3 in two Candida species physically interacts with the TEN domain of TERT and telomeric DNA. Proc Natl Acad Sci U S A. 2011;108:20370–20375. doi: 10.1073/pnas.1017855108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fisher TS, Taggart AK, Zakian VA. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- 156.Bertuch AA, Lundblad V. The Ku heterodimer performs separable activities at doublestrand breaks and chromosome termini. Mol Cell Biol. 2003;23:8202–8215. doi: 10.1128/MCB.23.22.8202-8215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pfingsten JS, et al. Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model. Cell. 2011;148:922–932. doi: 10.1016/j.cell.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gallardo F, Olivier C, Dandjinou AT, Wellinger RJ, Chartrand P. TLC1 RNA nucleocytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 2008;27:748–757. doi: 10.1038/emboj.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009;23:928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ferreira HC, et al. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat Cell Biol. 2011;13:867–874. doi: 10.1038/ncb2263. [DOI] [PubMed] [Google Scholar]
- 161.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 162.Mishra K, Shore D. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr Biol. 1999;9:1123–1126. doi: 10.1016/s0960-9822(99)80483-7. [DOI] [PubMed] [Google Scholar]
- 163.Lopez CR, et al. Ku must load directly onto the chromosome end in order to mediate its telomeric functions. PLoS Genet. 2011;7:e1002233. doi: 10.1371/journal.pgen.1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ribes-Zamora A, Mihalek I, Lichtarge O, Bertuch AA. Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat Struct Mol Biol. 2007;14:301–307. doi: 10.1038/nsmb1214. [DOI] [PubMed] [Google Scholar]
- 165.Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3' telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci U S A. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pedroso IM, Hayward W, Fletcher TM. The effect of the TRF2 N-terminal and TRFH regions on telomeric G-quadruplex structures. Nucleic Acids Res. 2009;37:1541–1554. doi: 10.1093/nar/gkn1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang H, Nora GJ, Ghodke H, Opresko PL. Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding. J Biol Chem. 2011;286:7479–7489. doi: 10.1074/jbc.M110.205641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Raghuraman MK, Cech TR. Effect of monovalent cation-induced telomeric DNA structure on the binding of Oxytricha telomeric protein. Nucleic Acids Res. 1990;18:4543–4552. doi: 10.1093/nar/18.15.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yanez GH, Khan SJ, Locovei AM, Pedroso IM, Fletcher TM. DNA structure-dependent recruitment of telomeric proteins to single-stranded/double-stranded DNA junctions. Biochem Biophys Res Commun. 2005;328:49–56. doi: 10.1016/j.bbrc.2004.12.134. [DOI] [PubMed] [Google Scholar]
- 171.Schaffitzel C, et al. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci U S A. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by Gquartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 173.Wang Q, et al. G-quadruplex formation at the 3' end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 2011;39:6229–6237. doi: 10.1093/nar/gkr164. [DOI] [PMC free article] [PubMed] [Google Scholar]