Abstract
ER-Golgi membrane transport and autophagy are intersecting trafficking pathways that are tightly regulated and crucial for homeostasis, development and diseases. Here, we identify UVRAG, a Beclin1-binding autophagic factor, as a PI(3)P-binding protein that depends on PI(3)P for its ER localization. We further show that UVRAG interacts with RINT-1, and acts as an integral component of the RINT-1-containing ER tethering complex, which couples phosphoinositide metabolism to COPI-vesicle tethering. Displacement or knockdown of UVRAG profoundly disrupted COPI cargo transfer to the ER and Golgi integrity. Intriguingly, autophagy caused the dissociation of UVRAG from the ER tether, which in turn worked in concert with the Bif-1-Beclin-PI(3)KC3 complex to mobilize Atg9 translocation for autophagosome formation. These findings identify a regulatory mechanism that coordinates Golgi-ER retrograde and autophagy-related vesicular trafficking events through physical and functional interactions between UVRAG, phosphoinositide, and their regulatory factors, thereby ensuring spatiotemporal fidelity of membrane trafficking and maintenance of organelle homeostasis.
Keywords: UVRAG, RINT-1, phospholipid, Golgi-to-ER traffic, COPI, Atg9, Autophagy
Membrane trafficking is essential for organelle homeostasis and basal cellular functions. Given numerous intersecting trafficking pathways in cells, they must be coordinated in time and space for targeted cargo delivery. The organelles and vesicles are coded with distinct phosphoinositides (PIs), which serve as signaling and/or binding platforms for effectors to direct the membrane flow1,2. Phosphatidylinositol 3-phosphate (PI(3)P) is an essential regulator of endocytic, phagocytic, and autophagic trafficking1,3. However, a recent study suggested that PI(3)P is also required for ER-related functions4. Using a HPLC method, another report further demonstrated that the ER contains more PI(3)P than previously expected5. In support of this view, both the class III PI(3)-kinase (PI(3)KC3) and the myotubularin-related protein 3 (MTMR3), a PI(3)-phosphatase, have been localized to the ER, where they presumably fine-tune PI(3)P synthesis and turnover in response to various stresses6–8. A few ER-associated proteins that bind PI(3)P have been recently identified, including DFCP-17, FYCO19, and WIPI-110, but they are mostly involved in autophagy-induced PI(3)P function. Little is known about the physiological role of steady-state PI(3)P in ER-related trafficking, and the effectors of PI(3)P important for ER function remain uncertain.
One important type of effectors for phosphoinositides is the tethering factors, which ensure fidelity in vesicle transport through their interaction with cargo carriers and regulation of the SNARE-mediated membrane fusion11,12. The ER tethering events requires the Dsl1p complex in yeast13–15. The Dsl1p complex, which includes Dsl1p, Tip20p, and Sec39p, mediates traffic of COPI-vesicles from the Golgi to the ER by capturing COPI-coated vesicles and facilitating SNARE-mediated membrane fusion15,16. The mammalian equivalents of Dsl1p, Tip20p, and Sec39p are ZW10, Rad50-interactor 1 (RINT-1), and neuroblastoma amplified gene (NAG), respectively15,17–19. Inhibition of the ER-tether precludes Golgi-ER transport and disrupts Golgi structure in mammalian as in yeast15,17–19. However, the regulation of the RINT-1-ZW10-NAG complex remains unclear. It is also not known whether any phosphoinositide-restricted membrane is involved to ensure spatiotemporal regulation of retrograde traffic, as has been observed in other traffic events1,2.
Autophagy, a tightly regulated membrane remodeling process for lysosome-dependent degradation of cytosolic contents, is distantly related to the Golgi and ER dynamics20. Functional disruption of the Golgi by depletion of COPI-proteins leads to the accumulation of LC3-marked vesicles and inhibition of autophagic degradation21. Moreover, blocking the exit from the Golgi impairs the membrane expansion of the autophagosome22. In line with this, vesicles containing Atg9, a membrane supplier for autophagosomes, are derived from the Golgi-like apparatus23–26. All these correlative events have led to the concept that autophagy is interconnected with the Golgi pathway.
UVRAG is a Beclin1-interacting protein through its coiled-coil domain, which activates PI(3)KC3 during autophagy27–32. Moreover, UVRAG has a unique activity in the endocytic pathway, where it promotes endocytic trafficking via its interaction with the class C vacuolar protein sorting (C-Vps) complex29,33. Our recent work further demonstrated that UVRAG preserves genomic stability by associating with the centrosome and DNA repair machinery in a manner independent of autophagy34. The differential localization and incorporation into a variety of functionally distinct complexes suggest UVRAG a multitasking protein than previously appreciated. In continuing effort to characterize the functions of UVRAG, we report here that UVRAG binds PI(3)P through its C2 domain, and subsequently be localized to the ER, where it interacts with the RINT-1-containing ER-tether to control Golgi-to-ER COPI-transport and the integrity of the Golgi-complex. Furthermore, UVRAG dissociates from the tether during autophagy, and works in concert with the Beclin1-containing complex and PI(3)P to mobilize Atg9 trafficking required for autophagy. Thus, our study revealed an unexpected role for PI(3)P at the ER, also extends the function of UVRAG from an autophagy promoter to an important coordinator of phosphoinositide regulation and of the two trafficking events in cells.
RESULTS
UVRAG is a phosphoinositide-binding protein
UVRAG contains an amino-terminal lipid-binding C2 domain (Fig. 1a)31, yet, its lipid-binding activity has not been determined. Using an Escherichia coli-purified UVRAG GST-fusion protein (Supplementary Fig. S1a), we tested its ability to bind phosphoinositides using a protein-lipid overlay assay. The UVRAG-C2 domain bound strongly to PI(3)P and PI(4)P, and more weakly to PI(5)P, but little to other phosphoinositides (Fig. 1b–c). No binding of GST to lipids was detected (Fig. 1b). Under similar conditions, untagged full-length UVRAG also preferentially bound PI(3)P and PI(4)P, while no lipid-binding was observed for control IgG (Fig. 1d).
Figure 1.
UVRAG interacts with phosphoinositides.
(a) Schematic representation of full-length UVRAG with the boundary of the C2 domain indicated above the diagram. C2, the phosphoinositide-binding domain; CC, the coiled-coil domain.
(b) The affinity of the UVRAG-C2 domain for phospholipids was assessed using a protein-lipid overlay assay. The left panel indicates the identity of lipid species on PIP strips. Bacterial purified GST-fusion of UVRAG-C2 (WT), but not GST, binds PI(3)P, PI(4)P, and PI(5)P. The K78A/R82A mutant of C2 is defective for phospholipid-binding, whereas the K87A/N88A mutant is partially impaired. Binding of the FYVE and PH domains of Hrs and FAPP1, respectively, to PI(3)P and PI(4)P served as quality controls (right).
(c) The relative affinity of the GST-fusion proteins for phosphoinositides was quantified by densitometric analysis of the GST immunoblots in (b) from five independent experiments (n = 5). Error bars represent standard deviation (SD). A.U., artificial unit.
(d) Binding of full-length UVRAG (untagged) to lipid blots. The same PIP strips (b) were used and immunoblotted with antibodies to IgG (right) and UVRAG (left).
(e) Multiple sequence alignment of the C2B domain from synaptotagmin-1 (Syt-1), and the C2 domains from phospholipase C beta (PLC-β) and protein kinase C-alpha (PKC-α), and UVRAG, using the ClustalW alignment program. Identical residues are highlighted in red, conserved and similar residues are highlighted in yellow and green, respectively. The two functionally critical residues, K78 and R82, are indicated by an asterisk. The secondary structure of the UVRAG-C2 domain is depicted below the alignment. The sequences of PLC-β, Syt-1, and PKC-α were obtained from Swiss-Prot with accession numbers Q00722, P21707, and P05696, respectively.
(f) Homology model of UVRAG-C2 with conserved PI(3)P-interacting residues highlighted with spheres. The crystal structures of PKC-α, PLC-β, and Syt-1 C2B were used as templates to develop a homology model of the UVRAG-C2 domain. Binding orientation of PI(3)P with the UVRAG-C2 domain was developed by molecular docking method and PI(3)P is highlighted with yellow sticks. Close view of the interaction pattern demonstrates that PI(3)P forms hydrogen-bond interactions with the K78 and R82 residue side chains. These two residues are depicted by sticks with dots and hydrogen bonds illustrated with dotted lines.
(g) Pull-down of UVRAG-C2 domain GST-fusion proteins by liposomes bearing the indicated phosphoinositides. GST-Hrs- FYVE and GST-FAPP1-PH are also shown.
To further understand its lipid-binding, the UVRAG-C2 domain was used to search for homologs by DELTA-BLAST. The C2 domains of PKC-α, PLC-β and synaptogamin-1 (C2B) were retrieved as best hits. Their sequence alignment revealed several conserved amino acids that may be required for UVRAG phospholipids binding (Fig. 1e). Using the crystal structures of PKC-α, PLC-β and synaptogamin-1, we built a homology model for UVRAG-C2, which comprises four antiparallel β-sheets connected by loops. The two PI(3)P-interacting conserved residues (K78 and R82) in the β2 strand are surface-exposed (Fig. 1f). Previous studies35 suggested that K211 of PKC-α that corresponds to the R82-residue of UVRAG-C2, interacts with phosphoinositol-4,5-bisphosphate. Similarly, synaptogamin-1-C2B K327 interacts with inositol hexakisphosphate36,37. Analyses of the UVRAG-C2 binding orientations revealed that one of the PO4 group of PI(3)P forms an hydrogen-bond interaction with K78 and the second PO4 interacts with the R82 side chains (Fig. 1f). Indeed, a double alanine (A) substitution (K78A/R82A) abrogated phospholipid-binding of UVRAG-C2 (Fig. 1b–c). Mutations of other conserved residues (K87 and N88) to alanine impaired, but did not completely eliminate phosphoinositide-binding (Fig. 1b). Consistently, GST-C2, but not GST, co-sedimented with liposomes containing PI(3)P or PI(4)P, but not with liposomes containing PC, PE, or PI (Fig. 1g). Moreover, UVRAG-C2 bound tightly to PI(3)P-containing liposomes at low concentrations (Supplementary Fig. S1b). Again, the K78A/R82A mutation abolished the liposome binding of UVRAG to PI(3)P and PI(4)P (Fig. 1g). Thus, UVRAG binds selectively to phosphoinositides through its C2 domain.
PI(3)P-binding is required for UVRAG localization to the ER
PI(3)P was thought to be located in endosomes38, but recently was also found in abundance in the ER4,5, while PI(4)P is enriched at the trans-Golgi network (TGN)39,40. As UVRAG binds preferentially to PI(3)P and PI(4)P, we re-examined its subcellular distribution by confocal microscopy using markers of the early secretory pathway. UVRAG displayed a reticular distribution with a variable number of vesicular structures in the cytoplasm (Fig. 2). Endogenous UVRAG staining extensively overlapped with that of the endogenous ER proteins calnexin and PDI, and the overexpressed DsRed-ER marker (Fig. 2a–c), but to a lesser extent with the cis-, medial-, or trans-Golgi markers, GM130 and p115, overexpressed GFP-mannosidase II (ManII), and endogenous TGN46, respectively (Fig. 2d–g), and surprisingly less with EGFP-FAPP1-PH39, a reporter of Golgi-PI(4)P (Fig. 2h). A partial overlap was also detected between UVRAG and coatomer proteins Sec31 and β'-COP (Fig. 2i–j). Consistent with our previous study29, UVRAG partially colocalized with the endosomal marker GFP-p40phox(PX)41 (Fig. 2k). Although UVRAG binds PI(4)P in vitro, only limited amount was found in the Golgi (Fig. 2l). One possible explanation is that PI(4)P in the Golgi is bound to unknown proteins, making it inaccessible to UVRAG; alternatively, UVRAG association with PI(4)P might need to be triggered by specific signals. Nevertheless, these results indicate that UVRAG is present in the ER, but far less in the Golgi.
Figure 2.
UVRAG localizes at the ER.
(a–k) Confocal analyses of the subcellular colocalization of endogenous UVRAG with ER markers, including endogenous Calnexin (red, a), endogenous PDI (red, b), and overexpressed DSred-ER (red, c); with Golgi markers, including endogenous GM130 and p115 (red, d–e, cis-Golgi), overexpressed GFP-Man II (green, f, medial-Golgi), endogenous TGN46 (red, g, trans-Golgi), and overexpressed PI(4)P probe, GFP-FAPP1-PH (green, h); with endogenous coatomer proteins, including Sec31 (red, i, COPII-related) and β'-COP (red, j, COPI-related); and with the endosome marker GFP-p40phox(PX) (green, k) in HeLa cells. Insets show high magnification of the selected areas. Scale bars, 10 μm.
(l) Confocal microscopic quantification of colocalization of UVRAG with indicated markers (data are mean ± SD, n = 150 cells obtained by gathering data of three independent experiments); **, p < 0.01; ***, p < 0.001.
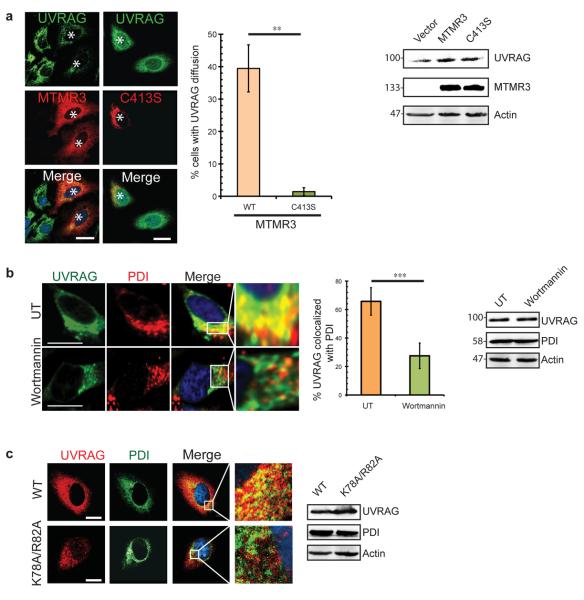
If UVRAG binds PI(3)P, decrease in PI(3)P should result in dissociation of UVRAG from the ER. Expression of MTMR3, an ER-related phosphatase that dephosphorylates PI(3)P without affecting other PI pools6,8, led to a drastic reduction of the reticular distribution of UVRAG (Fig. 3a). This was not observed in non-transfected cells or cells expressing the inactivated MTMR3-C413S mutant (Fig. 3a). As a control, the localization of the PI(4)P reporter GFP-FAPP1-PH was unaffected by MTMR3, and MTMR3 did not perturb the distribution of GFP-p40phox(PX) that localizes to PI(3)P-enriched endosomes (Supplementary Fig. S1c). Similarly, when cells were treated with wortmannin to block PI(3)P synthesis, we observed a partial release of UVRAG from the ER, although the ER remained intact and ER markers remained at the ER (Fig. 3b). In contrast, the relative distribution of UVRAG to the Golgi-associated PI(4)P was unaffected by wortmannin (Supplementary Fig. S1d). As expected, the K78A/R82A mutant defective for PI(3)P-binding almost completely lost its co-distribution with ER markers (Fig. 3c). Hence, UVRAG is a PI(3)P effector and localizes to the ER by binding PI(3)P in vivo.
Figure 3.
UVRAG association with the ER is PI(3)P-dependent.
(a) UVRAG is released from the ER membrane in cells overexpressing MTMR3. HeLa cells transfected with MTMR3-mStrawberry or the catalytically inactive mutant MTMR3-C413S were stained with anti-UVRAG antibody and processed for confocal microscopy. Asterisks indicate MTMR3-transfected cells (left). Scale bars, 10 μm. The percentage of transfected cells with diffused staining of UVRAG was quantified (middle). Data are mean ± SD; n = 240 cells collected from four independent experiments; ** p < 0.01. Western blot analysis shows the levels of endogenous UVRAG in the presence or absence of MTMRS (right). See Supplementary Fig. S9 for uncropped data.
(b) Wortmannin treatment releases UVRAG from the ER. HeLa cells were treated with 100 nM wortmannin for 1 hr, and processed for confocal microscopy using anti-UVRAG and anti-PDI antibodies. Insets highlight the relative localization of UVRAG at the ER. Scale bars, 10 μm. Confocal co-localization is indicated (middle panel). Data are mean ± SD; n = 75 cells for each group from three independent experiments; *** p < 0.001. Endogenous UVRAG and PDI expression before and after wortmannin treatment were confirmed by immunoblotting (right panel). See Supplementary Fig. S9 for uncropped data.
(c) Mutation of the C2 domain leaves UVRAG unable to associate with the ER. HeLa cells expressing Flag-UVRAG (wt) or its K78A/R82A mutant were fixed and processed for confocal microscopy for colocalization with the ER marker PDI. Insets highlight the relative distribution of UVRAG at the ER. UVRAG and endogenous PDI expression were confirmed by immunoblotting. See Supplementary Fig. S9 for uncropped data. Scale bars, 10 μm.
UVRAG interacts with RINT-1 and forms a complex with the RINT-1-ZW10-NAG ER tether
To further dissect the events downstream of PI(3)P-mediated ER association of UVRAG, we conducted a yeast two-hybrid screen to identify the ER-related interactors other than PI(3)P and Beclin1, using the C-terminal part (residues 270–699) as bait. RINT-1 was identified as a prominent candidate in our screening. This interaction was confirmed by co-immunoprecipitation (co-IP) in cells transiently expressing Flag-UVRAG and HA-RINT-1 (Fig. 4a). Moreover, endogenous RINT-1 and UVRAG co-immunoprecipitated (Fig. 4c). Using several UVRAG deletion mutants, we identified residues 270–442 of UVRAG as its major site for RINT-1 interaction, which is independent from its binding to Beclin1 or PI(3)P as aforenoted (Fig. 4a–b). Significant colocalization of endogenous UVRAG and RINT-1 was observed as a reticular pattern reminiscent of ER staining (Supplementary Fig. S2f).
Figure 4.
UVRAG interacts with the ER tethering complex.
(a) UVRAG C-terminal 270-442 region interacts with RINT-1. Cells (293T) were co-transfected with HA-RINT-1, together with Flag-UVRAG or its mutant derivatives, and whole cell lysates (WCLs) were immunoprecipitated (IP) with anti-Flag, followed by immunoblotting (IB) with an anti-HA antibody.
(b) Schematic representation of UVRAG (WT) and its deletion mutants, and summary of their interactions with RINT-1. Interaction was determined by coimmunoprecipitation of Flag-UVRAG with HA-RINT-1 from 293T cell lysates. +, strong binding; −, no binding.
(c) Interaction between endogenous UVRAG and RINT-1. WCLs of 293T cells were used for IP with control serum (control) or an anti-UVRAG (left) or anti-RINT-1 antibody (right), followed by immunoblotting with the indicated antibodies. The bottom panel shows endogenous protein expression.
(d) ER localization of RINT-1, ZW10 and UVRAG. HeLa cells were stained with anti-UVRAG (green), anti-PDI (ER marker, red), and anti-ZW10 or anti-RINT-1 (blue), followed by confocal microscopy. Insets highlight UVRAG co-localization with RINT-1 and ZW10 at the ER. Scale bars, 10 μm.
(e) Interaction between endogenous UVRAG and the RINT-1-ZW10-NAG tethering complex proteins. WCLs of 293T cells were used for IP with control serum (IgG) or an anti-UVRAG (1st panel), anti-RINT-1 (2nd panel), anti-ZW10 (3rd panel), and anti-NAG (4th panel), followed by immunoblotting with the indicated antibodies. 10% of the whole-cell lysates were used as input. Syn18, syntaxin 18; Syn6, syntaxin 6. See Supplementary Fig. S9 for uncropped data.
Previous work identified the N-terminal region (residues 1–256) of RINT-1 as the site for binding ZW1018. To examine whether RINT-1 interaction with UVRAG involves a separate region, we mapped the domain of RINT-1 interacting with UVRAG. The coiled-coil domain of RINT-1, which encompasses the ZW10-binding pocket, mediated UVRAG interaction (Supplementary Fig. S2a–c). We asked whether ZW10 and UVRAG compete for the same binding site on RINT-1. Increasing expression of either ZW10 or UVRAG did not affect their interaction with RINT-1 (Supplementary Fig. S2d–e). Indeed, endogenous UVRAG, RINT-1, and ZW10 readily co-immunoprecipitated, and all interacted with NAG (Fig. 4e). Of note, little, if any, Beclin1 co-precipitated with RINT-1 and ZW10, whereas it was readily recovered in the co-IP complex of UVRAG (Fig. 4e). We observed near-complete co-localization of endogenous UVRAG with RINT-1 and ZW10 and with the ER marker (Fig. 4d). Finally, knockdown of RINT-1 reduced the association of UVRAG with ZW10 and NAG, suggesting that RINT-1 mediates the interaction of UVRAG with the ER-tether (Supplementary Fig. S2g–h). In line with this, ER-localized syntaxin 18, but not Golgi-associated syntaxin 6, co-immunoprecipitated with UVRAG and other components of the ER-tether (Fig. 4e). Furthermore, β'-COP, an outer-shell protein of COPI-coat, co-immunoprecipitated with UVRAG, along with RINT-1, ZW10, and NAG (Fig. 4e). These results indicate that UVRAG primarily associates with the RINT-1-ZW10-NAG tether and may participate in the ER tether-mediated vesicle transport.
UVRAG is required for the coat-tether interaction at the ER
We next evaluated the effects of UVRAG on the ER-tether complex, and its interaction with the COPI-coat. As shown in Figure 5a, strikingly less β'-COP coimmunoprecipitated with RINT-1 and ZW10 upon UVRAG knockdown. Of note, depletion of autophagy-related Atg16L1 did not appreciably affect the ability of RINT-1 and UVRAG to pull-down β'-COP and other factors in the ER-tether, suggesting that this is not an outcome of autophagy deficiency (Fig. 5a). Moreover, UVRAG depletion did not affect the RINT-1-ZW10-NAG heterotrimer assembly, nor did it perturb their ER localization (Fig. 5a and Supplementary Fig. S3a). Consistent with decreased COPI-binding, depletion of UVRAG caused redistribution of β'- and δ-COP from a juxtanuclear to a diffused puncta pattern (Supplementary Fig. S3b–c). Nearly identical phenotypes were observed with RINT-1- and ZW10-knockdown cells (Supplementary Fig. S3c). These findings suggest that UVRAG mediates the interaction of the ER-tether with β'-COP, and by extension the COPI-coat, which is required for efficient fusion of retrograde vesicles to the ER.
Figure 5.
UVRAG interaction with the ER tether and PI(3)P is required for COPI-dependent retrograde transport to the ER.
(a) UVRAG knockdown impairs the interaction of the RINT-1-ZW10 complex with COPI in an autophagy-independent manner. Cells (293T) were transfected with control shRNA, UVRAG shRNA, or Atg16L1 shRNA for 72 hr. WCLs were used for IP with anti-RINT-1 (1st and 3rd panels), anti-ZW10 (2nd panel), or anti-UVRAG (4th panel), followed by immunoblotting with the indicated antibodies. Input represents 10% whole-cell lystates. Note that β'-COP interaction was suppressed by UVRAG knockdown but unaffected by Atg16L1 knockdown. See Supplementary Fig. S9 for uncropped data.
(b) Retrograde transport of ts045-VSVG-KDELR-YFP from Golgi to the ER is inhibited by UVRAG knockdown. HeLa cells serially transfected with UVRAG- or control-shRNA, then with VSVG-KDELR-YFP expression vector, were incubated at 32°C (permissive condition) overnight. Upon shifting to 40°C (non-permissive condition), the ER-like distribution pattern of VSVG-KDELR-YFP was registered. Representative confocal images of the distribution pattern of VSVG-KDELR at 40°C and its co-localization with PDI were shown (upper panel). Data are mean ± SD, n = 100 cells obtained from three independent experiments; ** p < 0.01. Scale bars, 10 μm.
(c) UVRAG interaction with PI(3)P and RINT-1 are both required for COPI-dependent retrograde transport, as shown by the redistribution of VSVG-KDELR from the Golgi to the ER. HeLa cells were transfected with control shRNA or UVRAG shRNA. The UVRAG-depleted cells were then complemented with empty vector, Flag-UVRAG, UVRAGΔ270-442, UVRAGK78A/R82A, or UVRAGΔC2, along with the transfection of VSVG-KDELR-YFP. The ER pattern for the chimeric KDELR was quantified. Data are the mean ± SD; n = 200 cells obtained from three independent experiments; * p < 0.05; ** p < 0.01. Endogenous and reconstituted UVRAG expression was confirmed by immunoblotting (right panel).
(d) PI(3)P depletion inhibits Golgi-to-ER retrograde transport of VSVG-KDELR. HeLa cells were transfected with empty vector, wild-type, the C413S mutant of MTMR3, or treated with wortmannin or rapamycin, and then the ER pattern of VSVG-KDELR was quantified. Data are the mean ± SD; n = 100 cells for each group obtained from three independent experiments; ** p < 0.01.
(e) Interaction between endogenous UVRAG and the ER tethering complex under wortmannin treatment. WCLs of 293T cells treated with wortmannin were used for IP with anti-UVRAG or anti-RINT-1 antibody, followed by immunoblotting with the indicated antibodies. The panel of input shows endogenous protein expression. See Supplementary Fig. S9 for uncropped data.
Physical association of UVRAG with RINT-1 and PI(3)P are required for Golgi-to-ER transport
To examine the effect of UVRAG on the Golgi-to-the ER COPI-transport, we tracked the redistribution of a COPI cargo, VSVG-KDELR (vesicular stomatitis virus G protein fused with the KDEL receptor)42,43. VSVG-KDELR was mainly present in the perinuclear Golgi at 32°C. Upon shifting to 40°C, the chimera moved out of the Golgi and eventually distributed as an ER-like structure (Supplemental Fig. S3d). Knockdown of UVRAG markedly inhibited the COPI delivery of VSVG-KDELR to the ER (Fig. 5b). In contrast, no inhibition was observed in Atg7−/−-deficient iMEF (immortalized MEF) and cells depleted of Beclin1 or Atg16L1 (Supplementary Fig. S3e). To further verify this, we reconstituted UVRAG-depleted cells with either wild-type, RINT-1-binding-defective UVRAGΔ270–442, or PI(3)P-binding-defective UVRAGK78A/R82A and UVRAGΔC2 mutants. Inhibition of VSVG-KDELR transport by UVRAG knockdown was reversed by WT, but not by UVRAGΔ270–442, UVRAGK78A/R82A and UVRAGΔC2 that are shRNA-resistant (Fig. 5c). These results indicate that UVRAG interaction with RINT-1 and PI(3)P is required for COPI-retrograde transport.
PI(3)P-dependent action of UVRAG may couple COPI-transport to the pathways regulating PI(3)P metabolism. To test this, PI(3)P levels in cells were altered by expression of MTMR3 or wortmannin. Expression of MTMR3 inhibited retrograde COPI-transport, which was abrogated by the enzyme-dead C413S mutant (Fig. 5d). Treatment with wortmannin also led to a reduced ER-pool of VSVG-KDELR (Fig. 5d). Moreover, the addition of wortmannin reduced UVRAG interaction with the RINT-1-complex, and precluded β'-COP from binding the ER-tether, but did not affect formation of the RINT-1-ZW10-NAG complex (Fig. 5e). These results are consistent with a model where UVRAG targets PI(3)P to recruit the ER-tether close to PI(3)P-sites, and its interaction with RINT-1 in turn promotes the tether-coat interaction and enhances overall COPI-transport to the ER.
UVRAG regulates cis-Golgi homeostasis
Given that the Golgi-ER transport is pivotal to Golgi homeostasis15,17,18, we next investigated the consequence of UVRAG suppression to the Golgi by electron microscopy (EM). As shown in Fig. 6a, knockdown of UVRAG induced dilated and scattered Golgi-cisternae as compared to the well-formed Golgi-stacks in control. Similar changes were observed in RINT-1-depleted cells (Supplementary Fig. S4a). In accord, UVRAG depletion disorganized GM130 and p115 into a punctuate pattern without affecting their overall levels as seen in several cell lines (Fig. 6b and Supplementary Fig. S4b–d). Furthermore, knockdown of RINT-1 or ZW10 caused a similar disruption of cis-Golgi (Supplementary Fig. S4e), as observed previously18,44. Little change was detected in the distribution of PDI, GalNac-TII, and TGN46, and Sec31a (Supplementary Fig. S4d). Because cis-Golgi was specifically disrupted, we examined whether the ER-to-the Golgi protein transport was affected by UVRAG depletion using the well-established transport assay of VSVG-GFP45. When the cells were shifted from 40°C to 32°C, VSVG was rapidly depleted from the ER and concentrated at the early-Golgi in control cells, whereas it remained dot-like at the cell periphery and failed to concentrate in the Golgi in UVRAG- and RINT-1-depleted cells (Fig. 6c). Notably, inhibition of UVRAG did not affect brefeldin A-induced Golgi tubulation and redistribution of Golgi proteins into the ER-network through direct tubular connection46 (Supplementary Fig. S5a–b). These results indicate that UVRAG and its ER-interactors are required for the ER-Golgi trafficking and the dynamic integrity of early-Golgi.
Figure 6.
UVRAG and its interaction with RINT-1 and PI(3)P are required for cis-Golgi maintenance.
(a) Golgi morphology upon UVRAG depletion. HeLa cells were transfected with control-or UVRAG-specific shRNA for 72 hr and subjected to electron microscopy (EM) analysis. Compared to control cells (left), the Golgi structure was swollen and fragmented in cells lacking UVRAG. The level of Golgi fragmentation was quantified (right). Data represents mean ± SD; n = 50 cells obtained by gathering data of two independent experiments; * p < 0.05. Scale bars, 500 nm.
(b) Depletion of UVRAG leads to cis-Golgi dispersion. HeLa cells were transfected with control- or UVRAG-specific shRNA expressing GFP as an expression marker and then stained for GM130 (red; highlighted in arrows). Nuclei were stained with DAPI (blue). The dispersed distribution of GM130 in shRNA-transfected cells was quantified (middle). Data are the mean ± SD; n = 500 cells obtained from five independent experiments; p = 0.0001. The expression of UVRAG and GM130 in treated cells is also shown (right). Scale bars, 10 μm.
(c) The effect of UVRAG on membrane trafficking from the ER to Golgi. HeLa cells serially transfected with UVRAG-, RINT-1-, or Control-shRNA, then with VSVG-GFP expression vector, were incubated at 40°C (non-permissive condition) overnight. The cells were then shifted to 32°C (permissive condition) for 30 min to allow cargo transit from the ER to the Golgi. Before and after the temperature shift, the cells were fixed and labeled with anti-PDI and anti-GM130 antibodies to define the ER and Golgi region, respectively. Representative confocal microscopy images of the distribution pattern of the retrograde cargo at 40°C and 32°C are shown (left). The Golgi-like distribution pattern of VSVG-GFP was registered (right). Data are mean ± SD; n = 100 cells from three independent experiments; ** p < 0.01. Scale bars, 10 μm.
(d–e) UVRAG interaction with PI(3)P and RINT-1 are required for Golgi integrity. HeLa cells were transfected with control shRNA (1st row) or UVRAG shRNA (2nd to 6th rows). The UVRAG-depleted cells, as indicated by GFP expression, were then transfected with empty vector (2nd row), Flag-UVRAG (3rd row), UVRAGΔ270-442 mutant (4th row), UVRAGK78A/R82A mutant (5th row), or UVRAGΔCCD mutant (6th row), followed by confocal microscopy using anti-GM130 (red) for cis-Golgi staining, anti-Flag (purple) for the ectopically expressed UVRAG protein expression, and DAPI for nuclei (blue). Asterisks indicate shRNA-transfected cells and arrows denote the cells reconstituted with UVRAG (wt or mutant) expression. The dispersed distribution of GM130 was quantified (e), and data represent the mean ± SD; n = 200 cells from four independent experiments; * p < 0.05; ** p < 0.01.
As observed in COPI-transport, wild-type UVRAG suppressed the disassembly of cis-Golgi induced by UVRAG depletion (Fig. 6d–e). However, expression of the shRNA-resistant UVRAGΔ270–442 and UVRAGK78A/R82A mutant failed to rescue Golgi morphology (Fig. 6d–e). Thus, UVRAG and its interaction with the RINT-1-complex and PI(3)P are required for intact Golgi structure. Notably, expression of UVRAGΔCCD, which is deficient in Beclin1-binding only31, rescued the Golgi morphology in UVRAG-depleted cells as seen with WT (Fig. 6d–e). Accordingly, removing beclin1 or the C-Vps proteins Vps16 and Vps18 had marginal effect on GM130 (Supplementary Fig. S5c–d), indicating that action of UVRAG in the early secretory pathway is independent of its previously recognized activities in autophagy and endosomal trafficking.
Autophagy induction triggers a shift of UVRAG from the RINT-1-complex to the Beclin1-complex and inhibits COPI transport to the ER
The data so far imply that UVRAG action with the ER-tether is functionally separable from its autophagic activity as part of a complex with Beclin1, Bif-1, and PI(3)KC329–31. An intriguing question rises then: how does UVRAG coordinate these two events to optimize cargo delivery in response to different stimuli? To this end, we examined endogenous interaction of UVRAG with the RINT-1-containing tethering complex and the Beclin1-containing autophagy complex during autophagy induced by rapamycin (mTOR-dependent autophagy), SMER28 (mTOR-independent autophagy)47,48, or HBSS (starvation) (Fig. 7a–b and Supplemental Fig. S6a). Under normal conditions, UVRAG forms a complex with RINT-1, ZW10, and NAG and interacts with β'-COP. During autophagy, however, minimal levels of UVRAG co-immunoprecipitated with the RINT-1-complex in autophagy-inducing cells, suggesting dissociation of UVRAG from the tether (Fig. 7a–b and Supplemental Fig. S6a). Accordingly, the ability of RINT-1 to interact with β'-COP was strikingly impaired (Fig. 7a–b), although the RINT-1-ZW10-NAG complex formation remained intact (Fig. 7a–b). Consistent with this, rapamycin inhibited retrograde transport of VSVG-KDELR to a level similar to that observed with wortmannin (Fig. 5d). These data indicate that autophagy causes disassembly of UVRAG from the RINT-1-complex and attenuated retrograde transport.
Figure 7.
UVRAG dissociates from the RINT-1-complex, and interacts with the Beclin1-complex during autophagy.
(a–b) Differential interaction of UVRAG with the RINT-1-ZW10-NAG tethering complex and the Beclin1-Bif-1-PI(3)KC3 autophagy complex after treatment with rapamycin (a) and HBSS (b). Cells (293T) treated with rapamycin (50 nM) in (a) or with HBSS in (b) were subjected to IP with anti-UVRAG (1st panel), anti-RINT-1 (2nd panel), or anti-PI(3)KC3 (3rd panel), followed by immunoblotting with the indicated antibodies. Input (10%) shows endogenous protein expression. See Supplementary Fig. S9 for uncropped data.
(c) Gel filtration analysis of UVRAG complex formation under normal conditions and after rapamycin-induced autophagy (50 nM). Affinity-purified UVRAG complexes from HCT116 cells stably expressing Flag-UVRAG were fractionated by Superose-6 gel filtration column and the eluates were analyzed by Western blotting for UVRAG, RINT-1, ZW10, Beclin1, PI3KC3, and Bif-1 proteins. Whole cell lysates (2.5%) were used as the input (lane-1). The elution profile of each protein was quantitated by densitometry analysis and normalized. Relative densitometry units were plotted against fraction number. Black arrows indicate the positions of the molecular weight size markers (kDa). Red arrows indicate the peak shift of the UVRAG eluates.
Concomitant with UVRAG dissociation from the tether, the interaction of UVRAG with Bif-1, Beclin1, and PI(3)KC3 was enhanced by autophagy (Fig. 7a–b and Supplemental Fig. S6a). Indeed, autophagy induction increased UVRAG-Beclin1 colocalization, whereas its co-staining with RINT-1 was reduced (Supplemental Fig. S6b). Notably, regardless of autophagy induction, Beclin1, Bif-1, and PI(3)KC3 were largely excluded from the RINT-1-complex, and likewise, the ER-tether proteins were largely excluded from the PI(3)KC3-complex (Fig. 7a–b). Thus, UVRAG associates with two distinct complexes in a mutually exclusive manner. Analogous results were obtained with exogenous UVRAG expression. A clear shift was observed in the binding affinity of Flag-UVRAG from the RINT-1-complex to the Beclin1-complex (Supplemental Fig. S6c). Interestingly, UVRAGK78A/R82A exhibits a similar shift as that of wild-type. Moreover, UVRAGΔ270-442 failed to bind the ER-tether under both conditions, but its association with the Beclin1-complex was similarly increased during autophagy (Supplemental Fig. S6c). This suggests that UVRAG recruitment to the Beclin1-complex is independent from its association with RINT-1. We also performed gel filtration analysis of purified UVRAG-complexes and found that RINT-1 and ZW10 co-elute with UVRAG as a large complex (~ 443kD), whereas Beclin1, PI3KC3, and Bif-1 derived from the UVRAG affinity eluate have a different elution profile (Fig. 7c). Autophagy induction, however, caused a clear shift of UVRAG peak elution from the RINT-1-complex to then elute with Beclin1, PI3KC3, and Bif-1 (Fig. 7c). Conversely, when cells underwent retrograde-traffic pulse induced by expressing VSVG-KDELR49, the association of UVRAG with the RINT-1-complex was increased, while its binding to the Beclin1-complex was concomitantly reduced (Supplemental Fig. S6d). Hence, UVRAG shifts between two complexes to cross-regulate retrograde traffic and autophagy.
The roles of Beclin1, RINT-1, and PI(3)P in UVRAG-mediated Atg9 trafficking during autophagy
The UVRAG-associated Bif-1-Beclin1-PI3KC3 complex was implicated in mediating Atg9 traffic26. We also found that a portion of Atg9 co-localized with GM130 (Supplementary Fig. S6e). Given that the UVRAG-RINT-1 association is required for the stability of cis-Golgi, we asked whether it is similarly essential in the dynamics of Atg9. Unlike cis-Golgi that dispersed upon UVRAG knockdown, depletion of UVRAG and/or expression of any of these mutants did not affect distribution of Atg9 under normal condition (Fig. 8a). Upon induction of autophagy, Atg9 displayed punctate structures throughout the cytoplasm, as shown in control cells and in previous reports23,25,50. Notably, depletion of UVRAG resulted in a dramatic reduction of Atg9 translocation, which was reversed by wild-type and UVRAGΔ270-442, but not by UVRAGΔCCD and UVRAGK78A/R82A (Fig. 8a–c). Consistently, UVRAG or Beclin1 depletion markedly inhibited Atg9-vesicle formation, and both the total number of autophagic vesicles and the overlapping Atg9 staining with LC3-puncta were severely suppressed (Fig. 8d and Supplemental Fig. S7a). In contrast, depletion of RINT-1 or Atg16L1 had no observable effect on Atg9 movement (Fig. 8d and Supplemental Fig. S7a). In agreement with the findings that Atg9 trafficking is linked to autophagosome51, the production of LC3-II was greatly increased in the WT- and Δ270-442-expressing cells compared with that in the ΔCCD- or K78A/R82A-expressing cells (Supplemental Fig. S7b). To verify that enhanced levels of LC3-II represent increased autophagy rather than an impaired degradation, we measured the levels of p62, an autophagic substrate52. As expected, expression of WT and Δ270-442 markedly reduced the levels of p62, compared with what was seen with vector, ΔCCD or K78A/R82A (Supplemental Fig. S7b). Thus, RINT-1-binding is dispensable for UVRAG-mediated autophagy, whereas both Beclin1 and PI(3)P interaction of UVRAG are required for efficient autophagy. Overall, UVRAG functions in two distinct but functionally-related trafficking events: one involving the RINT-1-complex in Golgi-ER COPI-transport and the other involving the Beclin1-complex in the Golgi-autophagosome Atg9-transport, both of which share a requirement for PI(3)P-binding of UVRAG and being thereof functionally interdependent (Supplemental Fig. S8).
Figure 8.
The role of Beclin1, RINT-1, and PI(3)P in UVRAG-mediated Atg9 trafficking during autophagy.
(a–c) UVRAG interaction with Beclin1 and PI(3)P, but not with RINT-1, is required for autophagy-induced Atg9 dispersal. HeLa cells were stably transfected with control shRNA (1st row) or UVRAG shRNA (2nd–6th rows). The UVRAG-depleted cells were then transfected with empty vector (2nd row), Flag-UVRAG (3rd row), UVRAGΔ270–442 mutant (4th row), UVRAGK78A/R82A mutant (5th row), or UVRAGΔCCD mutant (6th row), and incubated in complete medium (untreated, UT, 1st panel), starvation medium (HBSS 2 hr, 3rd panel), or treated with rapamycin (50 nM 2 hr, 2nd panel) or SMER28 (50 μM, 2 hr, 4th panel). The distribution pattern of endogenous Atg9 (green) in the UVRAG (wt or mutant)-transduced cells (red) was analyzed by confocal microscopy (a). Asterisks denote condensed (non-dispersed) Atg9 in UVRAG-depleted HeLa cells and arrows denote the cells complemented with the wild-type or mutant UVRAG. The percentage of cells with Atg9 translocation was quantified in (c) and the expression of UVRAG and actin were immunoblotted and shown in (b). Data represent the mean ± SD; n = 300 cells from three independent experiments; n.s., not significant; **, p < 0.01; ***, p < 0.001. Scale bars, 10 μm.
(d) Effect of knockdown of UVRAG, Beclin1, and RINT-1 on the translocation of Atg9 to the LC3-labelled autophagsomes during autophagy. Control-, UVRAG-, Beclin1-, or RINT-1-knockdown HeLa cells were incubated in complete medium (normal condition), starvation medium (HBSS, 2 hr), or treated with rapamycin (100 nM, 2 hr) or SMER28 (50 μM, 2 hr). The cells were then stained for endogenous Atg9 (green) and LC3 (red) followed by confocal microscopy analysis. The percentage of Atg9-vesicles colocalized with LC3+-autophagic puncta was quantified (bottom middle). Endogenous UVRAG, Beclin1, and RINT-1 protein expression are shown (bottom right). Data represents mean ± SD; n = 200 cells obtained from three independent experiments. n.s. not significant; * p < 0.05; ** p < 0.01; *** p < 0.001. Scale bars, 10 μm.
DISCUSSION
We report here that UVRAG, through its interaction with RINT-1, functions as a positive regulator of the ER-tether in Golgi-ER retrograde transport, and demonstrate the mechanism underlying the dynamic regulation of Golgi-ER transport and Atg9 trafficking in response to autophagy via a `partnering shift' and PI(3)P-association of UVRAG.
UVRAG is a versatile protein that localizes in different subcellular compartments to regulate cellular pathways in a coordinated fashion27,30,34,53,54. UVRAG associates with Bif-1 and Beclin1 through CCD and PR regions, respectively, promoting Beclin1-mediated autophagy30,31. Moreover, endosomal UVRAG binds C-Vps to facilitate endosome/autophagosome maturation (Supplementary Fig. S8a)29. Furthermore, UVRAG C-terminal region is implicated in genomic stability by targeting the centrosome and associating with the Ku-DNA-PK complex for DNA repair (Supplementary Fig. S8a)34. Apart from these findings, we herein established that loss of UVRAG impairs the ER tether-mediated Golgi-ER traffic and cis-Golgi, suggesting an important mechanism for UVRAG in the early secretory pathway. Notably, removing autophagy-related Beclin1, Atg16L1 or endosome-associated C-Vps activity failed to phenocopy UVRAG knockdown, suggesting a minor participant of other membrane defects that might arise due to UVRAG deficiency. Consistent with this, UVRAG directly binds RINT-1 and modulates the ER-tether in cargo delivery. The RINT-1-interacting domain of UVRAG is also implicated in centrosome targeting, however, the trafficking role of UVRAG is less likely to be dependent on the centrosome interaction as CEP63 can only be detected in complex with UVRAG at the centrosome as previously shown34. Likewise, the UVRAG mutant that lacks RINT-1-binding ability but remains competent for autophagy, perturbed COPI transport as a relic of UVRAG deficiency, suggesting that the UVRAG-ER tether interaction is required for complete Golgi-ER function.
Our study established that UVRAG is a downstream effector of PI(3)P, which acts as a scaffold to recruit the tether into PI(3)P-rich patches at the ER for site-specific COPI-vesicle docking. Depletion or blocking of PI(3)P, or point mutations in UVRAG-C2 decreased UVRAG association with the ER, which in turn impaired cargo delivery and Golgi homeostasis. Moreover, the region of UVRAG for PI(3)P-recognition is separable from that binding RINT-1. In fact, the interactions of PI(3)P and the RINT-1-complex with UVRAG are all necessary for Golgi-ER transport at the steady-state. These results imply that UVRAG functions through two successive activities in Golgi-ER transport: one involving PI(3)P to specify membrane identity and the other involving the ER-tether to capture COPI-vesicle. Thus, we postulate that basal PI(3)P has a direct role in ER functions that is not contingent on its inducible requirement for autophagy.
Although RINT-1-knockdown affects Golgi-ER dynamics, it has no discernible effect on Atg9 fission. A question then arises: how are the two processes hierarchically coordinated by UVRAG? We found that coordination relies on UVRAG ability to assemble with different protein complexes following induction of autophagy. Under basal conditions, UVRAG acts as a positive regulator of the ER-tether for fusion of COPI-vesicles with the ER (Supplementary Fig. S8b). When autophagy is induced, UVRAG assembles the Beclin1-complex, which appears critical for Atg9-membrane translocation and requires interaction with Beclin1 (Fig. 8b). Such recruitment coincides with the dissociation of UVRAG from the ER-tether and with negative control of Golgi-ER transport. Our findings indicate that the Golgi-ER transport and Atg9-trafficking pathways are interdependent and intimately coordinated. PI(3)P-bound UVRAG fulfills its dual roles by differential regulation of the Beclin1- and RINT-1-complex, underscoring that the multitasking functions of UVRAG are important to homeostasis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to acknowledge Drs. V. Hsu, Z. Yue, J. Lippincott-Schwartz, T. Yoshimori, W.M. Yuan, S. Firestein, T. Yoshimori, J. Laporte, and Y. Ohsumi for providing reagents, Drs. Hauser and E. Barron for performing electron microscopy. We thank all the members of the Liang laboratory for helpful discussion. The authors wish to thank Dr. M. Torres for her editorial assistance. This work was supported by American Cancer Society (RSG-11-121-01-CCG to C. Liang), National Institutes of Health grants (R01 CA140964 and R21 CA161436 to C. Liang), and core services performed through grant NIAID U19AI083025.
Footnotes
Author contributions
S.H., D.N., and B.M. performed most experiments of this study and analyzed the data. S.H. contributed extensively to the revision work. JH. L performed protein-lipid binding assay and confocal microscopy. T.Z. conducted autophagy-related assays. I.G., S.P., Z.Z., and S.O. helped with data collection. N.B., B.L., and HS. Y. conducted bioinformatics analysis. WH. L., YT., HG. W., VD., R.P., and M.T. provided critical constructs and antibodies. A.M. and P.F. advised on gel filtration analysis. C.L designed research, analyzed data and wrote the paper.
COMPETING FINANCIAL INTEREST STATEMENT
The authors declare no competing financial interests.
Reference
- 1.Jean S, Kiger AA. Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat Rev Mol Cell Biol. 2012;13:463–70. doi: 10.1038/nrm3379. [DOI] [PubMed] [Google Scholar]
- 2.Vicinanza M, D'Angelo G, Di Campli A, De Matteis MA. Phosphoinositides as regulators of membrane trafficking in health and disease. Cellular and molecular life sciences : CMLS. 2008;65:2833–41. doi: 10.1007/s00018-008-8353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burman C, Ktistakis NT. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett. 2010;584:1302–12. doi: 10.1016/j.febslet.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Amoasii L, et al. Myotubularin and PtdIns3P remodel the sarcoplasmic reticulum in muscle in vivo. Journal of Cell Science. 2013 doi: 10.1242/jcs.118505. [DOI] [PubMed] [Google Scholar]
- 5.Sarkes D, Rameh LE. A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem J. 2010;428:375–84. doi: 10.1042/BJ20100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taguchi-Atarashi N, et al. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 2010;11:468–78. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- 7.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. Journal of Cell Biology. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenzo O, Urbe S, Clague MJ. Analysis of phosphoinositide binding domain properties within the myotubularin-related protein MTMR3. J Cell Sci. 2005;118:2005–12. doi: 10.1242/jcs.02325. [DOI] [PubMed] [Google Scholar]
- 9.Pankiv S, et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–69. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proikas-Cezanne T, Ruckerbauer S, Stierhof YD, Berg C, Nordheim A. Human WIPI-1 puncta-formation: a novel assay to assess mammalian autophagy. FEBS Lett. 2007;581:3396–404. doi: 10.1016/j.febslet.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 11.Vicinanza M, D'Angelo G, Di Campli A, De Matteis MA. Function and dysfunction of the PI system in membrane trafficking. The EMBO journal. 2008;27:2457–70. doi: 10.1038/emboj.2008.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–82. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Ren Y, et al. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139:1119–29. doi: 10.1016/j.cell.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tripathi A, Ren Y, Jeffrey PD, Hughson FM. Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nature structural & molecular biology. 2009;16:114–23. doi: 10.1038/nsmb.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt HD. Dsl1p/Zw10: common mechanisms behind tethering vesicles and microtubules. Trends in Cell Biology. 2010;20:257–268. doi: 10.1016/j.tcb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Meiringer CT, et al. The Dsl1 protein tethering complex is a resident endoplasmic reticulum complex, which interacts with five soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptors (SNAREs): implications for fusion and fusion regulation. J Biol Chem. 2011;286:25039–46. doi: 10.1074/jbc.M110.215327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirose H, et al. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. Embo Journal. 2004;23:1267–1278. doi: 10.1038/sj.emboj.7600135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arasaki K, Taniguchi M, Tani K, Tagaya M. RINT-1 regulates the localization and entry of ZW10 to the syntaxin 18 complex. Mol Biol Cell. 2006;17:2780–8. doi: 10.1091/mbc.E05-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aoki T, et al. Identification of the Neuroblastoma-amplified Gene Product as a Component of the Syntaxin 18 Complex Implicated in Golgi-to-Endoplasmic Reticulum Retrograde Transport. Molecular Biology of the Cell. 2009;20:2639–2649. doi: 10.1091/mbc.E08-11-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraft C, Martens S. Mechanisms and regulation of autophagosome formation. Curr Opin Cell Biol. 2012;24:496–501. doi: 10.1016/j.ceb.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–21. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Molecular Biology of the Cell. 2010;21:2270–84. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young ARJ, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. Journal of Cell Science. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 24.Orsi A, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, is required for autophagy. Molecular Biology of the Cell. 2012 doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto H, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–33. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi Y, et al. Bif-1 regulates Atg9 trafficking by mediating the fission of Golgi membranes during autophagy. Autophagy. 2011;7:61–73. doi: 10.4161/auto.7.1.14015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Molecular Biology of the Cell. 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–U262. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–87. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang C, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 32.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 33.Sun Q, et al. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J Biol Chem. 2010;286:185–91. doi: 10.1074/jbc.M110.126425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Z, et al. A Dual Role for UVRAG in Maintaining Chromosomal Stability Independent of Autophagy. Dev Cell. 2012;22:1001–16. doi: 10.1016/j.devcel.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guerrero-Valero M, et al. Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6603–7. doi: 10.1073/pnas.0813099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joung MJ, Mohan SK, Yu C. Molecular level interaction of inositol hexaphosphate with the C2B domain of human synaptotagmin I. Biochemistry. 2012;51:3675–83. doi: 10.1021/bi300005w. [DOI] [PubMed] [Google Scholar]
- 37.Fukuda M, Kojima T, Aruga J, Niinobe M, Mikoshiba K. Functional diversity of C2 domains of synaptotagmin family. Mutational analysis of inositol high polyphosphate binding domain. J Biol Chem. 1995;270:26523–7. doi: 10.1074/jbc.270.44.26523. [DOI] [PubMed] [Google Scholar]
- 38.Gillooly DJ, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. The EMBO journal. 2000;19:4577–88. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godi A, et al. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- 40.Wang YJ, et al. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- 41.Kanai F, et al. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat Cell Biol. 2001;3:675–8. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- 42.Yang JS, et al. Key components of the fission machinery are interchangeable. Nat Cell Biol. 2006;8:1376–82. doi: 10.1038/ncb1503. [DOI] [PubMed] [Google Scholar]
- 43.Yang JS, et al. COPI acts in both vesicular and tubular transport. Nat Cell Biol. 2011;13:996–1003. doi: 10.1038/ncb2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirose H, et al. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 2004;23:1267–78. doi: 10.1038/sj.emboj.7600135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweizer A, et al. Identification of an intermediate compartment involved in protein transport from endoplasmic reticulum to Golgi apparatus. European journal of cell biology. 1990;53:185–96. [PubMed] [Google Scholar]
- 46.Hess MW, Muller M, Debbage PL, Vetterlein M, Pavelka M. Cryopreparation provides new insight into the effects of brefeldin A on the structure of the HepG2 Golgi apparatus. J Struct Biol. 2000;130:63–72. doi: 10.1006/jsbi.2000.4230. [DOI] [PubMed] [Google Scholar]
- 47.Sarkar S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nature chemical biology. 2007;3:331–8. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams A, et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nature chemical biology. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.San Pietro E, et al. Group IV phospholipase A(2)alpha controls the formation of inter-cisternal continuities involved in intra-Golgi transport. PLoS Biol. 2009;7:e1000194. doi: 10.1371/journal.pbio.1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webber JL, Young AR, Tooze SA. Atg9 trafficking in Mammalian cells. Autophagy. 2007;3:54–6. doi: 10.4161/auto.3419. [DOI] [PubMed] [Google Scholar]
- 51.Yen WL, Legakis JE, Nair U, Klionsky DJ. Atg27 is required for autophagy-dependent cycling of Atg9. Molecular biology of the cell. 2007;18:581–93. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. The Journal of cell biology. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang C, Sir D, Lee S, Ou JH, Jung JU. Beyond autophagy: the role of UVRAG in membrane trafficking. Autophagy. 2008;4:817–20. doi: 10.4161/auto.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peplowska K, Cabrera M, Ungermann C. UVRAG reveals its second nature. Nat Cell Biol. 2008;10:759–61. doi: 10.1038/ncb0708-759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.