Abstract
Trials of human immunodeficiency virus type 1 (HIV) pre- and postexposure prophylaxis show promise. Here, we describe a novel strategy for deciphering mechanisms of prophylaxis failure that could improve therapeutic outcomes. A healthcare worker began antiretroviral prophylaxis immediately after a high-risk needlestick injury but nonetheless became viremic 11 weeks later. Single-genome sequencing of plasma viral RNA identified 15 drug susceptible transmitted/founder HIV genomes responsible for productive infection. Sequences emanating from these genomes exhibited extremely low diversity, suggesting virus sequestration as opposed to low-level replication as the cause of breakthrough infection. Identification of transmitted/founder viruses allows for genome-wide assessment of molecular mechanisms of prophylaxis failure.
Keywords: HIV-1 transmission, occupational exposure, post-exposure prophylaxis, multiple virus transmission, single genome sequencing, virus sequestration
(See the editorial commentary by Kijak and Kim on pages 1542–4.)
Prevention of human immunodeficiency virus type 1 (HIV) infection is one of the most important and challenging goals of global health. Current efforts are broadly focused, and promising interventions under development include preexposure prophylaxis (PrEP) and postexposure prophylaxis (PEP), using antiretroviral drugs, topical microbicides, and vaccines. Recent clinical trials have demonstrated efficacies of 31%–67% for these prophylactic strategies in different risk groups [1]. There are no prospective randomized trials of PEP, but an earlier retrospective case-control study of occupational exposures suggested an 81% reduction in HIV acquisition [2]. Despite these encouraging findings, protection in all studies was incomplete. Methodologically, it has been challenging to pinpoint specific mechanisms of prophylaxis failure. Nonetheless, elucidation of mechanisms of prophylaxis failure could aid the development of more-effective regimens. Here, we describe a novel application of the single-genome sequencing (SGS) strategy to identify T/F viruses responsible for prophylaxis failure and implicate a novel mechanism of PEP treatment failure.
CASE REPORT
A 53-year-old Australian female healthcare worker (HCW) sustained a needlestick injury with a hollow-bore 21-gauge needle after drawing blood from a patient with advanced-stage HIV infection. She reported that the needle pierced the pulp of her gloved index finger and that she inadvertently struck the syringe plunger, such that a small amount of blood was likely injected. The HCW had no other risk factors for HIV acquisition. The source patient had been infected with HIV for >10 years, with prior treatment experience with 3 classes of antiretrovirals. At the time of the exposure, he was nonadherent to his prescribed regimen of zidovudine, lamivudine, and nevirapine. He had an HIV load of >100 000 copies/mL, a CD4+ T-cell count of 279 cells/mL, and a recent HIV genotype demonstrating a K103N mutation. The HCW initiated PEP with zidovudine, lamivudine, and indinavir within 2 hours of the exposure and completed 4 weeks of therapy (days 1–32) that included a 4-day interruption between days 22–25. At a routine visit 5 weeks after the needlestick exposure and several days after completing PEP, the HCW was asymptomatic and had a negative third-generation HIV antibody enzyme-linked immunosorbent assay (ELISA). Eleven weeks after the exposure (6 weeks after PEP discontinuation), the HCW presented with new-onset fever, arthralgias, and exudative pharyngitis. Laboratory examinations revealed a plasma HIV load of 2 200 000 copies/mL, an HIV–positive ELISA, and an indeterminate result of an HIV Western blot (faint <1+ gp160 band only). She received a diagnosis of acute HIV infection, Fiebig stage IV [3].
METHODS
Study Subjects
Blood samples from the index HCW and source patient were obtained after receiving informed consent under the Royal Perth Hospital Human Research Ethics Committee. Blood specimens were obtained from the HCW 81 days after exposure and from the source patient 8 days after the exposure.
Viral RNA and Genomic DNA Extraction, Complementary DNA (cDNA) Synthesis, Single-Genome Amplification, and Direct Sequencing
Viral RNA was extracted from plasma, DNA was extracted from peripheral blood mononuclear cells (PBMCs), cDNA was synthesized from env and pol viral RNA, and SGS performed as previously described [4–6]. Sequences from the index subject are identified throughout with the prefix 43 715, and those from the source subject are identified with the prefix 43 690.
Sequence Analysis
Sequences were aligned with ClustalW and were by hand using MacClade 4.08. Phylogenetic trees were generated by the neighbor-joining method, using ClustalW, or by maximum likelihood, using PhyML v.3. Sequences were assessed for APOBEC3G/F signatures and recombination (Supplementary Materials).
RESULTS
Phylogenetic Analyses
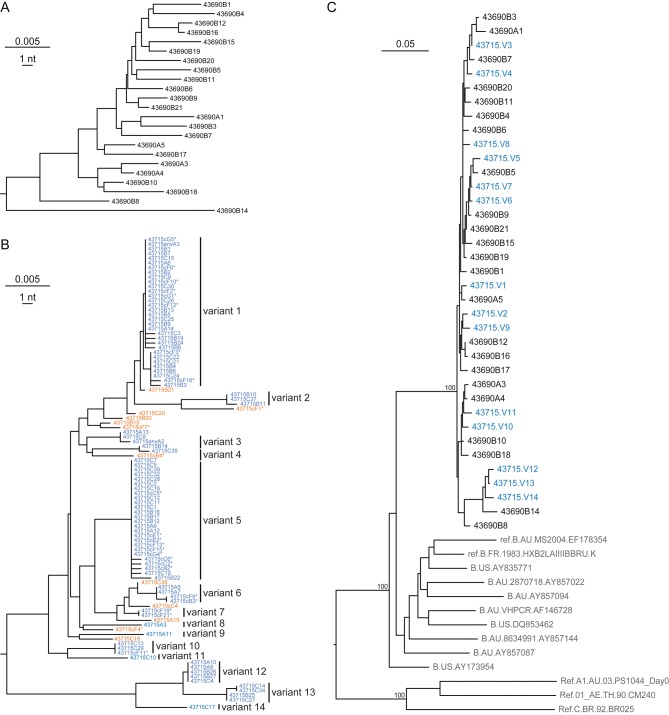
A total of 179 full-length pol and gp41 env sequences from the HCW's plasma and PBMCs and 61 full-length pol and gp41 env sequences from the source patient's plasma were determined. Figure 1A illustrates a neighbor-joining phylogeny of 23 gp41 env sequences from the source patient, revealing broad genetic diversity (maximum, 4.44%) and quasispecies complexity typical of chronic HIV infection [4, 7]. Figure 1B reveals a strikingly different pattern of env diversity in the recently infected HCW. Here, multiple sets of identical or nearly identical sequences are interspersed with sequences of recombinant origin. This phylogenetic pattern is characteristic of acute infection by multiple genetically diverse viruses [4–6], where each low diversity sequence lineage represents the progeny of a distinct T/F virus, and interspersed viruses generally represent discrete recombinants. Occasional T/F viruses represented by a single sequence can also be identified on the basis of their genetic distance from other sequences. The 2 largest lineages among the gp41 sequences had sufficient numbers of sequences to allow for robust modeling of sequence diversification [4, 8]. These sequences conformed to a model of neutral evolution, including a star-like phylogeny and a Poisson distribution of mutations (Supplementary Table 1). Sequences from all low diversity lineages coalesced to unambiguous T/F sequences. Thus, we can conclude that the env sequences from the index subject depicted in Figure 1B represent the progeny of at least 14 discrete T/F viruses.
Figure 1.
Neighbor-joining and maximum-likelihood phylogenies of human immunodeficiency virus type 1 (HIV) gp41 env sequences. A, Neighbor-joining phylogeny of single-genome sequencing (SGS)–derived gp41 env sequences from the chronically infected source patient's plasma viral RNA reveals unstructured wide genetic diversity typical of chronic HIV infection. B, Neighbor-joining phylogeny of SGS-derived sequences from the acutely infected index subject's plasma viral RNA and peripheral blood mononuclear cell (PBMC) DNA reveals multiple low diversity lineages typical of acute HIV infection (variants 1–14; blue), as well as interspersed interlineage recombinant sequences (orange). Sequences derived from PBMCs are indicated with asterisks. C, Maximum-likelihood phylogeny of sequences from the source patient (black) and the 14 T/F consensus genomes from the index subject (blue) interspersed within a monophyletic lineage separate from all reference sequences (gray), including subtype B sequences from Australia. Scale bars indicate nucleotide distance.
These 14 T/F genomes were next analyzed together with sequences from the source patient and unlinked reference subjects (Figure 1C). Source and index sequences formed a single monophyletic radiation supported by high bootstrap values (100%), indicating clear epidemiological linkage. Source and index sequences exhibited similar maximum diversities (4.44% and 4.49%, respectively) and were interspersed in the phylogenetic tree, again indicating epidemiological linkage and absence of obvious selection for particular variants in the transmission event.
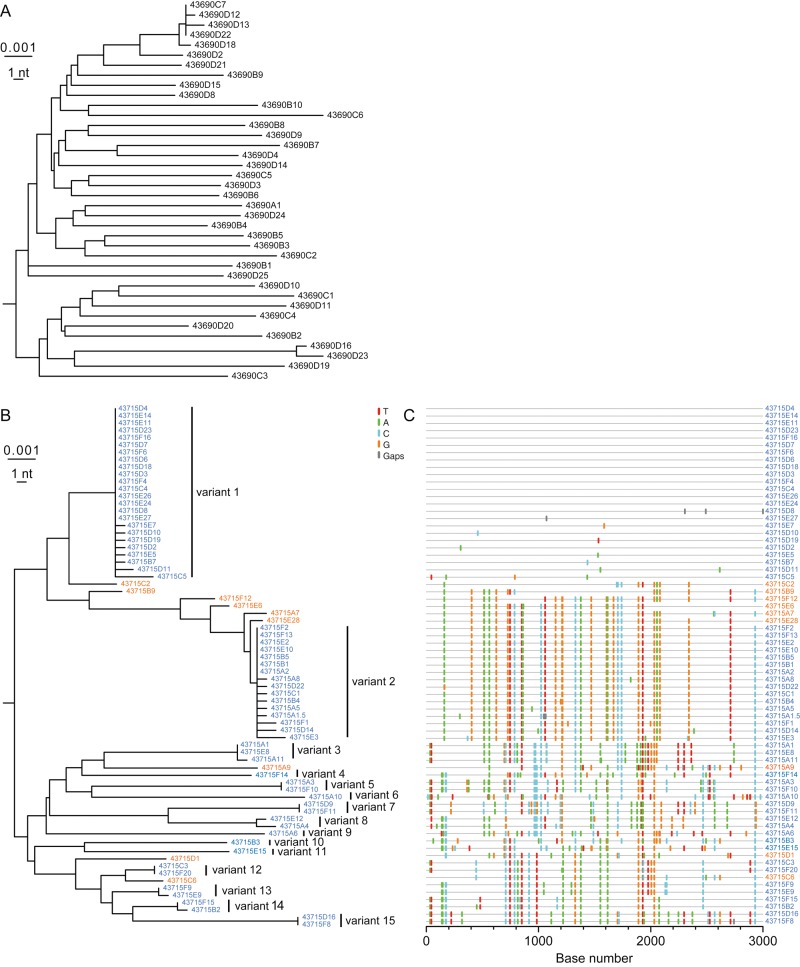
Figure 2 depicts the neighbor-joining phylogeny of the source patient's viral pol sequences and the phylogeny and Highlighter plot of the index subjects' viral pol sequences, which corroborate and extend findings from the env analyses. Again, the pattern of sequence diversity in the chronically infected source patient was strikingly different from that in the HCW, with sequences from the latter again consisting of multiple sets of identical or nearly identical sequences characteristic of T/F virus lineages. As with env, the pol sequences composing the most-populated lineages conformed to a model of random sequence evolution (Supplementary Table 1), with each lineage's sequences coalescing to unambiguous T/F sequences [4]. The enumerated T/F viruses represent minimum estimates, which are substantially affected by sampling depth (Supplementary Materials). Our estimates of at least 14-15 T/F viruses, based on env and pol analyses, are in good agreement.
Figure 2.
Neighbor-joining phylogenetic tree and Highlighter plot analysis of pol sequences. A, Neighbor-joining phylogeny of single-genome sequencing (SGS)–derived pol sequences from the chronically infected source patient's plasma viral RNA, displaying long branch lengths, with the exception of the single cluster of closely related sequences. Such sequence clusters of near identity have been reported as minor components of chronically infected patients' plasma virus quasispecies and reflect recent clonal expansion of a virus within a diverse chronic virus swarm [7]. B, Neighbor-joining phylogeny of SGS-derived pol sequences from the acutely infected index subject's plasma viral RNA reveals multiple low diversity lineages (variants 1–15; blue), as well as interspersed interlineage recombinant sequences (orange). C, A Highlighter plot of index case pol sequences represents as colored tics the nucleotide polymorphisms between the top sequence (43715D4) and the remaining pol sequences aligned below. The plot illustrates the homology of each of the T/F lineages (blue), the heterogeneity between T/F lineages, and the chimeric structure of the recombinant sequences (orange).
Mathematical Modeling of T/F Sequence Evolution
Sequence lineages with sufficient numbers of sequences for analysis conformed to a model of random HIV evolution [4], and we could thus use well-established parameters for HIV reverse transcriptase error rate and virus generation time to estimate the time to a most recent common ancestor (MRCA) [8]. Analysis of the most-populated T/F lineages in both env and pol revealed far shorter times to an estimated MRCA than the 81 days documented clinically. For example, for gp41 env lineages v1 and v5 (Figure 1B), the estimated times to a MRCA were 37 and 18 days, respectively; for pol lineages v1 and v2 (Figure 2B), MRCA estimates were 14 and 23 days, respectively (Supplementary Table 1). These low MRCA estimates suggest that each sequence lineage evolved from a discrete T/F virus that began to replicate only after PEP was discontinued. This finding implies a substantial period of virus sequestration and evolutionary arrest during PEP.
Drug Resistance Does Not Contribute to Virus Breakthrough
To determine whether transmission of drug-resistant viruses or selection for drug resistance could have contributed to PEP failure, we analyzed all sequences from the index and source patients for genotypic evidence of antiretroviral resistance. In the reverse transcriptase gene, a K103N mutation was detected in all source and index patient sequences (Supplementary Figure 1A). This mutation could be explained by the source patient's prior therapy but is irrelevant to the PEP regimen administered to the HCW. In protease, the HCW sequences contained a single M46I mutation in only 1 of 15 T/F lineages (2 of 72 sequences total; see variant 15 in Figure 2). Importantly, M46I confers resistance to protease inhibitors only in combination with other resistance-associated mutations [9]. Thus, drug resistance mutations relevant to the prescribed PEP regimen were neither transmitted nor evolved during or after the administration of PEP.
DISCUSSION
Understanding molecular mechanisms of HIV prophylaxis failure has important implications for public health and for the personal health of individuals exposed to HIV. Here, we apply a novel experimental strategy for characterizing and discriminating between potential mechanisms of PEP failure. Despite timely initiation of a potent 3-drug PEP regimen, the HCW demonstrated delayed viremia and seroconversion resulting from transmission of at least 15 T/F viruses. This is the first example of occupational HIV exposure and PEP failure in which the molecular identities and minimum numbers of actual T/F viral genomes responsible for infection have been unambiguously determined.
The surprising findings of 15 drug-sensitive T/F viruses, together with minimal diversification of their progeny and the absence of relevant transmitted or selected drug resistance-associated mutations, suggest virus sequestration and associated evolutionary arrest underlying virus breakthrough in this subject. Several mechanisms could potentially explain the sequestration and evolutionary stasis of transmitted viral genomes throughout PEP. These can be divided into (1) preintegration latency, (2) postintegration latency, (3) incomplete suppression of virus replication, and (4) virus trapping. HIV virion–associated RNA can enter cells and be arrested before proviral integration (preintegration latency); however, the lifespan of preintegration complexes is generally believed to be only a few days, far less than the 4-week period of PEP in this subject. Similarly, postintegration latency would require the unlikely occurrence of infection of many thousands, if not millions, of CD4+ T-cells in the first hours after virus exposure, to generate the 15 or more latently-infected memory CD4+ T-cells required to explain our findings. A third possibility is low-level viral replication via cell-to-cell spread, which has been postulated to be less sensitive to drug suppression than cell-free virus spread [10]. This mechanism also seems unlikely given the evolutionary stasis and absence of accumulated resistance mutations in our 15 T/F virus lineages. A fourth possibility, which we favor, is virus trapping and sequestration by follicular dendritic cells (DCs) or other antigen presenting cells. The initial WB immunoreactivity to gp160 (only) is consistent with this hypothesis. Follicular DCs are known to bind and retain infectious HIV in the form of immune complexes for at least 9 months in murine models following footpad inoculation. Such virus can prime B cells for antibody production and initiate productive virus infection upon coculture of follicular DCs and permissive human CD4+ T cells [11].
The literature contains many reports of combination PEP failure in occupational HCW exposures in resource-rich countries. Notably, the majority of these HCWs also experienced significant delays in symptoms of acute retroviral syndrome (mean, 46 days; range, 23–79 days) and antibody seroconversion (mean, 67 days; range, 23–97 days) [12]. Because clinical symptoms and systemic viremia typically develop within 10 days (95% confidence interval, 7–21 days) after virus exposure [3], this protracted time course suggests a scenario similar to our case, in which exponential virus replication began after completion of PEP. Similar delays in viremia and antibody seroconversion were observed in macaque studies in which tenofovir-based PEP failed to protect against intravenous inoculation [13]. Thus, delayed viremia, clinical symptoms, and antibody seroconversion may be common during HIV infection arising after failed PEP.
The remarkably high numbers of T/F genomes seen in this case have not been observed in sexual transmission of HIV [4] nor in the majority of transmissions in injection drug users [6]. The high numbers of T/F viruses in the index case, however, are indistinguishable from what has been observed in a minority of acutely infected injection drug users and in rhesus macaques inoculated intravenously with simian immunodeficiency virus [6, 14]. Thus, while surveys of occupational HIV exposures estimate a low risk of virus acquisition following percutaneous exposure (0.3%) [15], with currently available PEP regimens further diminishing this risk [2], there may be atypical exposures, like the one involving the HCW described here, in which the risk of virus acquisition is particularly high. In such cases, more-prolonged therapy with potent combination antiretroviral therapy may be warranted.
In summary, we have demonstrated the utility of SGS and T/F virus analyses to distinguish potential molecular mechanisms of PEP failure. Future clinical trials of PEP and PrEP by may benefit from a SGS approach to the elucidation of mechanisms of virus breakthrough and treatment failure.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgment. We thank Patricia Crystal for manuscript preparation.
Financial support. This work was supported by the National Institutes of Health Center for HIV/AIDS Vaccine Immunology (grant U19AI067854), the Center for HIV/AIDS Vaccine Immunology and Immunogen Development (grant UM1AI100645), and the Bill and Melinda Gates Foundation (grant 37874).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Padian NS, McCoy SI, Karim SS, et al. HIV prevention transformed: the new prevention research agenda. Lancet. 2011;378:269–78. doi: 10.1016/S0140-6736(11)60877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardo DM, Culver DH, Ciesielski CA, et al. A case-control study of HIV seroconversion in health care workers after percutaneous exposure. Centers for Disease Control and Prevention Needlestick Surveillance Group. N Engl J Med. 1997;337:1485–90. doi: 10.1056/NEJM199711203372101. [DOI] [PubMed] [Google Scholar]
- 3.Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–9. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 4.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, Bar KJ, Wang S, et al. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 2010;6:e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bar KJ, Li H, Chamberland A, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol. 2010;84:6241–7. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parrish NF, Wilen CB, Banks LB, et al. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. PLoS Path. 2012;8:e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giorgi EE, Funkhouser B, Athreya G, Perelson AS, Korber BT, Bhattacharya T. Estimating time since infection in early homogeneous HIV-1 samples using a Poisson model. BMC Bioinformatics. 2010;11:532. doi: 10.1186/1471-2105-11-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson VA, Calvez V, Gunthard HF, et al. 2011 update of the drug resistance mutations in HIV-1. Top Antivir Med. 2011;19:156–64. [PMC free article] [PubMed] [Google Scholar]
- 10.Sigal A, Kim JT, Balazs AB, et al. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature. 2011;477:95–8. doi: 10.1038/nature10347. [DOI] [PubMed] [Google Scholar]
- 11.Smith BA, Gartner S, Liu Y, et al. Persistence of infectious HIV on follicular dendritic cells. J Immunol. 2001;166:690–6. doi: 10.4049/jimmunol.166.1.690. [DOI] [PubMed] [Google Scholar]
- 12.Health Protection Agency Centre for Infections and Collaborators. 2005. Occupational transmission of HIV: summary of published reports. London: Health Protection Agency Centre for Infections and Collaborators. http://www.hpa.org.uk . [Google Scholar]
- 13.Tsai CC, Emau P, Follis KE, et al. Effectiveness of postinoculation (R)-9-(2-phosphonylmethoxypropyl) adenine treatment for prevention of persistent simian immunodeficiency virus SIVmne infection depends critically on timing of initiation and duration of treatment. J Virol. 1998;72:4265–73. doi: 10.1128/jvi.72.5.4265-4273.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keele BF, Li H, Learn GH, et al. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J Exp Med. 2009;206:1117–34. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell DM. Occupational risk of human immunodeficiency virus infection in healthcare workers: an overview. Am J Med. 1997;102:9–15. doi: 10.1016/s0002-9343(97)89441-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.