Abstract
Purpose
Chronic graft-versus-host disease (cGVHD) is a severe immunological complication that occurs after allogeneic hematopoietic stem cell transplantation (HSCT). Although oral cGVHD occurs in >25 % of cGVHD patients and leads to decreased quality of life, its etiology is poorly understood. The present retrospective cross-sectional analysis of oral cGVHD patients sought to (1) test the feasibility of liquid chromatography tandem mass spectrometry (LC-MS/MS) to identify protein biomarkers of oral cGVHD and (2) to gain a clearer understanding of salivary proteins impacted by oral cGVHD.
Methods
Using unstimulated whole saliva, we compared pooled saliva from five patients with a diagnosis of moderate or severe oral cGVHD, with a gender-and age- matched pool of five cGVHD patients with no oral mucosal findings. LC-MS/MS was used to identify salivary proteins, followed by Ingenuity Pathway Analysis (IPA). Selected mass spectrometric findings, including lactotransferrin, lactoperoxidase, and albumin, were confirmed by targeted label-free quantification.
Results
LC-MS/MS led to confident identification of 180 proteins. Of these proteins, 102 changed in abundance at least 2 fold, including 12 proteins identified only in the No oral cGVHD group. Downregulation of ~0.4 fold was confirmed for both lactotransferrin and lactoperoxidase in Oral cGVHD saliva using targeted label-free quantification. IPA analysis implicated pathways involved in cellular metabolism and immunoregulation.
Conclusions
Reduction of salivary lactoperoxidase, lactotransferrin, and several cysteine proteinase inhibitor family proteins suggests impaired oral antimicrobial host immunity in cGVHD patients. This shotgun proteomic analysis of oral cGVHD saliva using targeted label-free quantification of select proteins supports the use of mass spectrometry for future validation in a large patient population as noninvasive tests for screening, early detection, and monitoring of cGVHD.
Electronic supplementary material
The online version of this article (doi:10.1007/s10875-012-9738-4) contains supplementary material, which is available to authorized users.
Keywords: Saliva, oral, GVHD, biomarker, proteomics, label-free quantitation, transplant, graft versus host
Introduction
Chronic graft-versus-host disease (cGVHD) is a severe immunological complication that occurs after allogeneic hematopoietic stem cell transplantation (HSCT) and is the leading cause of late, non-relapse death in this patient group. GVHD and its debilitating sequelae are a barrier limiting the more widespread use of HSCT [1]. Chronic GVHD can affect nearly every organ system through a donor-origin cellular response directed against host tissue leading to immune dysregulation, immunodeficiency, and impaired organ function [2]. The end organ effects of cGVHD often resemble autoimmune and other immunologic disorders, suggesting a common pathophysiology, and are the direct cause of decreased patient quality of life, impaired physical status, and early mortality [3].
Oral manifestations are present in at least 25 % of patients with cGVHD. The oral cavity is a highly affected organ system in cGVHD, second only to skin involvement [1, 2]. The spectrum of oral cGVHD includes mucosal lesions, salivary gland dysfunction, and restricted mouth opening, leading to pain, xerostomia, and loss of oral function [2–4]. Current diagnosis is based on clinical exam in conjunction with histopathological features.[5, 6].
Regardless of etiology, changes in salivary function and the composition of saliva affect oral homeostasis leading to dental, periodontal, and oral mucosal disease [7, 8]. Whole saliva (oral fluid) is a complex mixture of salivary gland secretions, gingival crevicular fluid, plasma protein transudates, oral keratinocyte products, and microbial flora, which serve myriad protective antimicrobial, immunomodulatory, and anti-inflammatory functions [9]. Changes in salivary function and mucosal breakdown seen in cGVHD could alter salivary composition and directly influence oral manifestations including pain, dysfunction, avoidance of nutritional food, infections, and oral health [2, 10]. Mucosal damage seen in oral cGVHD would further disrupt the local oral environment and facilitate the entry of microbial pathogens into the systemic circulation of patients, potentially contributing to sepsis after HSCT [11].
As cGVHD often mimics the findings of autoimmune disorders [12], the consideration of autoimmune diseases known to affect salivary glands and/or oral mucosa, such as Sjögren’s syndrome, scleroderma, and systemic lupus erythematosus, may provide insight into the pathophysiology and organ involvement of cGVHD in the oral cavity [13]. The proteomic analysis of saliva has shown great promise as a tool for the identification of biomarkers for primary Sjögren’s syndrome [14, 15]. Further, saliva could reflect the local oral consequence of the systemic process of cGVHD, and could help to elucidate the underlying pathophysiology of this disease process.
In a longitudinal study, Imanguli et al. first reported changes in the salivary proteome in the early post-HSCT period (1–6 months), some of which persisted though 6 months post-HSCT [16]. Considerable advances in detection and identification of the salivary proteome in the past several years have enabled finer analysis of the contents of saliva, and more detailed connections to systemic pathways [17, 18]. Consequently, the aim of the present study was twofold. First, to identify protein markers of oral cGVHD which could lead to improved diagnosis and monitoring after therapeutic intervention. Currently, the diagnostic gold standard for oral cGVHD is a clinical exam paired with biopsy of the buccal mucosa. Given that serial mucosal biopsy is not recommended for monitoring of the oral cavity, it would be preferable from the standpoint of both patient and practitioner to identify salivary markers for non-invasive serial testing. Second, a clearer understanding of salivary proteins impacted by oral cGVHD will elicit information about the mechanisms and etiological process of oral cGVHD which, in turn, could lead to development of more specific therapy.
In this retrospective cross-sectional pilot study, we compared the whole saliva of 5 patients with a diagnosis of moderate or severe oral cGVHD, as measured using the Oral Mucositis Rating Scale (ORMS) [19], with a gender-and age-matched control group of 5 cGVHD patients with no oral mucosal findings. Liquid chromatography tandem mass spectrometry (LC-MS/MS) led to confident identification of 180 proteins. Of these proteins, 102 changed in abundance at least 2 fold, including 12 proteins identified only in the with No oral cGVHD group. Selected mass spectrometric findings, including lactotransferrin, lactoperoxidase, and albumin, were confirmed by targeted label-free quantification.
Methods
Study Groups and Oral cGVHD Diagnosis
Patients in this study are enrolled subjects of ongoing transplant and cGVHD studies at the National Institutes of Health, National Cancer Institute (NCI) (clinicaltrials.gov #NCT00331968 and #NCT00520130). All data involving human subjects comply with the guiding principles for experimental procedures found in the Declaration of Helsinki of the World Medical Association, and all subjects signed NCI Institutional Review Board approved informed consents.
All patients underwent allogeneic HSCT, had a clinical diagnosis of cGVHD in an organ system, and were referred to the NIH Dental Clinic for comprehensive evaluation of the oral cavity. From these referrals, a study group of five male patients was selected, as described below, from patients who were diagnosed with oral cGVHD (Oral cGVHD) and compared with gender and age-matched male patients diagnosed as not having oral cGVHD (No oral cGVHD). Only males were chosen for the pilot study to identify bonafide changes between cGVHD patients and control GVHD patients, as sex hormones related to puberty and menstrual cycle have been reported to alter the abundance of proteins in female subjects [20].
Patients referred to the NIH Dental Clinic with cGVHD undergo a detailed examination by a dentist with expertise in oral cGVHD using the ORMS scale, a ratings scale (total score with a range from 0 to 270) designed to evaluate oral mucosal changes in HSCT treatments [19]. An ORMS score of above 34 was considered moderate to severe oral cGVHD for this study. In some but not all cases, a punch biopsy of the right buccal mucosa was taken and sent to pathology for a histopathology review of possible oral cGVHD findings (formalin fixed, paraffin embedded, sectioned, and hematoxylin and eosin stained).
Statistical analysis of the characteristics of the study groups was performed using a Mann–Whitney test to determine the difference in the two groups for continuous variables, with p ≤ 0.05 being considered statistically significant.
Saliva Collection and Preparation
Whole unstimulated saliva samples were collected on ice into sterile tubes at the time of oral evaluation. Immediately after collection, a 1/20 volume of protease cocktail inhibitor was added to the saliva and frozen at −80 °C. The protease cocktail inhibitor contained 0.1 M Tris–HCl, pH7.4, 0.1 M epsilon amino caproic acid, 0.05 M sodium EDTA, 25 mg/L of pepstatin A, 0.025 M benzamidine-HCl, 0.25 mg/L of leupeptin and 0.5 M phenylmethylsulfonyl fluoride. To perform mass spectrometry analysis, saliva samples were thawed and centrifuged at 2,600 x g for 15 min at 4 °C to separate the supernatant from the pellet. Subsequently, protein concentration was determined by bicinchoninic acid (BCA) and aliquot was prepared by pooling equal amount of protein from each subject within a group and pooled to normalize the difference between subjects and reduce individual variation for mass spectrometry (MS). The remaining saliva samples were subsequently stored at −80 °C until further analysis.
Reduction, Alkylation, and Trypsin Digestion
Fifty micrograms of pooled whole saliva from each group (No oral GVHD and Oral GVHD) was precipitated overnight by trichloroacetic acid (TCA). The protein pellet was dissolved and denatured with 6 M urea, reduced with 200 mM diothiothretol for 60 min at room temperature followed by alkylation with 200 mM iodoacetamide at room temperature in the dark for 60 min. The urea concentration was diluted with ammonium bicarbonate prior to digestion by trypsin (Promega, Madison, WI) overnight at 37 °C with gentle shaking at a protein-to-trypsin ratio of 30:1(w/w). Peptides were desalted using Oasis HLB-1 (1 mg) reversed phase cartridge (Waters, Milford, MA) and vacuum concentrated to dryness. Subsequently, digests were divided into two aliquots, with one aliquot used for peptide/protein identification and relative protein quantification by spectral counting and the second aliquot for label-free “targeted” quantification of select differentially expressed proteins.
Protein Identification by Liquid Chromatography Coupled Tandem Mass Spectrometry (LC-MS/MS)
Five micrograms of tryptic peptide mixtures from No oral cGVHD and oral cGVHD subjects were loaded separately onto Zorbax C18 trap column (Agilent Tech., Santa Clara, CA) for 8.3 min at a flow rate of 6.0 μL/min for further desalting of the peptide mixture. The peptides were then separated on a 10 cm Picofrit Biobasic C18 analytical column (New Objective, Woburn, MA) using an on-line Eksigent (Dublin, CA) nano-LC ultra HPLC system. The peptides were eluted using a 120 min acetonitrile gradient (5–35 %) at flow rate of 250 nL/min. Peptides were ionized using electrospray ionization (ESI) in positive ion mode and detected on a LTQ-Orbitrap Velos (Thermo Fisher Scientific, San Jose, CA). Precursor ions were selected for MS/MS using a data-dependent method in which the top 6 most intense ions from the MS1 precursor scan were selected. All precursor ions were measured in the Orbitrap with the resolution set at 30,000 (m/z 400). Precursor ions were fragmented by collision-induced dissociation (CID) with normalized collision energy of 35 %, and all fragment ions were measured in the LTQ. The targeted LC-MS/MS runs were 1 h, while the discovery runs were 2 h.
Data Analysis
All LC-MS/MS data were searched using the MASCOT algorithm within Proteome Discoverer 1.3 (Thermo Electron Corp, San Jose, CA) against human Swissprot protein database (Version Sprot_101911) to obtain peptide and protein identifications. For all searches, trypsin was specified as the enzyme for protein cleavage allowing up to 2 missed cleavages. Oxidation (M) and carbamidomethylation (C) were set as dynamic and fixed modifications. Mass tolerance of 20 ppm and 0.8 Da were set for precursor and fragment ions, respectively.
Ingenuity Pathway Analysis (IPA)
The data set containing down-regulated proteins (≤0.5) were analyzed by Ingenuity Pathway Analysis version 9.0 (Ingenuity® Systems, www.ingenuity.com, Mountain View, CA). Statistically significant protein identification by IPA is based on mapping input proteins with a continuously curated database of published literature to a range of function, cellular location, canonical pathways and disease inter-relationships. The association between proteins in the dataset and canonical pathways in the Ingenuity Pathways Knowledge Base was measured as a ratio of the number of molecules from the data that maps to a pathway divided by the total number of molecules that map to the canonical pathway. A right-tailed Fisher’s Exact Test was used to calculate the p-value of the probability that the association between each protein in the dataset and the canonical pathway is random. Pathways with p value < 0.05 and at least 2 associated proteins were selected as potential pathways affected by oral cGVHD [21].
Targeted Label-Free Quantification
For targeted analysis, all nano-LC parameters were the same as LC-MS/MS. However, MS parameters were adjusted to target only a specific set of peptides. An inclusion list was prepared, consisting of the accurate m/z values of tryptic peptides from a select group of differentially expressed proteins to confirm the differential expressed observed by LC-MS. Peptides were selected for the inclusion list based on 3 criteria. The tryptic peptides selected: 1) contained no missed cleavages, 2) had a charge state of +2, +3, or +4, and 3) contained no methionine residues. Five μg of peptide was injected into the MS. All precursors were detected in the Orbitrap at a resolution of 30,000 (m/z 400) and fragment ions were measured in the LTQ. Once a precursor m/z from the inclusion list was detected in the MS1 scan, a subsequent MS/MS spectrum was acquired. In order to account for potential co-eluting peptides, 3 subsequent MS/MS scan events were included in the method.
For confirmation of peptides, automated label-free quantification was carried out using in-house developed software, QUOIL [21]. For MS/MS data visualization, MASCOT results were imported into Scaffold 3Q + (Proteome Software, Portland, OR).
Results
Demographic and Clinical Characteristics of Study Subjects
Table 1 summarizes the clinical and oral cGVHD characteristics of the study groups. Both groups include similarly aged males; the majority had matched, unrelated peripheral blood HSCT, and all were evaluated for oral cGVHD at least 100 days after transplant. Systemic immunosuppression was scored according to the Intensity of Immunosuppression Scale, which is used for classification of the level of systemic immunosuppression in post-transplant studies: 1 = None, 2 = Mild (single agent prednisone < 0.5 mg/kg/day), 3 = Moderate (Prednisone ≥ 0.5 mg/kg/day and/or any single agent/modality), and 4 = High (2 or more agents/modalities ± prednisone ≥ 0.5 mg/kg/day [22]. Importantly, there was no difference in mean systemic immunosuppression between No oral cGVHD (2.60 ± 1.52) and Oral cGVHD (3.20 ± 1.30) subjects in this study. There was also no significant difference in salivary flow rate between groups (No oral cGVHD 0.445 ± 0.126 ml/min; oral GVHD 0.394 ± 0.142, p-value 0.8). The Oral cGVHD group had an ORMS score above 34, while the No oral cGVHD group had very low ORMS scores (ORMS = 49.00 ± 17.06 versus 7.80 ± 4.82, p = 0.01). Figure 1 shows a patient from the oral cGVHD group, highlighting the clinical and histological findings seen in Oral cGVHD, (right panels A−D) and compares these with a patient in the No oral cGVHD group. (left panels E−H) The Oral cGVHD patients presented clinically with intraoral atrophy, erythema, hyperkeratosis, lichenoid reactions, ulcerations, and edema, and histologically with copious lymphocytic infiltrate and the presence of apoptotic bodies in the oral buccal mucosa.
Table I.
Clinical and oral cGVHD characteristics of No oral cGVHD and Oral cGVHD study groups
| No oral cGVHD group | Oral cGVHD group | P value**** | |
|---|---|---|---|
| N = 5 | N = 5 | ||
| Age (years) | 51.4 ± 5.59, 45–59* | 52.2 ± 8.50, 42–64* | 1 |
| ORMS | 7.80 ± 4.82, 0–13 | 49.00 ± 17.06, 34–77 | 0.01 |
| Days from transplant | 592 ± 582, 132–1573 | 1396 ± 1345, 180–3667 | 0.3 |
| Disease** | 3 CML/Lymphoma | 3 CML/Lymphoma | - |
| 1 AML | 2 AML | ||
| 1 CLL | - | ||
| Cell source | 4 Peripheral Blood | 5 Peripheral Blood | - |
| 1 Bone Marrow | - | ||
| Donor relationship | Unrelated | 4 Unrelated | - |
| - | 1 Related | ||
| HLA match | 5 Matched | 3 Matched | - |
| - | 2 Mismatched | ||
| Oral histopathology | 4 of 4 biopsies taken showed no oral cGVHD | 2 of 2 oral biopsies taken showed oral cGVHD | - |
| Intensity of immunosuppression*** | 2.60 ± 1.52 | 3.20 ± 1.30 | 0.4 |
| Unstimulated salivary flow rate (ml/min) | 0.445 ± 0.126 | 0.394 ± 0.142 | 0.8 |
* Mean ± Standard Deviation, range
**Abbreviations: CML, chronic myeloid leukemia; AML, acute myelogenous leukemia; CLL, chronic lymphocytic leukemia
*** 1 = None, 2 = Mild (single agent prednisone < 0.5 mg/kg/day), 3 = Moderate (Prednisone ≥ 0.5 mg/kg/day and/or any single agent/modality), 4 = High (2 or more agents/modalities ± prednisone ≥ 0.5 mg/kg/day)
**** Mann–Whitney test
Fig. 1.
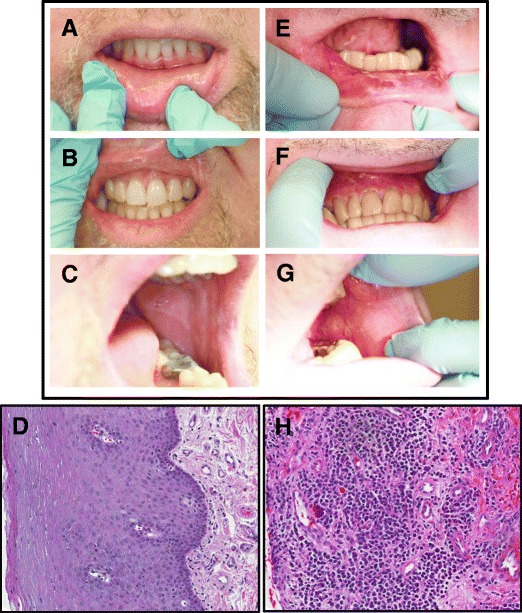
Clinical and histological presentation of no oral cGVHD (a–d) versus oral cGVHD (e–h). Clinical presentation of oral cGVHD includes atrophy, erythema, hyperkeratosis, lichenoid reactions, ulcerations, and edema, here seen on the lower lip (e), maxillary anterior gingiva (f), and buccal mucosa (g) of a patient with oral cGVHD, as compared with the normal intraoral mucosa of a patient with no oral cGVHD (a–c). H&E staining of the oral mucosa from the above-matched patients demonstrates characteristic GVHD-like changes in the buccal mucosa compared with No oral cGVHD tissue (d), including prevalent lymphocytic infiltrate and apoptotic bodies (h). Buccal biopsy sections are shown in similar orientation at 10x magnification
Selection of Differentially Expressed Proteins
A quantitative proteomics study was performed to identify change in abundance of proteins between the oral GVHD and the No oral cGVHD groups. Figure 2 shows the work flow schematic for the identification and confirmation of the differentially expressed proteins. Equal amounts of protein were pooled from each subject within a group and pooled for identification by mass spectrometry and subsequent targeted label-free quantitation. No significant difference was noted in mean total protein per ml for individual samples in either group (No oral cGVHD 0.97 ± 0.61 μg/ml; Oral cGVHD 1.08 ± 1.1 μg/ml, p = 0.85). A protein was deemed a confident match if at least two unique peptides were detected for a given protein, which resulted in the identification of 180 proteins. A ≤ 0.5 or ≥ 2 fold change cut-off [23]was applied to the above list leading to identification of 102 differentially expressed proteins, including 12 proteins identified only in the No oral cGVHD group. The differentially expressed proteins are listed in Supplementary Table 1. The spectral counts for several of the 12 proteins are relatively high (e.g. 27 for BPI1A_HUMAN), indicating that the identified difference is real and not part of an instrumental issue. Of note were several members of the cysteine proteinase inhibitor family that were identified in this study. Cystatin-SA, cystatin-C, cystatin-B, cystatin-S and cystatin-D were down-regulated in oral cGVHD, while cystatin-SN did not change in abundance.
Fig. 2.
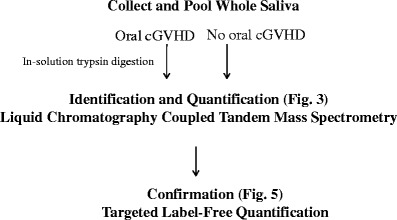
Study Workflow. The diagram overviews the procedures used for the identification and quantification of differentially expressed salivary proteins for No oral cGVHD and Oral cGVHD subjects
Distribution of Differentially Expressed Proteins
Of the 102 differentially expressed proteins found in Oral cGVHD or No oral cGVHD whole saliva, 12 proteins were identified only in the No oral cGVHD group while 90 proteins were down-regulated in the Oral cGVHD as compared with the No oral cGVHD group (≤0.5). Among down-regulated proteins, ~31 % changed 0.5–0.41 fold (n = 28), ~33 % changed 0.4–0.31 fold (n = 30), ~22 % of the proteins changed 0.3–0.21 fold (n = 20), and ~13 % changed ≤0.2 fold (n = 12). In contrast, 78 proteins showed little or no change in abundance, based on a fold ratio change cut-off >0.5 or <2, as shown in Fig. 3.
Fig. 3.
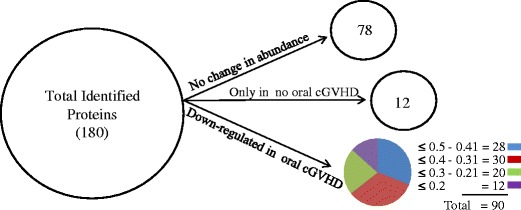
Distribution of differentially expressed proteins. Differentially expressed proteins (n = 180) were categorized using an arbitrary fold ratio change cut-off of ≤ 0.5 or ≥ 2 into proteins that did not change in abundance (n = 78), proteins that were identified only in the No oral cGVHD group (n = 12), and proteins that were down-regulated in the Oral cGVHD samples (n = 90). The number of proteins in each pathway is given above each bar
Ingenuity Pathways Analysis
Differentially expressed proteins (down-regulated) were mapped using Ingenuity Pathway Analyses (IPA) software (Version 9.0) to determine statistically significant (p < 0.05) canonical pathways. Based on published literature to date, in the oral cGVHD group, 83 proteins mapped to 12 different pathways (Fig. 4). Among the different pathways, glycolysis (n = 5) had the largest number of involved proteins while four other pathways involved three proteins.
Fig. 4.
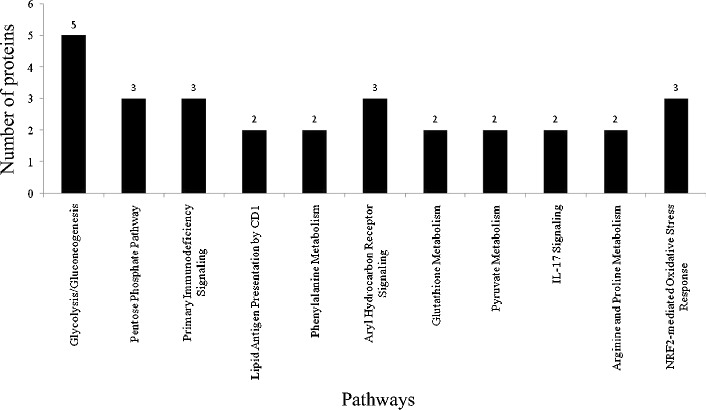
Canonical Pathways identified by Ingenuity Pathway Analyses (IPA). Significant pathways (n = 12) were mapped via identified down-regulated proteins. Values on each the y-axis indicate the number of involved proteins in each pathway
Targeted Label-Free Confirmation
Of the 180 proteins detected, 78 had little or no change in abundance, while 102 proteins were detected as either identified only in the No oral cGVHD group or were down-regulated (≤0.5). Of the down-regulated proteins, 2 proteins with antimicrobial activity were chosen for confirmation by targeted label-free approach: lactotransferrin, lactoperoxidase, Serum albumin, which did not change significantly, was included as an internal control.
Confirmation of lactotransferrin and lactoperoxidase by the targeted label-free approach is shown in Fig. 5. For each protein, two different peptides were used for confirmation and were deemed confident if both peptides showed similar differential expression. Figure 5 shows the chromatograms for one of the two peptides for lactotransferrin (NLLFNDNTECLAR) and lactoperoxidase (VGPLLACLLGK). Quantification of these three proteins in Oral cGVHD and No oral cGVHD samples was analyzed by the fold change values of single precursor ion chromatograms for lactotransferrin (~0.4) and lactoperoxidase (~0.4). The abundance of each peptide is calculated from the height of the peak intensity as shown on the y-axis, while the x-axis shows the time at which the peptides eluted. The retention times for all three peptides were comparable indicating little variation between peptide elution between Oral cGVHD and No oral cGVHD experimental runs.
Fig. 5.
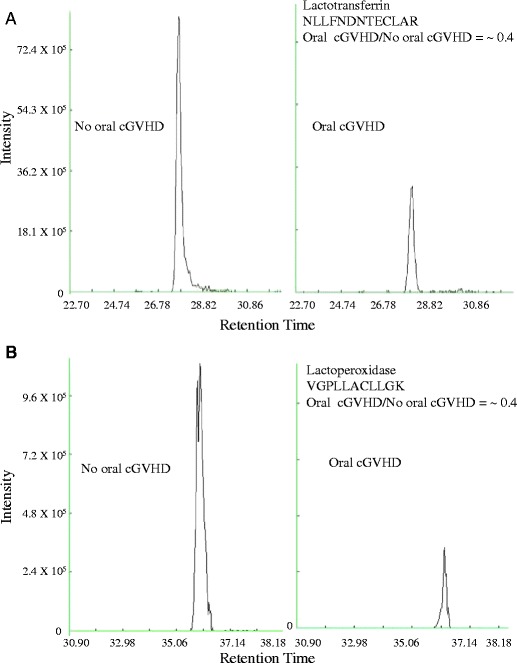
Label-Free Quantification of Selected Differentially Expressed Proteins between No oral cGVHD and Oral cGVHD groups. Shown are the extracted ion chromatograms showing “peptide elution plotted against intenity” for (a) lactotransferrin, and (b) lactoperoxidase by oral cGVHD classification to illustrate downregulation of each protein in the Oral cGVHD group
The top chromatograms in Fig. 5 panels A and B represent No oral cGVHD, while the bottom panels of A and B are Oral cGVHD chromatograms. There are stark differences in peptide abundance for lactotransferrin and lactoperoxidase (Fig. 5a and b, respectively), as shown by a fold change of ~0.4 in each case. Albumin showed no change in abundance.
Discussion
Salivary proteomic technology is an emerging science whose advancement is driving the field of salivary diagnostics to identify and understand disease processes. In the current pilot study, we applied cutting-edge salivary proteomic techniques to the identification of proteins in the pooled saliva of age- and gender-matched cGVHD patients with and without oral cGVHD with the overarching goal of determining the potential of these techniques for GVHD saliva analysis. In this study, a total of 180 proteins were confidently identified. Amongst these proteins, hemoglobin subunit alpha and hemoglobin subunit beta were identified, which may be due to contamination of saliva samples from hemolysis of red blood cells. The alpha and beta hemoglobin subunits have also been detected in whole ductal saliva, collected directly from a cannulated parotid gland, which strongly suggests that these 2 subunits of hemoglobin may be an intrinsic part of saliva. It is postulated that these blood proteins may enter normal saliva through the leaky epithelium in the parotid and potentially other salivary glands. Their detected levels did not differ between groups in this study, so blood contamination of cGVHD saliva is not considered to be a major confounding factor. A ≥2 or ≤ 0.5 fold change cut-off [23] was applied to the above list, resulting in the identification of 102 differentially expressed proteins. A two-fold cut-off of differentially expressed proteins indicates doubling, or 100 % increase in the expression ratio. Ingenuity Pathway Analysis (IPA) analysis was performed on 102 differentially expressed proteins leading to identification of 12 canonical pathways involved in the Oral cGVHD population compared to No oral cGVHD controls. Targeted free label confirmation focused on 3 proteins: lactotransferrin, lactoperoxidase and albumin, and detected significant downregulation of both lactotransferrin and lactoperoxidase, but no change in albumin levels, in the saliva of GVHD subjects.
Among the identified pathways, a majority of the proteins were related to cellular metabolism (glycolysis/gluconeogenesis, inositol, glutathione, pyruvate, argnine and proline metabolism), and immune signaling (IL-17 and primary immunodeficiency signaling). Although pathway analysis using IPA on these proteins does not provide stringent evidence for their direct association in cGVHD pathophysiology, it may provide new insight into potentially important networks for in-depth mechanistic analysis to validate their involvement in disease pathogenesis.
Lactoperoxidase, one of two proteins present in lower amounts in GVHD saliva that was confirmed, is part of the IPA-identified phenylanine metabolism pathway. There, lactoperoxidase catalyzes oxidation of certain intermediates within the phenyalanine pathway. However, lactoperoxidase is also a natural antimicrobial compound found in mammalian saliva, mucus and milk [24]. The natural acceptor molecule for lactoperoxidase in the body is thiocyanate, whose oxidation product, hypothiocyanate, is a potent antibacterial compound that inhibits the growth of an array of bacteria. At high concentrations, lactoperoxidase functions to directly kill certain bacteria, including Escherichia coli, Pseudamonas aeruginosa and Haemophilus influenza, and to limit replication of human immunodeficiency virus [24, 25].
Of particular interest is the reduction in detectable lactotransferrin in saliva of cGVHD patients. Lactotransferrin is an iron-binding glycoprotein found in high concentrations in epithelial secretions including saliva and human milk. This multi-functional antimicrobial compound is found in the secondary granules of neutrophils and also acts as a primary defense against bacteria via sequestration of iron that results in release of lipopolysaccharide (LPS) from the outer microbial membrane and thereby stymies bacterial growth [26, 27]. Furthermore, low levels of lactotransferrin act synergistically with other antimicrobial compounds to increase overall potency and rate of bacterial killing [27]. This protein acts not only as an antimicrobial compound, but also has mitogenic capacity, and plays a role in the activation of the cellular immune system through TLR4-dependant and independent macrophage activation [28, 29]. Lactotransferrin involvement has been previously implicated in oral and gut cGVHD [16, 30]. A 2001 case report detailed successful use of supplemental lactoferrin to treat gut GVHD refractory to conventional immunosuppressive therapy [30]. A 2007 saliva survey from our group found increased salivary levels of lactotransferrin through 6-months post-transplant in GVHD patients. In contrast, the present study and included a post-transplant age-matched patient population that was much later, on average, 45-months [16]. Involvement in both screens, however, clearly implicates lactotransferrin in an altered salivary proteome following HSCT.
Several members of the cysteine proteinase inhibitor family were identified in this study. Cystatin-SA, cystatin-C, cystatin-B, cystatin-S and cystatin-D were down-regulated in oral cGVHD, while cystatin-SN did not change in abundance. These proteins are secreted extracellularly, where they inhibit proteinases, thereby limiting proteolysis and tissue damage [31, 32]. Cystatin D is a potent inhibitor of human coronavirus at physiologic salivary concentrations [33]. Thus, lower levels of these proteins may be lead to increased tissue damage and lower defenses against common viral infections.
Taken together, reduction of lactoperoxidase, lactotransferrin, and several cysteine proteinase inhibitor family proteins suggest impaired oral antimicrobial host immunity in cGVHD patients. Xerostomia is reported in 77 % of oral cGVHD patients [4]. There was no statistical difference in mean salivary flow rate between groups in this study, however, the flow rate for both groups was slightly below the reported unstimulated flow rate in healthy individuals of 0.47 ± 0.29 ml/min[34]. This is in concordance with our clinical experience that this patient group is susceptible to frequent oral infections and irritations, though there is no published data comparing rates of oral infection (fungal, viral, bacterial) in post-HSCT patients with respect to cGVHD status.
Previous salivary analyses of cGVHD patients have reported differential expression of ions and proteins using assorted methods (flame photometry, spectrophotometry, atomic absorption) and differential gel electrophoresis (2DE-DIGE) techniques followed by identification of proteins by mass spectrometry [16, 35]. Although 2DE-PAGE/MS can be an effective tool, it has limitations including restricted dynamic range, poor identification of membrane proteins and poor reproducibility [36, 37]. To overcome this limitation, in-solution digestion of proteins was utilized for this study, and then molecules were identified using liquid chromatography coupled tandem mass spectrometry (LC-MS/MS).
To date, confirmation of potential salivary markers in cGVHD proteomic studies have been performed using Western blots and ELISA [16, 38, 39]. These techniques give valuable results; however, they are limited by the availability, sensitivity and specificity of antibodies for proteins [40]. Thus, it is often critical to use non-antibody based methods such as multiple reaction monitoring (MRM) [41] or accurate inclusion mass screening (AIMS) [42]. In this study, we used targeted label-free confirmation of three potential markers (lactotransferrin, lactoperoxidase and albumin) for successful development of quantitative assays.
Conclusions
Collectively, this is the first shotgun proteomic analysis of cGVHD subjects using mass spectrometry. We controlled for gender, age, and the intensity of immunosuppression. Our study confidently identified 180 proteins, 102 of which were differentially expressed, including 12 proteins uniquely expressed only in the No oral cGVHD group. Formal validation of specific findings requires a larger study with additional information to be gained from longitudinal sampling. Targeted label-free quantification of select proteins supports the use of mass spectrometry for validation in large patient population as noninvasive tests for screening, early detection, and monitoring of cGVHD. Patients with oral cGVHD have lower innate salivary defenses against oral microbial and viral insults, which calls for greater clinical diligence to maintain the oral and systemic health of patients with oral cGVHD.
Acknowledgments
We gratefully acknowledge Dr. Frances Hakim for thoughtful discussions during the course of this study, and sincerely thank the patients who were willing to participate in this research. This work was supported by the Intramural Research Programs of the NIDCR, NIH and the NCI, NIH. The authors declare no conflict of interest.
Electronic Supplement Materials
Below is the link to the electronic supplementary material.
Differentially expressed proteins identified in whole saliva with a minimum of two unique peptides in No oral cGVHD and Oral cGVHD groups. (XLSX 22 kb)
Footnotes
Carol W Bassim and Kiran S. Ambatipudi contributed equally.
References
- 1.Schubert MM, Correa MEP. Oral graft-versus-host disease. Dent Clin N Am. 2008;52:79–109. doi: 10.1016/j.cden.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Fall-Dickson, J.M., et al., Oral symptom intensity, health-related quality of life, and correlative salivary cytokines in adult survivors of hematopoietic stem cell transplantation with oral chronic graft-versus-host disease. Biology of Blood and Marrow Transplantation, 2010;16(7):948–56. [DOI] [PMC free article] [PubMed]
- 3.Imanguli MM, et al. Oral graft-versus-host disease. Oral Diseases. 2008;14:396–412. doi: 10.1111/j.1601-0825.2008.01448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imanguli M.M., et al. Salivary gland involvement in chronic graft-versus-host disease: Prevalence, clinical significance, and recommendations for evaluations. Biology of Blood and Marrow Transplantation. 2010;16(10):1362–69. doi: 10.1016/j.bbmt.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shulman HM, et al. Histopathologic diagnosis of chronic graft-versus-host disease: National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: II. Pathology Working Group Report. Biol Blood Marrow Transplant. 2006;12(1):31–47. doi: 10.1016/j.bbmt.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 6.Filipovich AH, et al. National Institutes of Health consensus development project on criteria for clnical trials in chronic graft-versus-host disease: I. Diagnosis and staging working group report. Biology of Blood and Marrow Transplantation. 2005;11:945–955. doi: 10.1016/j.bbmt.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Amerongen A, Veerman E. Saliva- the defender of the oral cavity. Oral Diseases. 2002;8:12–22. doi: 10.1034/j.1601-0825.2002.1o816.x. [DOI] [PubMed] [Google Scholar]
- 8.Dodds MW, Johnson DA, Yeh CK. Health benefits of saliva: A review. J Dent. 2005;33(3):223–33. doi: 10.1016/j.jdent.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Bassim C, et al. Salivary procalcitonin and periodontitis in diabetes. J Dent Res. 2008;87:630–4. doi: 10.1177/154405910808700707. [DOI] [PubMed] [Google Scholar]
- 10.Lenssen O, et al. Prevalence of nutrition-related problems among long-term survivors of allogeneic marrow transplantation. J Am Diet Assoc. 1990;90:835–842. [PubMed] [Google Scholar]
- 11.Ruescher T, et al. The impact of mucositis on alpha-hemolytic streptococcal infection in patients undergoing autologous bone marrow transplantation for hematologic malignancies. Cancer. 1998;82:2275–2281. doi: 10.1002/(SICI)1097-0142(19980601)82:11<2275::AID-CNCR25>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 12.Choi S, Levine J, Ferrara J. Pathogenesis and Management of Graft-versus-Host Disease. Immunol Allergy Clin N Am. 2010;30:75–101. doi: 10.1016/j.iac.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambatipudi K, et al. Quantitative analysis of age specific variation in the abundance of human female parotid salivary proteins. J Proteome Res. 2009;8:5093–5102. doi: 10.1021/pr900478h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baldini C, et al. Proteomic analysis of saliva: a unique tool to distinguish primary Sjögren’s syndrome from secondary Sjögren’s syndrome and other sicca syndromes. Arthritis Res Ther. 2011;13:R194. doi: 10.1186/ar3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferraccioli G, et al. Proteomic approaches to Sjögren’s syndrome: A clue to interpret the pathophysiology and organ involvement of the disease. Autoimmun Rev. 2010;9:622–626. doi: 10.1016/j.autrev.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Imanguli M, et al. Changes in salivary proteome following allogeneic hematopoietic stem cell transplantation. Exp Hematol. 2007;35:184–192. doi: 10.1016/j.exphem.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baum BJ, et al. Scientific frontiers: Emerging technologies for salivary diagnostics. Adv Dent Res. 2011;23(4):360–8. doi: 10.1177/0022034511420433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loo JA, et al. Comparative human salivary and plasma proteomes. J Dent Res. 2010;89(10):1016–23. doi: 10.1177/0022034510380414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schubert MM, et al. Clinical assessment scale for the rating of oral mucosal changes associated with bone marrow transplantation. Cancer. 1992;69:2469–2477. doi: 10.1002/1097-0142(19920515)69:10<2469::AID-CNCR2820691015>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 20.Ambatipudi KS, et al. Quantitative analysis of age specific variation in the abundance of human female parotid salivary proteins. J Proteome Res. 2009;8(11):5093–102. doi: 10.1021/pr900478h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, et al. Label-free protein Quantification using LC-coupled ion trap or FT mass spectrometry: Reproducibility, linearity, and application with complex proteome. J Proteome Res. 2006;5:1214–1223. doi: 10.1021/pr050406g. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell SA, et al. Determinants of functional performance in long-term survivors of allogeneic hematopoietic stem cell transplantation with chronic graft-versus-host disease (cGVHD) Bone Marrow Transplant. 2010;45(4):762–9. doi: 10.1038/bmt.2009.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao H, et al. Use of mass spectrometry to identify protein biomarkers of disease severity in the synovial fluid and serum of patients with rheumatoid arthritis. Arthritis Rheum. 2004;50(12):3792–803. doi: 10.1002/art.20720. [DOI] [PubMed] [Google Scholar]
- 24.Wijkstrom-Frei C, et al. Lactoperoxidase and human airway host defense. Am J Respir Cell Mol Biol. 2003;29(2):206–12. doi: 10.1165/rcmb.2002-0152OC. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi Y, et al. Virucidal effects of glucose oxidase and peroxidase or their protein conjugates on human immunodeficiency virus type 1. Antimicrob Agents Chemother. 1993;37(1):26–31. doi: 10.1128/AAC.37.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He J, Furmanski P. Sequence specificity and transcriptional activation in the binding of lactoferrin to DNA. Nature. 1995;373(6516):721–4. doi: 10.1038/373721a0. [DOI] [PubMed] [Google Scholar]
- 27.Travis SM, Singh PK, Welsh MJ. Antimicrobial peptides and proteins in the innate defense of the airway surface. Curr Opin Immunol. 2001;13(1):89–95. doi: 10.1016/S0952-7915(00)00187-4. [DOI] [PubMed] [Google Scholar]
- 28.Curran CS, Demick KP, Mansfield JM. Lactoferrin activates macrophages via TLR4-dependent and -independent signaling pathways. Cell Immunol. 2006;242(1):23–30. doi: 10.1016/j.cellimm.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Grey A, et al. The low-density lipoprotein receptor-related protein 1 is a mitogenic receptor for lactoferrin in osteoblastic cells. Mol Endocrinol. 2004;18(9):2268–78. doi: 10.1210/me.2003-0456. [DOI] [PubMed] [Google Scholar]
- 30.Inoue M, et al. Lactoferrin for gut GVHD. Bone Marrow Transplant. 2001;28(11):1091–2. doi: 10.1038/sj.bmt.1703283. [DOI] [PubMed] [Google Scholar]
- 31.Al-Hashimi I, Dickinson D, Levine M. Purification, molecular cloning, and sequencing of salivary cystatin SA-1. J Biol Chem. 1997;263:9381–9987. [PubMed] [Google Scholar]
- 32.Freije JP, et al. Human cystatin D. cDNA cloning, characterization of the Escherichia coli expressed inhibitor, and identification of the native protein in saliva. J Biol Chem. 1993;268(21):15737–44. [PubMed] [Google Scholar]
- 33.Collins AR, Grubb A. Cystatin D, a natural salivary cysteine protease inhibitor, inhibits coronavirus replication at its physiologic concentration. Oral Microbiol Immunol. 1998;13(1):59–61. doi: 10.1111/j.1399-302X.1998.tb00753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawes C, Macpherson LM. Effects of nine different chewing-gums and lozenges on salivary flow rate and pH. Caries Res. 1992;26(3):176–82. doi: 10.1159/000261439. [DOI] [PubMed] [Google Scholar]
- 35.Nagler RM, Nagler A. Sialometrical and sialochemical analysis of patients with chronic graft-versus-host disease–a prolonged study. Cancer Invest. 2003;21(1):34–40. doi: 10.1081/CNV-120016401. [DOI] [PubMed] [Google Scholar]
- 36.Washburn M, Wolters D, Yates JI. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 37.Nilsson T, et al. Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods. 2010;7(9):681–5. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- 38.Baum BJ, et al. Therapy-induced dysfunction of salivary glands: implications for oral health. Spec Care Dentist. 1985;5(6):274–7. doi: 10.1111/j.1754-4505.1985.tb00593.x. [DOI] [PubMed] [Google Scholar]
- 39.Izutsu KT, et al. Disordered salivary immunoglobulin secretion and sodium transport in human chronic graft-versus-host disease. Transplantation. 1983;35(5):441–6. doi: 10.1097/00007890-198305000-00010. [DOI] [PubMed] [Google Scholar]
- 40.Addona T, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keshishian H, et al. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaffe J, et al. Accurate Inclusion Mass Screening. Mol Cell Proteomics. 2008;7:1952–1962. doi: 10.1074/mcp.M800218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]


