Abstract
We studied the transmissibility of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) and healthcare-associated methicillin-resistant S. aureus (HA-MRSA) strains and the association of MRSA colonization pressure and MRSA transmission in critically ill children. Importantly, we found that in hospitalized children MRSA colonization pressure above 10% increases the risk of MRSA transmission 3-fold, and CA-MRSA and HA-MRSA strains have similar transmission dynamics.
Keywords: colonization pressure, methicillin-resistant Staphylococcus aureus, healthcare-associated infections, intensive care unit, transmission
Methicillin-resistant Staphylococcus aureus (MRSA) causes 8.5% of reported healthcare-associated infections in the United States [1]. MRSA strains USA300 and USA400, often referred to as community-associated MRSA (CA-MRSA) strains, initially were associated with community-onset infections. However, CA-MRSA strains encroached into healthcare settings and may be replacing the conventional healthcare-associated MRSA strains (HA-MRSA) [2]. It is not clear whether this trend is due to a greater prevalence of CA-MRSA strains and increased burden or to a greater transmissibility of CA-MRSA strains compared to HA-MRSA strains. Children are increasingly being hospitalized for community-onset MRSA infections [3]. Higher colonization prevalence is a known driver of nosocomial MRSA transmission in hospitalized adults, but this association has not been evaluated in hospitalized children. The objectives of this study were to determine the transmission dynamics of CA-MRSA and non–CA-MRSA strains and to evaluate the impact of colonization pressure on MRSA transmission in hospitalized children.
MATERIALS AND METHODS
Setting and Design
The Johns Hopkins Hospital is a tertiary healthcare center with an embedded 175-bed Children's Hospital. Since 1 March 2007, anterior nares swabs were obtained at the time of pediatric intensive care unit (PICU) admission and weekly thereafter and cultured for MRSA. Children admitted between 1 April 2007 and 10 June 2011 were eligible for inclusion. Children who had a surveillance or clinical culture grow MRSA contributed to overall prevalence.
Definitions
MRSA colonization at the time of PICU admission was defined as having an admission nares culture grow MRSA or any clinical culture grow MRSA within ≤3 days after PICU admission. PICU-acquired MRSA cases met the following criteria: (1) had surveillance or clinical cultures obtained >3 days after admission to the PICU grow MRSA, (2) had no previous history of MRSA, and (3) had a previous negative MRSA surveillance culture during the current admission. CA-MRSA strains included pulsed-field gel electrophoresis (PFGE) genotypes USA300 and USA400 [4]. HA-MRSA strains included all other PFGE USA genotypes and nontypeable strains. MRSA colonization pressure was defined as the proportion of total patient-days that were MRSA-positive patient-days [5].
Data Collection and Laboratory Methods
We searched a computerized surveillance support system (Theradoc, Hospira, Lake Forest, Illinois) to identify patients with surveillance cultures and cultures sent during clinical care that grew MRSA. Administrative databases and the electronic medical records were searched to obtain patient characteristics. As previously described, nasal surveillance swabs were plated on selective and differential media to detect MRSA (BBL CHROMagar MRSA plates, BD, Franklin Lakes, New Jersey, prior to 2008 and MRSASelect agar, Bio-Rad Laboratories, Hercules, California) [6]. We performed PFGE on available stored isolates including a control strain (S. aureus subspecies NCTC 8325) and USA PFGE strain types. We considered isolates to be related if their patterns had ≤3 band differences and to be unrelated if they had >3 band differences.
Statistical Analysis
Data were maintained in Microsoft Access 2007 (Microsoft Corporation, Redmond, Washington) and analyzed using Stata software, version 11.0 (StataCorp, College Station, Texas). Some MRSA isolates (mostly ones sent during clinical care) were not available for strain typing. To account for missing strain type data, we predicted strain type by using (1) strain typing data from a patient's prior or subsequent isolate, (2) antibiotic susceptibility of a MRSA clinical isolate [7], or (3) previous intensive care unit (ICU) admission in the preceding 1 year [6]. Comparisons of proportions were made using the Pearson χ2 test. For each day of the study, we calculated colonization pressure as a pooled average of the preceding 7 days [5]. We used log binomial regression models to examine the effect of colonization pressure both as a continuous variable and as quartile-derived categories on the risk of MRSA transmission. A 2-tailed P value of <.05 was considered significant for all regression models and statistical tests.
RESULTS
During the study period, there were 7365 admissions. Among 6388 patient visits in which MRSA was screened for on admission, 56% of the patients were male, the median age was 5.4 years, and 353 patients (5.5%) were either colonized with MRSA or had MRSA recovered from a clinical culture. We found no significant change in MRSA admission prevalence in our cohort during the study period (5.9%, 4.8%, 5.2%, and 4.2% in years 1–4, respectively). Of 353 patients who had a culture grow MRSA, 239 (3.2%) had isolates available for strain typing. Of these 239, 124 (51.9%) had CA-MRSA strains. After predicting the missing strain types, 167 of 353 patients (51.1%) had CA-MRSA strains.
Twenty-five patients acquired MRSA infection or colonization in the PICU, and 20 isolates were available for strain typing. To determine whether there was a difference in transmissibility between the strain types, we compared the proportions of CA-MRSA and HA-MRSA strains among patients with MRSA on admission versus patients who acquired MRSA in the unit during the entire study period. CA-MRSA strains accounted for 124 of 239 (51.9%) MRSA strains identified on admission and 7 of 20 (35.0%) unit-acquired strains (P = .15). After predicting missing strain types, 167 of 353 MRSA strains identified on admission and 13 of 25 unit-acquired strains were CA-MRSA (51.1% and 52.0%, respectively).
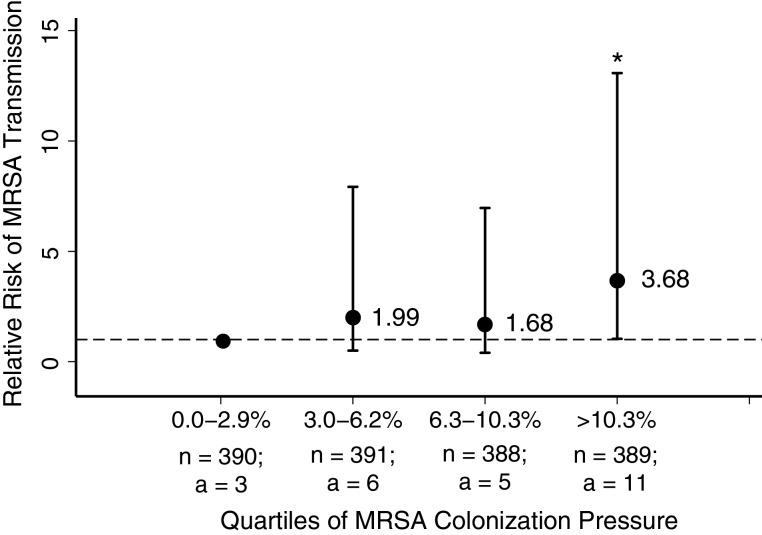
Median colonization pressure during the study period was 6.3% and ranged from 0.0% to 25.7%. We examined the effect of colonization pressure on the risk of MRSA transmission. Overall, risk of MRSA acquisition marginally increased with each percentage rise in colonization pressure (relative risk [RR], 1.08; 95% confidence interval [CI], 1.00–1.16). There was no significant difference in risk of MRSA acquisition when colonization pressure was between 3% and 6.2% (RR, 1.99; 95% CI, .5–7.92) or between 6.3% and 10.2% (RR, 1.68; 95% CI, .4–6.96), compared to when colonization pressure was <3.0% (Figure 1). When colonization pressure increased above 10.3%, the risk of MRSA transmission increased 3-fold (RR, 3.68; 95% CI, 1.03–13.08).
Figure 1.
Relative risk of methicillin-resistant Staphylococcus aureus (MRSA) transmission by increasing quartiles of colonization pressure. MRSA colonization pressure was defined as the average proportion of total patient-days that were MRSA-positive patient-days during the preceding 7 days and was categorized into quartiles. Relative risk was calculated from log binomial regression models specifying the first quartile of colonization pressure as the reference category (broken line, RR = 1.0). *P < .05; n = number of days in each quartile; a = number of MRSA acquisitions in each quartile.
DISCUSSION
We found similar proportions of CA-MRSA strains acquired by children in the PICU and colonizing children at time of admission, suggesting similar transmission dynamics of CA-MRSA and HA-MRSA strains. Similar to findings in critically ill adults [5], among critically ill children, as MRSA colonization pressure increases, so does the risk of MRSA transmission.
There has been speculation in the literature that CA-MRSA strains may be more transmissible than traditional HA-MRSA strains [8]. MRSA-colonized patients are a known reservoir for MRSA transmission in ICU settings. Therefore, if CA-MRSA strains have greater transmission capacity than HA-MRSA strains, then the proportion of unit-acquired CA-MRSA should exceed that of prevalent-on-admission CA-MRSA strains. Although our cohort had relatively few episodes of MRSA transmission, importantly, our data did not suggest that CA-MRSA strains were more transmissible than HA-MRSA strains, as evidenced by similar proportions of CA-MRSA strains acquired in the unit and harbored on admission to the unit.
To our knowledge, our study is the first to evaluate the impact of colonization pressure on MRSA transmission in hospitalized children. This study confirms reports that in adults, MRSA colonization pressure correlates with MRSA transmission [5, 9]. When the MRSA colonization pressure exceeded 10.3%, the risk of MRSA transmission was >3 times greater than when MRSA colonization pressure was <3%. One strength of this study was the inclusion of data from weekly surveillance and clinical cultures [10]. Williams et al reported a marked increase in transmission when colonization pressure in an adult general medicine unit exceeded the monthly median value of 6.7% [9]. Merrer and colleagues, in a study of adult ICU patients, found that risk of MRSA acquisition was nearly 6 times greater when weekly colonization pressure was >40% compared to when it was <10%. Interestingly, MRSA transmission only correlated with MRSA colonization pressure >31% [5]. Our lower threshold for increased transmission may be in part explained by differences in patient characteristics, baseline colonization pressure, and sample size.
Monitoring colonization pressure of MRSA in healthcare environments may provide a useful metric to assess risk of MRSA transmission and guide infection control measures. Our study estimates a critical point above which colonization pressure appears to have a clear impact on MRSA transmission in hospitalized children. Interventions, such as targeted or universal MRSA decolonization, can reduce the burden of MRSA and may reduce transmission and infections. Colonization pressure, which can be either estimated based on daily data or as a periodic assessment, may provide a marker for when units should consider special infection prevention measures.
There were several limitations to our study: the small sample size reduced our ability to determine whether strain-specific MRSA colonization pressure impacts strain-specific MRSA transmission; predicting missing strain types may have caused misclassification of MRSA isolates; the imperfect sensitivity of nares cultures may have led to misclassification of MRSA-colonized patients as acquired MRSA cases; and other strain typing methods such as spa typing and SCCmec typing may identify slightly different proportions of CA-MRSA strains.
This study highlights that colonization pressure is one of several risk factors for MRSA acquisition; larger studies should confirm whether different MRSA strains are more transmissible, as this has important infection control implications. Interventions that decrease colonization pressure could provide a safer environment for children by reducing MRSA transmission and subsequent infections.
Notes
Acknowledgments. We thank the JHH Microbiology laboratory staff, the JHH PICU nursing staff, and the JHH Department of Hospital Epidemiology and Infection Control for their support of this study.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (1 K23 AI081752-03 to A. M. M.).
Potential conflicts of interest. K. C. C. serves on the scientific advisory boards of NanoMR and Quidel and has received grant support from Curetis and Nanosphere. T. M. P. receives grant support from Sage Products, MedImmune, and Merck, was on advisory boards for Hospira and Pfizer, and has received travel expenses from bioMérieux. A. M. M. has received grant support from Sage Products and bioMérieux. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Sievert DM, Ricks P, Edwards JR, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 2.Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis. 2008;46:787–94. doi: 10.1086/528716. [DOI] [PubMed] [Google Scholar]
- 3.Gerber JS, Coffin SE, Smathers SA, Zaoutis TE. Trends in the incidence of methicillin-resistant Staphylococcus aureus infection in children's hospitals in the United States. Clin Infect Dis. 2009;49:65–71. doi: 10.1086/599348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–20. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merrer J, Santoli F, Appere de Vecchi C, Tran B, De Jonghe B, Outin H. Colonization pressure” and risk of acquisition of methicillin-resistant Staphylococcus aureus in a medical intensive care unit. Infect Control Hosp Epidemiol. 2000;21:718–23. doi: 10.1086/501721. [DOI] [PubMed] [Google Scholar]
- 6.Milstone AM, Carroll KC, Ross T, Shangraw KA, Perl TM. Community-associated methicillin-resistant Staphylococcus aureus strains in pediatric intensive care unit. Emerg Infect Dis. 2010;16:647–55. doi: 10.3201/eid1604.090107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milstone AM, Goldner BW, Ross T, Shepard JW, Carroll KC, Perl TM. Methicillin-resistant Staphylococcus aureus colonization and risk of subsequent infection in critically ill children: importance of preventing nosocomial methicillin-resistant Staphylococcus aureus transmission. Clin Infect Dis. 2011;53:853–9. doi: 10.1093/cid/cir547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bootsma MC, Bonten MJ. Unraveling the dynamics of community-associated methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2013;56:1075–7. doi: 10.1093/cid/cit013. [DOI] [PubMed] [Google Scholar]
- 9.Williams VR, Callery S, Vearncombe M, Simor AE. The role of colonization pressure in nosocomial transmission of methicillin-resistant Staphylococcus aureus. Am J Infect Control. 2009;37:106–10. doi: 10.1016/j.ajic.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Ajao AO, Harris AD, Roghmann M, et al. Systematic review of measurement and adjustment for colonization pressure in studies of methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, and Clostridium difficile acquisition. Infect Control Hosp Epidemiol. 2011;32:481–89. doi: 10.1086/659403. [DOI] [PMC free article] [PubMed] [Google Scholar]