DNA repair pathways can enable tumor cells to survive DNA damage induced by chemotherapy and thus provide prognostic and/or predictive value. In this study, the authors sought to assess the differential expression, bimodal distribution, and prognostic and predictive role of DNA repair genes in individual breast cancer molecular subtypes including estrogen receptor-positive/ HER2-negative, estrogen receptor-negative/HER2-negative, and HER2-positive cancers. The predictive value of DNA repair gene expression was assessed in breast cancer patients treated with neoadjuvant taxane/anthracycline- or anthracycline-containing regimens, and gene set analyses were performed by grouping DNA repair genes according to biological pathways.
Keywords: Neoadjuvant therapies, DNA repair pathways, Predictive factors, Breast cancer subtypes, DNA damaging agents
Abstract
DNA repair pathways can enable tumor cells to survive DNA damage induced by chemotherapy and thus provide prognostic and/or predictive value. We evaluated Affymetrix gene expression profiles for 145 DNA repair genes in untreated breast cancer (BC) patients (n = 684) and BC patients treated with regimens containing neoadjuvant taxane/anthracycline (n = 294) or anthracycline (n = 210). We independently assessed estrogen receptor (ER)-positive/HER2-negative, HER2-positive, and ER-negative/HER2-negative subgroups for differential expression, bimodal distribution, and the prognostic and predictive value of DNA repair gene expression. Twenty-two genes were consistently overexpressed in ER-negative tumors, and five genes were overexpressed in ER-positive tumors, but no differences in expression were associated with HER2 status. In ER-positive/HER2-negative tumors, the expression of nine genes (BUB1, FANCI, MNAT1, PARP2, PCNA, POLQ, RPA3, TOP2A, and UBE2V2) was associated with poor prognosis, and the expression of one gene (ATM) was associated with good prognosis. Furthermore, the prognostic value of specific genes did not correlate with proliferation. A few genes were associated with chemotherapy response in BC subtypes and treatment-specific manner. In ER-negative/HER2-negative tumors, the MSH2, MSH6, and FAN1 (previously MTMR15) genes were associated with pathological complete response and residual invasive cancer in taxane/anthracycline-treated patients. Conversely, PMS2 expression was associated with residual invasive cancer in treatments using anthracycline as a single agent. In HER2-positive tumors, TOP2A was associated with patient response to anthracyclines but not to taxane/anthracycline regimens. In genes expressed in a bimodal fashion, RECQL4 was significantly associated with clinical outcome. In vitro studies showed that defects in RECQL4 impair homologous recombination, sensitizing BC cells to DNA-damaging agents.
Implications for Practice:
The identification of molecular mechanisms and biomarkers of sensitivity to chemotherapy in breast cancer is still controversial. In this context, the cellular DNA repair machinery is expected to play an important role in response to different types of chemotherapy. The differential expression of many DNA repair genes between ER-positive and ER-negative breast tumors could contribute to the different clinical behavior of these two breast cancer subtypes. We demonstrated that specific DNA repair genes are prognostic and predictive of chemotherapy response in a molecular subtype and treatment specific manner suggesting their contribution in the risk of tumor recurrence and response to chemotherapy. Such prognostic and predictive value warrant further exploration in clinical trials for optimizing treatment tailoring to improve patient outcome. Furthermore, defects of RECQL4 impair homologous recombination and sensitize breast cancer cells to DNA damaging agents, e.g., PARP inhibitors and platinum agents, which allows for selecting patients who more likely will benefit from these agents.
Introduction
Breast cancer (BC) represents a molecularly and clinically heterogeneous disease comprising different molecular subtypes harboring different genetic alteration patterns [1–5]. Distinct gene pathways and biological processes are associated with prognosis and chemotherapy sensitivity in different BC subtypes [6–8]. DNA repair pathways maintain genomic integrity and are among the targets most frequently altered in cancer [9, 10]. The DNA repair machinery allows normal cells to repair DNA damage or induce apoptosis and cell cycle arrest if repair is not possible [11]. Anticancer drugs are highly influenced by the cellular DNA repair capacity, and specific alterations in DNA repair pathways have been reported to be associated with differences in patient response to chemotherapy in several cancers [12]. Drugs that inhibit a single specific DNA repair pathway in these tumors could prove useful as single-agent therapies, harboring the advantages of selectively targeting tumor cells and having fewer negative side effects. Several BC subtypes such as triple-negative BC (TNBC) are characterized by a higher degree of genomic instability because of defects in the DNA repair machinery [13, 14]. Moreover, TNBCs possessing BRCA mutations or having BRCAness features and therefore higher genomic instability have been shown to respond better to DNA-damaging agents such as platinum-containing compounds; to PARP inhibitors; and, in some cases, to chemotherapy. The role of DNA repair pathways in BC subtypes, however, has not been well established.
In this study, we sought to assess the differential expression, bimodal distribution, and prognostic and predictive role of DNA repair genes in individual BC molecular subtypes including estrogen receptor (ER)-positive/HER2-negative, ER-negative/HER2-negative, and HER2-positive cancers. A large series of publicly available gene expression datasets was collected. The predictive value of DNA repair gene expression was assessed in BC patients treated with regimens containing neoadjuvant taxane/anthracycline or anthracycline. We performed gene set analyses by grouping DNA repair genes according to biological pathways.
We also selected one relevant DNA repair gene that arose as a result of our in silico analysis for (a) in vitro assessment of the biological functions of this gene within the DNA repair pathways and (b) exploration of the potential mechanisms of the cell response to DNA-damaging chemotherapy in BC.
Materials and Methods
Patient Cohorts
Clinical characteristics of the patients included in the various datasets used in this study are presented in supplemental online Table 1. Response to chemotherapy was dichotomized as pathological complete response (pCR) or residual invasive cancer (RD). To further validate the differential expression of DNA repair-related genes in BC subtypes, we examined a BC cell line dataset that included 51 cell lines [15].
Gene Expression Normalization, Molecular Subtype Definition, and Molecular Markers
All gene expression data were generated with Affymetrix U133A and U133 Plus2 gene chips (Affymetrix, Santa Clara, CA, http://www.affymetrix.com) and normalized using the MAS5 algorithm (http://www.bioconductor.org) with the mean expression centered to 600 and log2-transformed. ER and HER2 status was defined as previously reported [16]. To take into account the scaling factor existing between the U133A and U133 Plus2 platforms [17], slightly different cutoff values were adopted (ESR1 >10.6 as ER-positive and ERBB2 >13.04 as HER2-positive in U133 Plus2 chips). Molecular subgroups were defined according to ER and HER2 status. A proliferation marker (mitosis kinase score) was used to assess the independence of the prognostic value for the DNA repair genes relative to proliferation. The proliferation marker was calculated as previously reported [18].
Identification of Differentially Expressed DNA Repair Pathway Genes
Based on a literature review and on reported DNA repair pathways assembled from the PubMed and Gene Ontology databases, we selected the most functionally relevant annotated DNA repair genes. We identified a set of 145 DNA repair-related genes with documented roles in several DNA repair pathways.
We assessed differential expression of DNA repair-related genes between ER-positive and ER-negative tumors (after adjusting for HER2 status) using the Student's t test. To control for the potentially confounding effects of HER2 status in this comparison, the randomized block design included in BRB-Array Tools (National Cancer Institute, Division of Cancer Treatment and Research, Biometric Research Branch, Bethesda, MD, http://linus.nci.nih.gov/BRB-Array-Tools.html) was used. The randomized block design fits two linear models to the expression data for each gene. The full model included the ER class variable and the blocking variable (i.e., HER2), whereas the reduced model included only the blocking variable. Likelihood ratio statistics were used to test the significance of the differences between ER-negative and ER-positive tumors. A similar analysis was performed for HER2-positive and HER2-negative tumors using ER status as a blocking variable.
Statistical analyses were performed using BRB-Array Tools v 3.7.0 Patch_1 and R software (R Foundation, Vienna, Austria, http://www.r-project.org). All statistical tests were two-sided. To provide more robust and reproducible findings, the strategy adopted to identify differentially expressed prognostic and predictive DNA repair genes was used on different datasets as independent discovery cohorts rather than combining them in a larger series. This approach allowed us to claim significant findings based not only on statistical significance but also on consistency across independent discovery cohorts, similar to other studies [7, 16, 18]. Consistent observations are more likely to represent true associations.
Distant metastasis-free survival (DMFS), defined as the time from diagnosis to distant metastasis, was the primary endpoint used to assess the prognostic value of DNA repair gene expression. DMFS was available in all prognostic datasets. To make the results comparable across the datasets, survival was censored at 10 years. Univariate Cox regression analysis was used to correlate genes as continuous variables with DMFS in various clinical subgroups. To identify differentially expressed genes between cases based on pCR and RD, we performed unequal variance t tests on each probe set.
Because we used different discovery datasets to assess the consistency of our findings, we also calculated combined p values to use the data more efficiently. To calculate the combined p value for the pooled prognostic and predictive datasets, we let pij denote the p value for gene i (where i = 1–TOT) in the prognostic or predictive dataset j (where j = 1 or 2) for disease subtype k (where k = 1 for an ER-positive tumor and 2 for an ER-negative tumor). For each gene i, we calculated a Fisher's combined probability p value over the two or three datasets with 4 or 6 df (i.e., two or three times the number of p values combined) according to the following formula, in which ln is the natural logarithm: Pik = −2(ln(pi1k) + ln(pi2k)).
We assessed bimodal index [19] to identify gene expression with bimodal distribution that could be biologically relevant. Briefly, identifying genes with bimodal expression patterns from large-scale expression profiling data is an important task. Bimodal expression patterns can result naturally from differential expression with the two modes centered on the mean expression of a gene in two distinct subgroups of samples. In the context of cancer, bimodal expression patterns can result from genomic lesions that occur in some patients but not in others.
Functional Evaluation of RECQL4 in Cellular Response to Chemotherapy and DNA Damaging Agents
Immunohistochemistry
The formalin-fixed, paraffin embedded BC tissues (39 ER-negative/HER2-negative and 61 ER-positive/HER2-negative) were deparaffinized and rehydrated. Antigen retrieval was performed by using EnVision Flex Target Retrieval solution (Dako, Glostrup, Denmark, http://www.dako.com). Immunohistochemistry was performed with RECQL4 monoclonal antibody (Abnova, Taipei, Taiwan, http://www.abnova.com/) using an automated staining workstation (DakoCytomation, Glostrup, Denmark, http://www.dakocytomation.com) with diaminobenzidine chromogen as substrate. Slides were counterstained with hematoxylin (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com). Immunostaining results were scored semiquantitatively, and only nuclear staining was taken into account. The scores for immunohistochemistry were 0, no staining; 1, few nuclear-stained tumor cells (<5% of positive tumor cells); 2, 5%–20% of positive tumor cells; and 3, >20% positive tumor cells. For pairwise comparisons, the scores were collapsed to low (score 1–2) versus high (score 3) expression.
RECQL4 Functional Studies
RECQL4 was targeted with two distinct small interfering RNAs (siRNAs; Thermo Scientific Molecular Biology, Waltham, MA, http://www.thermoscientificbio.com/Dharmacon/), which were applied to the MDA-MB-231 BC cell line. MDA-MB-231 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, http://www.atcc.org/). We optimized the transfection conditions for MDA-MB-231 cells as previously described [18]. Gene silencing was performed in three replicates in 96-well plates using predetermined optimal transfection conditions. Plates were incubated at 37°C for 96 hours.
Results
Differentially Expressed DNA Repair Genes by Molecular Tumor Subtype
Even in HER2-positive tumors, large-scale molecular differences exist according to ER status [20]; therefore, we compared the expression of DNA repair genes between ER-positive and ER-negative tumors after adjusting for HER2 status. Then we compared the expression of DNA repair genes between HER2-positive and HER2-negative tumors after adjusting for ER status. In two discovery datasets, we identified 27 genes that were differentially expressed between ER-positive and ER-negative tumors with a p value ≤.01 in each dataset (Table 1, supplemental online Table 2A, and supplemental online Fig. 1) (expected false discovery rate ∼0.2 gene). These differences in gene expression were validated using three independent BC datasets and a series of 51 BC cell lines (Table 1, supplemental online Table 2A). All 27 genes were confirmed as significantly differentially expressed in each dataset, supporting the consistency and reliability of our findings. Conversely, in the cell line-derived data, only 9 of 27 genes were differentially expressed (p ≤ .05). Using the same stringent criteria, no genes were differentially expressed between HER2-positive and HER2-negative tumors in the two discovery datasets (supplemental online Table 2B).
Table 1.
DNA repair genes overexpressed in ER-negative and ER-positive breast tumors
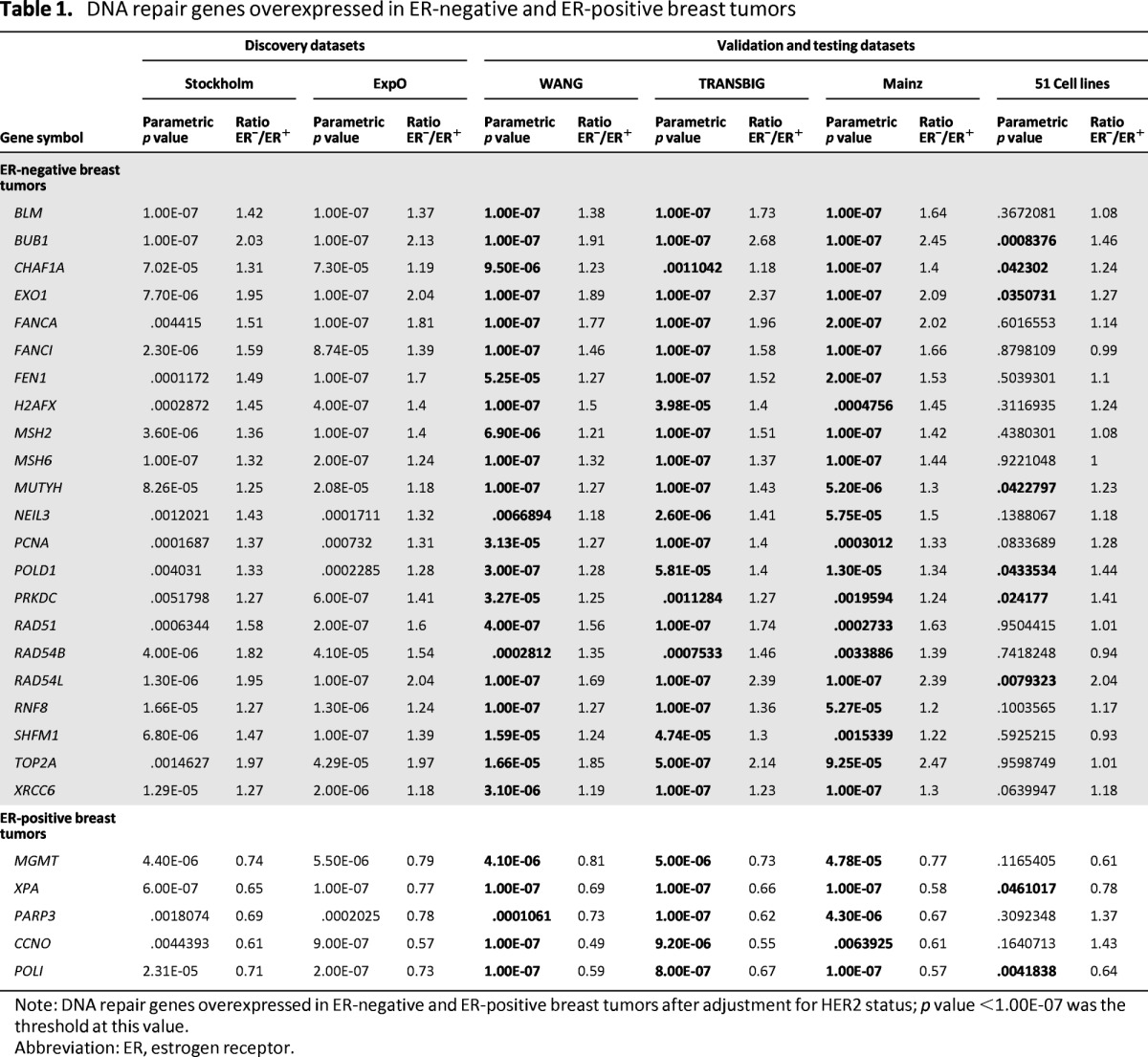
Note: DNA repair genes overexpressed in ER-negative and ER-positive breast tumors after adjustment for HER2 status; p value <1.00E-07 was the threshold at this value.
Abbreviation: ER, estrogen receptor.
DNA Repair Genes as Prognostic Factors in Each Molecular Tumor Subtype
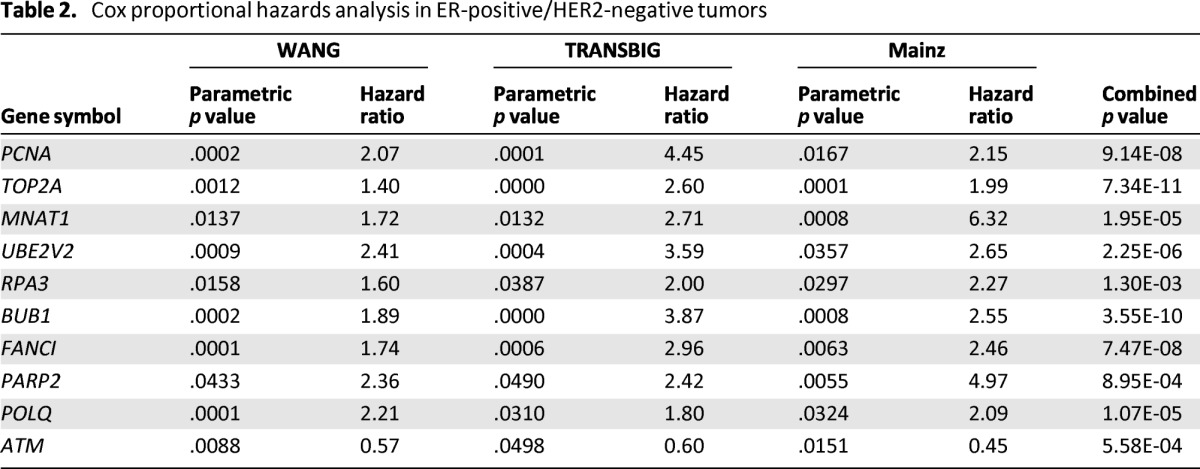
We assessed the prognostic value of DNA repair gene expression in untreated node-negative BC patients. The analysis was performed separately in each prognostic dataset based on the three molecular BC subtypes. Genes that correlated with patient outcome at a p value <.05 in all datasets were defined as prognostic (expected false discovery rate ∼0.2 gene). We found that none of the 145 DNA repair genes analyzed showed prognostic value in HER2-positive and ER-negative/HER2-negative tumor subtypes (supplemental online Tables 3B and 3C). In ER-positive/HER2-negative tumors, we found nine genes that were significantly associated with poor prognosis and one gene (ATM) that was associated with good prognosis (Table 2, supplemental online Table 3A). Interestingly, some of these genes weakly correlated or did not correlate (ATM, MNAT1) with proliferation markers (supplemental online Fig. 2). We assessed the prognostic value of each of these genes in a multivariate model including a proliferation marker [18] and progesterone receptor (PGR). TOP2A, POLQ, and MNAT1 expression retained significance after adjustment for these markers (p = .031, p = .012, p = .0006, respectively) (supplemental online Table 4). Similar results were obtained using the genomic grade index [21] as a proliferation marker (data not shown). These markers seem to provide prognostic information independent of proliferation.
Table 2.
Cox proportional hazards analysis in ER-positive/HER2-negative tumors
DNA Repair Genes as Predictive Factors of Response to Chemotherapy in Each Molecular Tumor Subtype
We assessed the predictive value of DNA repair gene expression according to specific treatment regimens (anthracycline- or taxane/anthracycline-containing regimens) in each of the predictive datasets according to molecular tumor subtype (supplemental online Tables 5A and 5B). Genes with a combined p value <.05 and a consistent correlation with the regimen are shown in Tables 3 and 4. Significant associations were defined as having a p value ≤.05 in two of the independent datasets. Despite the less stringent statistical criteria adopted relative to our assessment for prognostic value, few genes were consistently associated with pCR.
Table 3.
Anthracycline sensitivity related to DNA repair genes
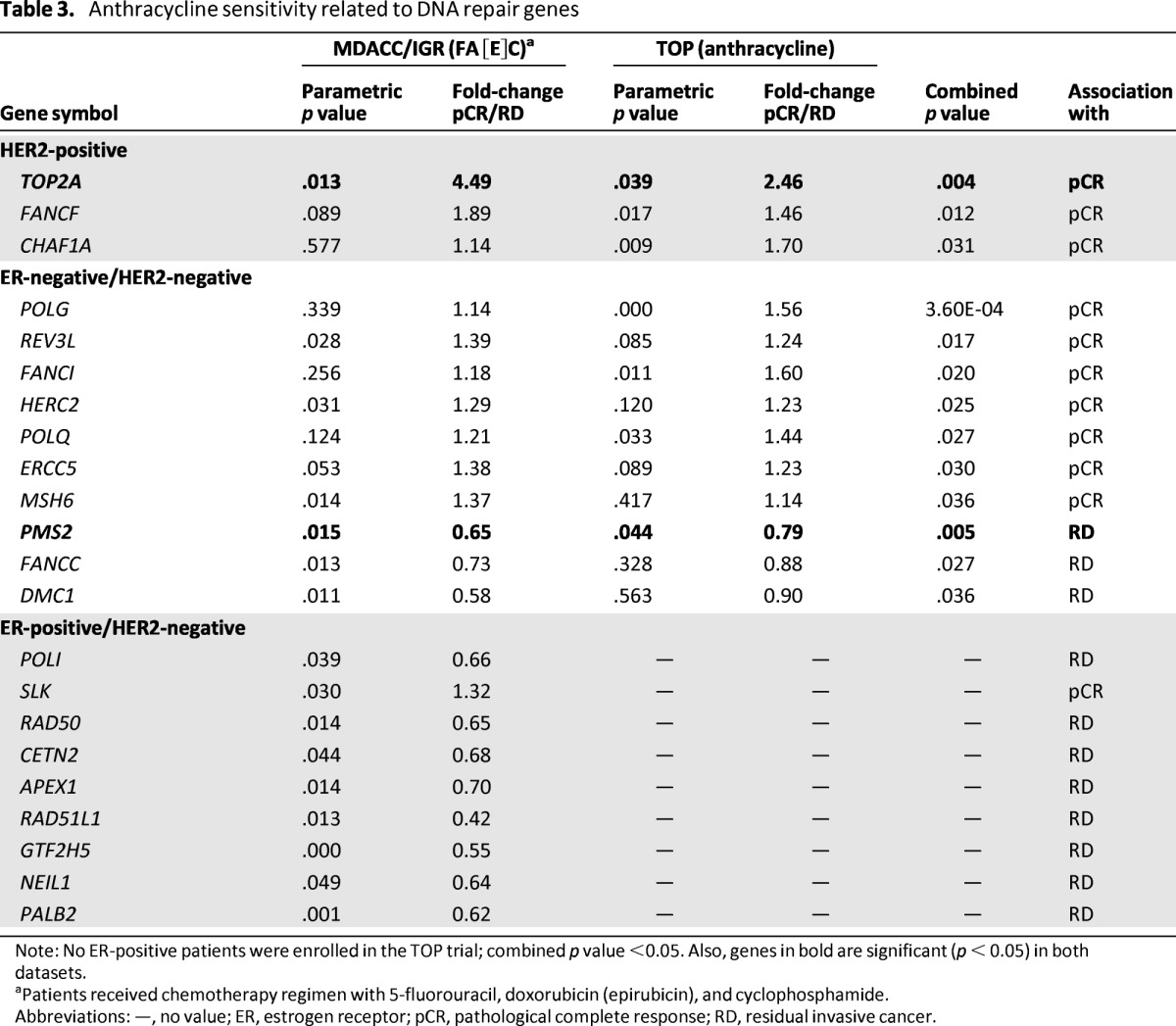
Note: No ER-positive patients were enrolled in the TOP trial; combined p value <0.05. Also, genes in bold are significant (p < 0.05) in both datasets.
aPatients received chemotherapy regimen with 5-fluorouracil, doxorubicin (epirubicin), and cyclophosphamide.
Abbreviations: —, no value; ER, estrogen receptor; pCR, pathological complete response; RD, residual invasive cancer.
Table 4.
Taxane sensitivity-related DNA repair genes
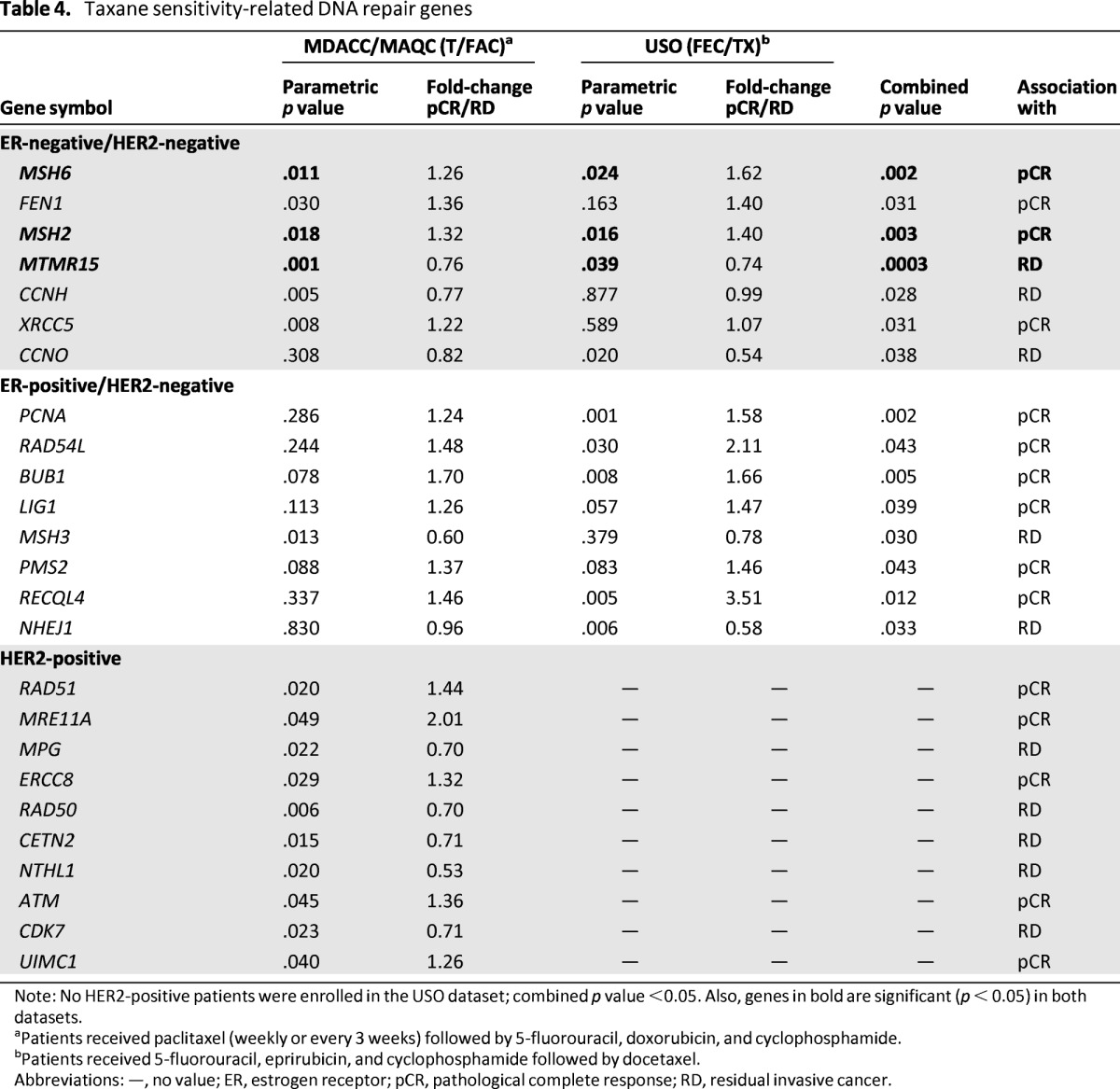
Note: No HER2-positive patients were enrolled in the USO dataset; combined p value <0.05. Also, genes in bold are significant (p < 0.05) in both datasets.
aPatients received paclitaxel (weekly or every 3 weeks) followed by 5-fluorouracil, doxorubicin, and cyclophosphamide.
bPatients received 5-fluorouracil, eprirubicin, and cyclophosphamide followed by docetaxel.
Abbreviations: —, no value; ER, estrogen receptor; pCR, pathological complete response; RD, residual invasive cancer.
In patients receiving anthracycline-based regimens, only TOP2A was significantly associated with pCR in HER2-positive samples (combined p = .004), whereas PMS2 was significantly associated with RD in ER-negative/HER2-negative tumors (combined p = .005) (Table 3). Notably, TOP2A expression did not show a significant association with anthracycline-containing regimens when other chemotherapeutic agents were used in combination.
In taxane/anthracycline regimens for ER-positive/HER2-negative and HER2-positive subtypes, none of the DNA repair genes was consistently associated with pCR or RD (Table 4). In ER-negative/HER2-negative tumors, however, mismatch repair (MMR) genes MSH2 and MSH6 were associated with a higher pCR rate (combined p = .003 and p = .002, respectively), and FAN1 (previously MTMR15) gene expression was associated with RD (combined p = .0003) (Table 4).
RECQL4 Impairs Homologous Recombination Sensitizing Breast Cancer Cell Lines to DNA Damaging Agents
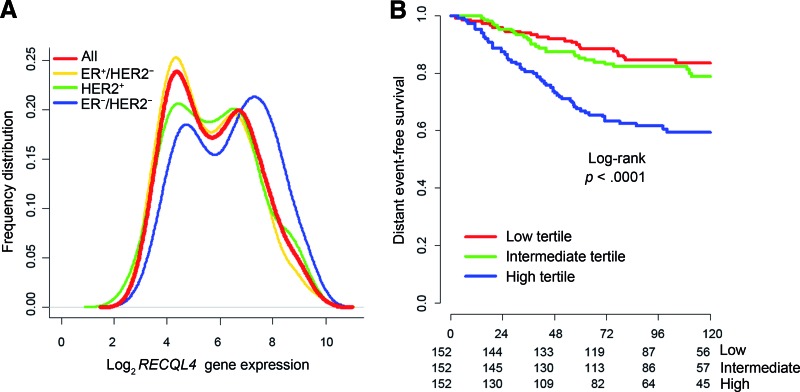
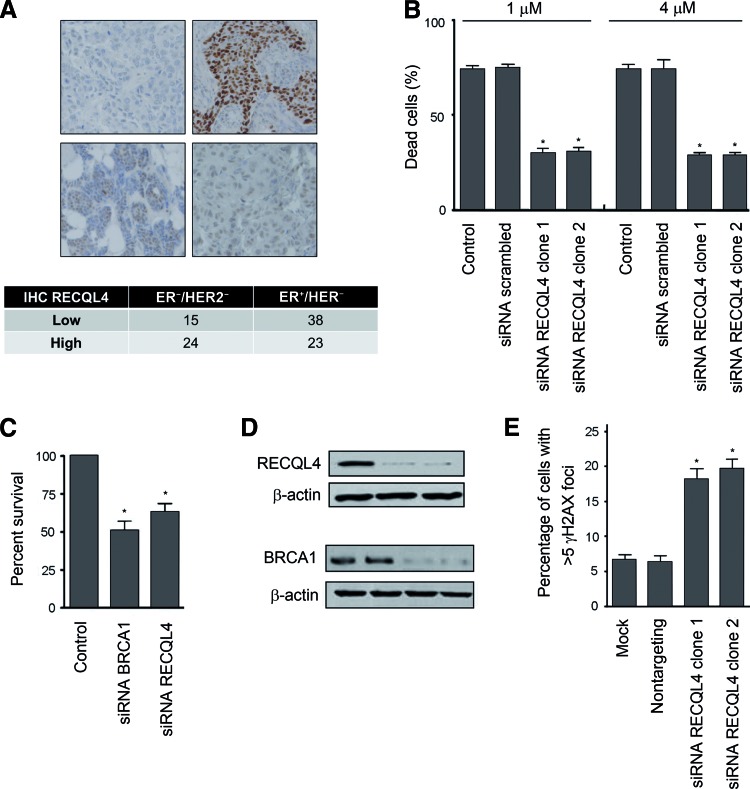
We hypothesized that genes with consistent bimodality among tumor subtypes could be enriched in biologically relevant genes. We defined bimodal genes (supplemental online Table 6) and assessed their prognostic and predictive value. Only RECQL4 showed bimodal distribution, and its expression was significantly associated with clinical outcomes (Fig. 1). RECQL4 was prognostic in ER-positive/HER2-negative tumors (supplemental online Table 3) after adjusting for proliferation and PGR expression (p = .046) (supplemental online Table 4). RECQL4 was also significantly associated (ER-positive/HER2-negative, p = .012) or trended toward a significant association (ER-negative/HER2-negative) with patient response to taxane/anthracycline-containing regimens (supplemental online Table 5B). Moreover, RECQL4 is an important helicase involved in the regulation of several DNA repair pathways. Consequently, we assessed the involvement of RECQL4 in DNA double-strand break (DSB) mechanisms and treatment responses. Overexpression of RECQL4 in ER-negative/HER2-negative tumors (supplemental online Table 2A) was confirmed by immunohistochemical detection of RECQL4 performed on a series of BC samples including 39 cases of ER-negative/HER2-negative tumors and 61 cases of ER-positive/HER2-negative tumors (p = .0264) (Fig. 2A).
Figure 1.
RECQL4 shows bimodal distribution, and its expression is significantly associated with clinical outcomes. (A): The frequency distribution calculated by the kernel function for RECQL4 in each molecular subtype and in the overall population is presented. (B): The Kaplan-Meier estimates for distant event-free survival are shown according to the tertiles of RECQL4 expression in ER-positive/HER2-negative tumors.
Figure 2.
Loss of RECQL4 is associated with resistance to taxanes and spontaneous DNA damage response activation. (A): IHC performed with anti-RECQL4 antibody in ER-positive/HER2-negative tumors (61 cases) and ER-negative/HER2-negative tumors (39 cases). RECQL4 was expressed to higher levels in ER-negative/HER2-negative tumors (p = .0264). (B): Death of RECQL4-silenced cells compared with scrambled or untransfected control cells after taxane treatment (1 μM or 4 μM) (*p < .001). Error bars indicate the standard deviation (SD; n = 3). (C): Survival of RECQL4-silenced cells compared with untransfected or BRCA1 siRNA control cells after PARP inhibition (*p < .001). (D): Immunoblot analysis of protein knockdown 96 hours after transfection of control, RECQL4, and BRCA1 siRNA constructs. (E): Nontargeting and RECQL4 siRNA were transfected into MDA-MB-231 cells, and γH2AX foci were counted 96 hours after transfection (mock, transfection with no siRNA) (*p < .001). Error bars represent the SD (n = 3).
Abbreviations: IHC, immunohistochemistry; siRNA, small interfering RNA.
We examined the effect of RECQL4 gene silencing on chemosensitivity to taxanes (1 μM and 4 μM paclitaxel) in the MDA-MB-231 cell line. Our results show that the downregulation of RECQL4 expression by two independent siRNAs led to an increase in taxane resistance in MDA-MB-231 cells (Fig. 2B).
We expected that RECQL4 likely functions in homologous recombination (HR) repair, and we predicted that HR deficiencies would cause synthetic lethality on PARP inhibition; therefore, we examined whether RECQL4 silencing could cause hypersensitivity to PARP inhibitors (AG14361 and olaparib). We found that RECQL4 silencing resulted in decreased cellular survival after treatment with PARP inhibitors AG14361 (1 μM) (Fig. 2C) and olaparib (data not shown). Of note, RECQL4 silencing yielded sensitivity similar to that observed when BRCA1 is silenced (Fig. 2C). Western blot analysis was used to confirm RECQL4 and BRCA1 silencing in MDA-MB-231 cells (Fig. 2D). RECQL4 silencing caused a fourfold increase in the percentage of cells staining for γH2AX foci in the absence of any added genotoxic stress agent when compared with nontargeting siRNA-transfected cells (Fig. 2E).
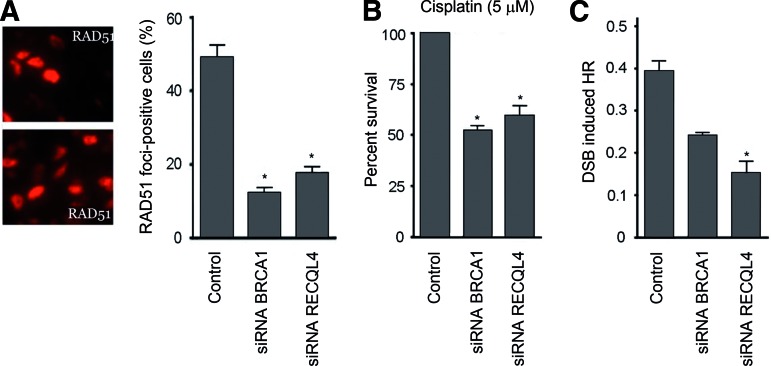
We next examined whether PARP inhibitor sensitivity and spontaneous DNA damage response activation in RECQL4-silenced cells was the result of defective DSB sensing and repair (Fig. 3). We performed immunofluorescence to measure the formation of RAD51 foci 8 hours after ionizing radiation, which is used as an indirect measure of HR repair (Fig. 3A). In contrast to control cells, RECQL4-silenced cells demonstrated complete absence of RAD51 foci in response to ionizing radiation (Fig. 3A). The reduced RAD51 foci in the RECQL4-silenced cells combined with the increased PARP inhibitor sensitivity suggested that these cells might harbor a reduced capability to repair DSBs by HR in the absence of RECQL4. Consistently, we found that RECQL4-silenced cells are hypersensitive to cisplatin (Fig. 3B), which induces DNA damage repair via HR-dependent pathways. In all cases, RECQL4 silencing yielded sensitivity similar to that observed in siBRCA1 cells.
Figure 3.
RECQL4 silencing impairs HR repair at DSBs. (A): RAD51 foci were scored by immunofluorescent imaging 8 hours after treatment with IR. Quantification of nuclei containing RAD51 foci was determined based on 200 cells. Error bars indicate the standard deviation (SD; n = 3; *p < .001). (B): Control, BRCA1- and RECQL4-silenced MDA-MB-231 cells were treated with cisplatin (5 μM) for 96 hours and then were measured for viability (*p < .05). Error bars indicate the SD (n = 5). (C): Control, BRCA1- and RECQL4-silenced MDA-MB-231 cells containing an integrated HR repair substrate were transfected with constructs expressing I-SceI to induce a DSB in the GFP reporter gene. The percentage of GFP-positive cells was scored by flow cytometry. Error bars indicate the SD (n = 3; *p < .05).
Abbreviations: DSB, double-strand break; HR, homologous recombination; IR, ionizing radiation; siRNA, small interfering RNA.
We then directly assessed HR repair in RECQL4-silenced cells using MDA-MB-231 cells expressing a dislocation-dependent reconstituted green fluorescent protein reporter plasmid, in which the GFP gene is interrupted by a single SceI restriction site [22]. Transient expression of the SceI endonuclease introduces a site-specific DSB within the reporter. Cells that are proficient in HR repair display restoration of GFP gene expression and increased fluorescence. Cells were analyzed by two-color flow cytometry to quantify GFP-positive cells relative to the total number of cells. We found that RECQL4-silenced cells have a significant defect in HR repair activity compared with control and BRCA1-silenced cells (Fig. 3C). Taken together, these results indicate that RECQL4 acts similar to BRCA1 and that RECQL4 silencing contributes to chemoresistance to taxanes and hypersensitivity to DNA damaging agents; this phenomenon most likely occurs through defects in HR repair.
Discussion
Different clinical subtypes of BC are characterized by different gene expression profiles, DNA copy number alterations, and mutation distributions [1–5, 23, 24]. In this study, we examined the expression of 145 DNA repair-related genes independently in each of the main molecular subtypes of BC. We focused our comparisons on ER-negative and ER-positive cancers after adjusting for HER2 status because these groups showed large-scale differences regardless of HER2 status [20]. Moreover, more complex molecular classification methods have not yet been standardized [1].
We found that 5 DNA repair genes were significantly overexpressed in ER-positive tumors and that 22 DNA repair genes were significantly overexpressed in ER-negative tumors. All of the overexpressed DNA repair gene candidates were confirmed (p < .05) using three independent validation datasets. The ER-negative overexpressed genes are involved in several DNA repair pathways including MMR (MSH2 and MSH6), HR (RAD51, RAD54L, RAD54B, and SHFM1), and Fanconi anemia (FANCA and FANCI) pathways as well as in other cellular processes that cooperate with DNA repair pathways such as chromatin remodeling (H2AFX and CHAF1A) and DNA polymerization, synthesis, editing, and processing (PCNA, POLD1, TOP2A, BLM, FEN1, and EXO1). These genes could play important biological functions in the ER-negative group contributing to the ER-negative-associated clinical characteristics of higher rate of early recurrence, younger age at diagnosis, higher histological grade, and higher chemotherapy benefit [25, 26]. Notably, the same genes were not similarly differentially expressed in a series of 51 BC cell lines, underscoring that the expression of some genes in cancer cell lines does not mimic the corresponding human BC.
DNA Repair Genes as Prognostic Factors in Breast Cancer Subtypes
The prognostic value of DNA repair genes has been assessed in untreated, node-negative BCs. We found that nine genes (BUB1, FANCI, MNAT1, PARP2, PCNA, POLQ, RPA3, TOP2A, and UBE2V2) were significantly associated with poor prognosis in ER-positive/HER2-negative cancers and that one gene (ATM) was significantly associated with good prognosis in ER-positive/HER2-negative cancers. In ER-positive/HER2-negative cancers, several markers of proliferation have already been shown to provide clinically relevant prognostic information (e.g., Oncotype DX, Genomic Health, Redwood City, CA, http://www.genomichealth.com/; MammaPrint, Agendia, Amsterdam, the Netherlands, http://www.agendia.com/; MapQuant Dx Genomic Grade, Qiagen Marseille, Marseille, France, http://www.qiagenmarseille.com/) [1, 27]. We assessed whether these markers were associated with proliferation. Only ATM and MNAT1 did not correlate or only weakly correlated with proliferation markers. We also assessed whether these prognostic genes provide independent information in multivariate models including proliferation marker and PGR expression, which are other established prognostic factors in this group of cancers [28]. Interestingly, MNAT1 retained significant prognostic value (p = .0006) in the multivariate model, suggesting potential value in assessing prognosis over conventional proliferation markers; however, the value of MNAT1 expression requires independent confirmation. Expression levels of TOP2A and BUB1 closely reflect the proliferative activity of cells [18, 29] and have been previously associated with recurrence in ER-positive/HER2-negative BC [30, 31]. PCNA and POLQ are involved in genomic replication and DNA repair [32, 33], and their overexpression has been correlated with poor clinical outcome in BC patients [34, 35]. Another study has shown that the DNA repair gene UBE2V2 contributes to the overall resistance of cancer cells to genotoxic agents [36].
Even if ATM expression does not provide independent prognostic information in multivariate analyses, ATM expression is not associated with proliferation, which could indicate that ATM expression has a relevant biological function in cell dedifferentiation. In fact, low ATM expression in BC tissue has been correlated with a higher rate of DNA mutations, a progressive cancer phenotype, and increased angiogenesis [37]. The expression of ATM has been associated with favorable patient outcomes [37], which is in agreement with our results. None of the DNA repair genes considered in our study was consistently associated with prognosis in both ER-negative/HER2-negative and HER2-positive tumors. These findings suggest that by including the expression of the MNAT1 gene in the clinical assessment, it could be possible to refine the risk of relapse prediction performed using established molecular markers of proliferation and PGR expression.
DNA Repair Genes as Predictors of Likelihood of Response to Chemotherapy
We also examined the association between expression of DNA repair genes and the likelihood of achieving a pCR in patients treated with preoperative taxane/anthracycline- or anthracycline-based chemotherapy regimens. In BC patients treated with anthracycline-based chemotherapy, we found that only higher expression of TOP2A was consistently associated with pCR, and this association was found only in the HER2-positive tumor group. This finding confirms previous data regarding the association between TOP2A expression and response to anthracycline-based regimens in HER2-positive tumors [38]. This consistency indirectly confirmed the validity of the discovery approach adopted in our study. The chromosomal region that incorporates HER2 is a gene-rich region of genetic instability that is pivotal to tumor progression and poor prognosis in BCs [39]. These data reinforce the clinical need to explore the inclusion of the assessment of TOP2A gene expression for tailoring the group of patients in the HER2-positive molecular subtype at higher likelihood to benefit from anthracycline administration [38, 40]. The amplification of HER2 is associated with important amplifications and/or deletions of neighboring TOP2A DNA repair genes and require further investigation for the identification of additional therapeutic targets. Notably, TOP2A expression was not associated with pCR when other drugs were administered concurrently with taxanes. This may suggest that the group of patients who show lower expression of TOP2A and less benefit from anthracycline-based therapy could derive substantial benefits from other drugs. This difference, correlated with TOP2A expression and chemotherapy regimen, also underscores the inherent limitation of assessing predictive markers for polychemotherapy regimens because of the potential different predictive value of each marker according to the drugs administered. Overall, these findings provide a supportive explanation for the additional clinical benefit of taxanes to anthracyclines in HER2-positive BCs [41]. In fact, our biomarker analysis in HER2-positive tumors indirectly suggests that different molecular subgroups of tumors, such as high and low TOP2A expression, may derive diverse benefits from anthracyclines or taxanes, respectively. Consequently, our data contribute to the current active debate about whether anthracycline should be abandoned as adjuvant treatment for HER2-positive disease [42, 43]. Because it seems that not all patients will benefit from the association of both drugs, our data also support the need for use of specific biomarkers to tailor selection of the appropriate regimen, which will account for greater advantages during adjuvant treatment.
The novel gene PMS2 is consistently associated with RD in ER-negative/HER2-negative tumors treated with anthracycline-based regimens. PMS2 is a major component of the DNA MMR system that heterodimerizes with MLH1 to form MutLα [44]. Sporadic BCs demonstrate microsatellite instability, reflecting the presence of MMR-deficient cells in approximately one-fourth of cases at the time of diagnosis [45]. MMR-deficient cells show resistance to doxorubicin, suggesting a role for MMR in topoisomerase II poison-mediated cell killing [45]. Although much of the current data have supported the involvement of MLH1 in topoisomerase II poison-mediated cytotoxicity, our results suggest that the MLH1 binding partner PMS2 warrants further investigation and should be considered as a potential biomarker for anthracycline resistance.
To date, several biomarkers have been evaluated for their potential value in the prediction of BC patient responses to taxanes such as p53, the microtubule-associated protein tau, and others, although different studies have yielded inconsistent results [46]. In our analysis, we found that higher expression of the MMR genes MSH2 and MSH6 is significantly associated with pCR in ER-negative/HER2-negative BC patients receiving neoadjuvant taxane/anthracycline-containing regimens. MSH2 and MSH6 could reside in a protein complex with BRCA1 and other DNA repair proteins as well as checkpoint signaling proteins (BASC) [45]. The members of this complex may regulate DNA repair and p53-mediated apoptosis following DNA damage. Although BRCA1 is a DNA damage response gene, BRCA1 also appears to regulate mitosis by interacting with the spindle microtubule protein γ-tubulin and thus playing a role in modulating the response to spindle poisons such as taxanes. Cells lacking a functional MMR system are unable to undergo G2/M arrest in response to several genotoxic agents, suggesting a link between MMR and G2/M arrest. The association between MMR proteins and BASC could contribute to the BRCA1-mediated modulation of apoptosis in response to spindle damage and could participate in BRCA1-dependent taxane sensitivity [45]. The expression of these biomarkers could help rationalize therapy decisions, although further prospective and independent evaluations are needed.
We hypothesized that DNA repair gene expression with bimodal distribution can indicate biologically relevant pathways that can be targeted. Within bimodal genes, only RECQL4 showed a significant association with clinical outcome. High RECQL4 expression is associated with poor prognosis and higher pCR after taxane/anthracycline regimens in ER-positive/HER2-negative tumors, and a similar trend was observed in ER-negative/HER2-negative tumors. Mutations in the RECQL4 gene are associated with the Rothmund-Thomson syndrome, a genetic disorder characterized by genomic instability, including trisomy, aneuploidy, and chromosomal rearrangements [47], suggesting a role for RECQL4 in preventing tumorigenesis and maintaining genome integrity in humans. RECQL4 has important biological functions in several DNA repair pathways and has been shown to form complexes with RAD51, which is a crucial factor for DSB repair through the HR pathway [47]. We confirmed that RECQL4 is involved in the HR pathway at DNA DSBs (Fig. 2D and 3A). ER-negative/HER2-negative tumors tend to be BRCA1 defective [1, 4]. Accordingly, loss of BRCA1 function leads to a tumor-specific dysfunction in DSB repair by HR, and agents that cause an increase in DSBs should selectively affect HR-deficient cells. We demonstrated that RECQL4 knockdown sensitizes BC cell lines to DNA damaging agents such as platinum and PARP inhibitors. These results demonstrate a novel mechanism of sensitivity to DNA damaging agents because of defects of RECQL4-mediated HR in BC cells, suggesting that RECQL4-deficient tumors may be hypersensitive to platinum-based or PARP inhibitor-based chemotherapy. PARP inhibitors target HR-based DNA repair defects similarly to cisplatin chemotherapy [48], and studies support the efficacy for the PARP inhibitor olaparib in BRCA1- and BRCA2-deficient BCs that have been previously treated with chemotherapy [49]. Despite their promise to make chemotherapy treatments more effective for patients with chemoresistant cancers, PARP inhibitors currently available through clinical trials are based on a single gene or biomarker that targets only those patients with BRCA1/2-mutated and basal-like TNBCs, which accounts for roughly 5% of all BC cases. RECQL4/HR-deficient tumors may benefit from PARP inhibitors that specifically target this DNA repair defect. Consequently, the new identified biomarker may broaden the scope of eligible patients.
We performed biomarker assessments based on molecular tumor subgroups and chemotherapy regimens to reduce the heterogeneity and increase the reliability of our results; however, the small sample size and small number of events in each tumor subset increased the risk of both false-negative and false-positive findings. Furthermore, multidrug regimens reduced the possibility of linking single markers to specific drug activity. Despite these limitations, our observations are consistent with the hypothesis that prognosis and chemotherapy response are at least partially associated with differential expression of some DNA repair-related genes. We confirmed the association of TOP2A gene expression with pCR to anthracyclines in HER2-positive tumors, but we also identified new genes associated with the likelihood of response to anthracycline- or taxane/anthracycline-based regimens. These prognostic and predictive markers warrant further investigation and independent validation in prospective studies. Because some DNA repair pathways can enable tumor cells to survive DNA damage induced by chemotherapy [50], inhibitors of specific DNA repair pathways could prove efficacious when used in combination with DNA-damaging chemotherapeutic drugs [50].
See www.TheOncologist.com for supplemental material available online.
Acknowledgments
L.S., T.I., and G.P.B. contributed equally to this work. This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC, Grant 6251 to L.S.), the Sandro Pitigliani and the Michelangelo Foundations (to L.S. and G.B.). We thank Dr. G. Pacini for assistance with data collection and for providing technical assistance. Presented at the 48th Annual Meeting of the American Society of Clinical Oncology, Chicago, Illinois, June 1–5, 2012.
Author Contributions
Conception/Design: Libero Santarpia, Giampaolo Bianchini
Provision of study material or patients: Libero Santarpia, Lajos Pusztai
Collection and/or assembly of data: Libero Santarpia, Angelo Di Leo, Naoki Hayashi, Giulia Bottai, Martha Stampfer, Fabrice André, Nicholas C. Turner, W. Fraser Symmans, Gabriel N. Hortobágyi, Lajos Pusztai, Giampaolo Bianchini
Data analysis and interpretation: Libero Santarpia, Takayuki Iwamoto, Giampaolo Bianchini
Manuscript writing: Libero Santarpia, Takayuki Iwamoto, Giampaolo Bianchini
Final approval of manuscript: Libero Santarpia, Takayuki Iwamoto, Angelo Di Leo, Naoki Hayashi, Giulia Bottai, Martha Stampfer, Fabrice André, Nicholas C. Turner, W. Fraser Symmans, Gabriel N. Hortobágyi, Lajos Pusztai, Giampaolo Bianchini
Disclosures
W. Fraser Symmans: Nuvera Biosciences, Inc. (IP); Nuvera Biosciences, Inc. (OI). Gabriel N. Hortobágyi: AstraZeneca, Allergan, Novartis, Genentech, SanofiAventis (C/A); Novartis (RF). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 2.Gatza ML, Lucas JE, Barry WT, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A. 2010;107:6994–6999. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santarpia L, Qi Y, Stemke-Hale K, et al. Mutation profiling identifies numerous rare drug targets and distinct mutation patterns in different clinical subtypes of breast cancers. Breast Cancer Res Treat. 2012;134:333–343. doi: 10.1007/s10549-012-2035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerji S, Cibulskis K, Rangel-Escareno C, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 7.Iwamoto T, Bianchini G, Booser D, et al. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J Natl Cancer Inst. 2011;103:264–272. doi: 10.1093/jnci/djq524. [DOI] [PubMed] [Google Scholar]
- 8.Ignatiadis M, Singhal SK, Desmedt C, et al. Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: A pooled analysis. J Clin Oncol. 2012;30:1996–2004. doi: 10.1200/JCO.2011.39.5624. [DOI] [PubMed] [Google Scholar]
- 9.Foster SS, De S, Johnson LK, et al. Cell cycle- and DNA repair pathway-specific effects of apoptosis on tumor suppression. Proc Natl Acad Sci U S A. 2012;109:9953–9958. doi: 10.1073/pnas.1120476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badura M, Braunstein S, Zavadil J, et al. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc Natl Acad Sci U S A. 2012;109:18767–18772. doi: 10.1073/pnas.1203853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer. 2012;12:587–598. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- 13.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 14.Rottenberg S, Jaspers JE, Kersbergen A, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bianchini G, Qi Y, Alvarez RH, et al. Molecular anatomy of breast cancer stroma and its prognostic value in estrogen receptor-positive and -negative cancers. J Clin Oncol. 2010;28:4316–4323. doi: 10.1200/JCO.2009.27.2419. [DOI] [PubMed] [Google Scholar]
- 17.Symmans WF, Hatzis C, Sotiriou C, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol. 2010;28:4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bianchini G, Iwamoto T, Qi Y, et al. Prognostic and therapeutic implications of distinct kinase expression patterns in different subtypes of breast cancer. Cancer Res. 2010;70:8852–8862. doi: 10.1158/0008-5472.CAN-10-1039. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Wen S, Symmans WF, et al. The bimodality index: A criterion for discovering and ranking bimodal signatures from cancer gene expression profiling data. Cancer Inform. 2009;7:199–216. doi: 10.4137/cin.s2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaz-Luis I, Winer EP, Lin NU. Human epidermal growth factor receptor-2-positive breast cancer: Does estrogen receptor status define two distinct subtypes? Ann Oncol. 2013;24:283–291. doi: 10.1093/annonc/mds286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Willers H, Feng Z, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pusztai L, Ayers M, Stec J, et al. Gene expression profiles obtained from fine-needle aspirations of breast cancer reliably identify routine prognostic markers and reveal large-scale molecular differences between estrogen-negative and estrogen-positive tumors. Clin Cancer Res. 2003;9:2406–2415. [PubMed] [Google Scholar]
- 24.Andre F, Job B, Dessen P, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15:441–451. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- 25.Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 26.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desmedt C, Ruíz-García E, André F. Gene expression predictors in breast cancer: Current status, limitations and perspectives. Eur J Cancer. 2008;44:2714–2720. doi: 10.1016/j.ejca.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Prat A, Cheang MC, Martín M, et al. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal a breast cancer. J Clin Oncol. 2013;31:203–209. doi: 10.1200/JCO.2012.43.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sparano JA, Goldstein LJ, Davidson NE, et al. TOP2A RNA expression and recurrence in estrogen receptor-positive breast cancer. Breast Cancer Res Treat. 2012;134:751–757. doi: 10.1007/s10549-012-2112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perreard L, Fan C, Quackenbush JF, et al. Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay. Breast Cancer Res. 2006;8:R23. doi: 10.1186/bcr1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naryzhny SN. Proliferating cell nuclear antigen: A proteomics view. Cell Mol Life Sci. 2008;65:3789–3808. doi: 10.1007/s00018-008-8305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yousefzadeh MJ, Wood RD. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair (Amst) 2013;12:1–9. doi: 10.1016/j.dnarep.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemée F, Bergoglio V, Fernandez-Vidal A, et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc Natl Acad Sci U S A. 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Xiao W, McCormick JJ, et al. Identification of a protein essential for a major pathway used by human cells to avoid UV- induced DNA damage. Proc Natl Acad Sci U S A. 2002;99:4459–4464. doi: 10.1073/pnas.062047799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye C, Cai Q, Dai Q, et al. Expression patterns of the ATM gene in mammary tissues and their associations with breast cancer survival. Cancer. 2007;109:1729–1735. doi: 10.1002/cncr.22592. [DOI] [PubMed] [Google Scholar]
- 38.Di Leo A, Desmedt C, Bartlett JM, et al. HER2/TOP2A Meta-analysis Study Group. HER2 and TOP2A as predictive markers for anthracycline-containing chemotherapy regimens as adjuvant treatment of breast cancer: A meta-analysis of individual patient data Lancet Oncol. 2011;12:1134–1142. doi: 10.1016/S1470-2045(11)70231-5. [DOI] [PubMed] [Google Scholar]
- 39.Arriola E, Marchio C, Tan DS, et al. Genomic analysis of the HER2/TOP2A amplicon in breast cancer and breast cancer cell lines. Lab Invest. 2008;88:491–503. doi: 10.1038/labinvest.2008.19. [DOI] [PubMed] [Google Scholar]
- 40.Press MF, Sauter G, Buyse M, et al. Alteration of topoisomerase II–alpha gene in human breast cancer: Association with responsiveness to anthracycline-based chemotherapy. J Clin Oncol. 2011;29:859–867. doi: 10.1200/JCO.2009.27.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayes DF, Thor AD, Dressler LG, et al. HER2 and response to paclitaxel in node-positive breast cancer. N Engl J Med. 2007;357:1496–1506. doi: 10.1056/NEJMoa071167. [DOI] [PubMed] [Google Scholar]
- 42.Slamon D, Eiermann W, Robert N, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365:1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burstein HJ, Piccart-Gebhart MJ, Perez EA, et al. Choosing the best trastuzumab-based adjuvant chemotherapy regimen: Should we abandon anthracyclines? J Clin Oncol. 2012;30:2179–2182. doi: 10.1200/JCO.2012.42.0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo Y, Lin FT, Lin WC. ATM-mediated stabilization of hMutL DNA mismatch repair proteins augments p53 activation during DNA damage. Mol Cell Biol. 2004;24:6430–6444. doi: 10.1128/MCB.24.14.6430-6444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kennedy RD, Quinn JE, Mullan PB, et al. The role of BRCA1 in the cellular response to chemotherapy. J Natl Cancer Inst. 2004;96:1659–1668. doi: 10.1093/jnci/djh312. [DOI] [PubMed] [Google Scholar]
- 46.von Minckwitz G, Martin M, Wilson G, et al. Optimizing taxane use in MBC in the emerging era of targeted chemotherapy. Crit Rev Oncol Hematol. 2013;85:315–331. doi: 10.1016/j.critrevonc.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 47.Petkovic M, Dietschy T, Freire R, et al. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- 48.Turner NC, Tutt AN. Platinum chemotherapy for BRCA1-related breast cancer: Do we need more evidence? Breast Cancer Res. 2012;14:115. doi: 10.1186/bcr3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tutt A, Robson M, Garber JE, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 50.Aziz K, Nowsheen S, Pantelias G, et al. Targeting DNA damage and repair: Embracing the pharmacological era for successful cancer therapy. Pharmacol Ther. 2012;133:334–350. doi: 10.1016/j.pharmthera.2011.11.010. [DOI] [PubMed] [Google Scholar]