Investigating recovery from hepatitis C virus (HCV) infection is like solving a difficult puzzle. Even after considerable progress, too many pieces are missing to see a complete picture. The study by Rauch et al fills in a key new piece and provides a focal point for future research on HCV recovery.
There are major differences in how people respond to HCV infection and its treatment. About 30% of persons who acquire HCV infection resolve viremia, leaving only the antibody response as a marker of prior exposure. These so-called spontaneous resolvers have no long-term consequences of HCV infection and can be distinguished readily from others with chronic hepatitis C and life-long viremia. Similarly, there are marked differences in the degree to which interferon alfa treatment suppresses HCV replication among those with chronic infection. Even after a single dose of interferon alfa, >2-log differences in HCV RNA reduction are evident among persons with genotype 1 HCV infection.1 Like spontaneous clearance, treatment-related resolution of chronic hepatitis C (sustained virologic response) is associated with clearance of viremia and reduction in the risk of long-term consequences of infection.2
There must be a host genetic basis for these disparate clinical outcomes. Aside from viral or environmental factors, some racial groups spontaneously recover from HCV infection more frequently than others. In 1 study, Caucasians were 5 times more likely to recover spontaneously from acute HCV infection than African Americans.3 Even when persons have been infected accidentally with the same HCV inoculum, spontaneous recovery occurs in some but not others, indicating that host genetics determines the outcome.4 Likewise, African Americans and probably Hispanics infected with genotype 1 HCV infection are less likely to respond to interferon alfa treatment than Caucasians.5–7
Convinced that there must be something different about the DNA of those who recovered and those with chronic infection, investigators have been working the puzzle using genetic tools. Logically, the initial genetic approach was to look for variability in “candidate” genes that encode key elements of the antiviral immune response. Persons possessing particular human leukocyte antigen types were more likely to recover from HCV, presumably owing to better processing and presentation of HCV-derived peptides or through differential interaction with other immunologic mediators such as KIR receptors.8–11 Differences (eg, polymorphisms) in genes encoding cytokines and other immunologic mediators also seemed to explain some HCV recovery.12,13
Some differences were found also in candidate genes of persons who responded to interferon alfa and ribavirin treatment compared with so-called nonresponders.14–16 In most instances, these studies were completely distinct from those focused on spontaneous resolvers. The expectation was that these were 2 biologically distinct processes—2 separate puzzles. However, for both forms of HCV resolution, many of the prime candidate genes were either not polymorphic or the frequency of the polymorphisms was not different in resolvers compared with those with persistent infection.17 Too many pieces were missing to see a complete picture.
An alternative to the candidate-gene approach for solving the puzzle is the genome-wide association study (GWAS), in which there is no a priori hypothesis about involved genes. In a GWAS the frequencies of 500,000 to 2 million single nucleotide polymorphisms (SNPs) that are distributed throughout the human genome are compared in cases and controls. This approach presents special challenges, such as how to account for ethnic and geographic differences in the prevalence of SNPs, as well as the issue of multiple comparisons.18 The latter situation refers to the probability that SNP frequencies in cases and controls would differ simply because of chance, which naturally increases when there are hundreds of thousands of “chances.” This issue typically requires that the study be very large or detect a SNP with a very strong association.19
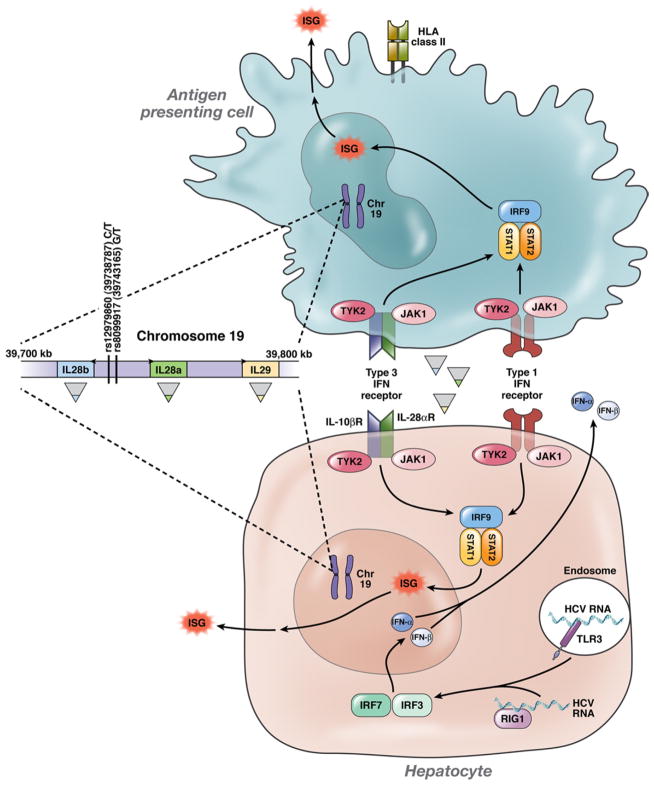
The study by Rauch et al in this issue of Gastroenterology is an example of using GWAS to detect a very strong genetic association in a study of a relatively genetically homogenous population.20 They compared the frequency that each of approximately 500,000 SNPs occurred in DNA from 347 Caucasian Swiss patients with spontaneous HCV resolution and 1,015 with persistent infection. Remarkably, although the differences in the frequencies of nearly all the 500,000 SNPs followed the expected normal distribution, 7 stood out as more than an order of magnitude less likely to have occurred by chance. All 7 strongly associated SNPs were located on chromosome 19, within 80 kb of the genes for a family of lambda interferons. The strongest association with spontaneous recovery was detected for rs8099917, a T/G SNP whose distribution was exceedingly unlikely to have occurred by chance (P = 6 × 10−9) and was located nearest to IL28b, the gene for interferon lambda 3. The investigators found that the frequency of the minor G allele was overrepresented among those with chronic hepatitis C (compared with those with spontaneous recovery) and was over-represented in the subset of those with chronic infection who failed to respond to pegylated interferon and ribavirin (compared with those who achieved a sustained virologic response).
Very recently, other groups also have found pieces of the HCV recovery puzzle near IL28b.21–24 Two of them also found that the T allele in rs8099917 was associated with sustained virologic response to treatment.21,22 Also using GWAS, Ge et al23 compared the frequencies of about 600,000 SNPs in DNA from 1,137 persons with persistent hepatitis C according to their response to peginterferon alfa and ribavirin therapy. Persons who were homozygous for the major C (vs T) allele at the rs12979860 SNP near IL28b were 2-fold more likely to respond to treatment than those homozygous for the alternative nucleoside (T). Likewise, by testing persons with spontaneous resolution of HCV, our group reported a >2.5-fold increased likelihood of recovery in persons homozygous for C at rs12979860 compared with controls with persistent HCV infection.24 In addition, we showed that the global distribution of the protective CC allele correlated strongly with ethnic differences in spontaneous resolution of HCV, just as Ge et al23 had reported for treatment-related response.
It is important to note that rs12979860 is just 4378 bases from rs8099917 and in this Swiss population their linkage was high (D′ 0.98; r2 = 0.5; Figure 1). Likewise, Ge et al reported high linkage between these 2 SNPs in Caucasians (r2 = 0.52) but very low linkage in African Americans (r2 = 0.07). Interestingly, data from the Hap-Map Project (www.hapmap.org)25 show that, in 2 African populations, the risk (G) allele in rs8099917 is infrequent (6.1% in Kenyans and 1.8% in Nigerians), which is lower than the 17% in the Swiss. Because blacks are more likely to have viral persistence and less likely to respond to treatment, this allele would not explain the racial differences in HCV outcomes. By contrast, the risk (T) allele in rs12979860 is more common in blacks than in Caucasians and was present in 50% and 69% of the Kenyans and Nigerians in HapMap, respectively.24 Thus, further study of this gene is needed to determine whether it explains racial differences in the outcome of this global infectious disease.
Figure 1.
HCV RNA triggers production of type 1 interferons (such as interferon β and interferon α) and probably lambda interferons by hepatocytes, and these molecules (and probably the virus itself) stimulate transcription of interferon stimulated genes (ISGs) in antigen presenting cells such as dendritic cells as well as in hepatocytes. Exogenous (therapeutic) interferon α and lambda interferon signal similarly, but in a “steady-state” environment in which HCV replication has been sustained despite ongoing expression of ISGs. Polymorphisms just upstream of the gene for IL-28b (interferon lambda 3) are strongly associated with spontaneous and treatment-associated resolution of HCV infection, but the mechanism is unknown.
At least 5 independent studies provide overwhelming evidence for the role of interleukin (IL)-28b in the pathogenesis of HCV infection. However, still more pieces of data are needed to complete the mechanistic picture. IL-28b (also known as interferon lambda 3) and other type 3 interferons like IL-28a or IL-29 trigger an antiviral cascade via JAK-STAT that is similar to and probably synergistic with type 1 interferons (such as interferon alfa), although using distinct receptors (Figure 1).26 Like type 1 interferons, lambda interferons have activity against HCV and other viral infections in vitro and in vivo. However, in vitro exogenous interferon lambda induces a slower, more sustained abundance of interferon stimulated genes than alfa interferon.27 Nonetheless, although these findings explain why interferon lambdas might play a role in HCV recovery, they do not answer why certain base sequences located upstream of the IL-28b start codon are associated with spontaneous and treatment-associated resolution of HCV infection.
Unfortunately, at this point, very few possible explanations have been excluded, and there are few strong mechanistic clues. One possibility is that these SNPs are markers of another DNA sequence that modifies IL-28b. Both rs12979860 and rs8099917 are strongly associated with a nonsynonymous IL28b mutation; the favorable SNP haplotype usually occurs in association with an arginine instead of a lysine at position 70 (rs8103142). This change is distant from the putative receptor binding location and its mechanistic significance remains unknown.28
Because these SNPs are located upstream of IL28b, it is plausible that these mutations correlate with the regulation of IL-28b transcription. Using the SNPExpress database, Ge et al23 reported no difference in IL-28b expression in PBMC from 80 HCV-uninfected persons homozygous for a proxy allele for rs12979860 (see the article’s supplemental material). On the other hand, 2 studies found that those who carried the G risk allele at rs8099917 had lower PBMC mRNA expression of IL-28b.21,29 It is likely that regulation will differ in the infected tissue and even between cell types within the liver, as has been reported recently for some interferon stimulated genes.30 IL-28b attenuates IL-13; it is also possible that the cytokine produced in persons with the protective allele diminishes IL-13 to a lesser extent than the molecule with the risk allele, similar to the protective effect of the least inhibitory interactions between KIR and HLA-C.10,31
The degree to which these and other pieces fit the puzzle is being examined. In the meantime, the work by Rauch et al and the other related studies published elsewhere provide incontrovertible genetic evidence for a role of IL-28b in spontaneous and treatment-related recovery from HCV infection. The work also underscores the power of large-scale genetic studies to reveal why there are ethnic differences in the clinical expression of global infectious diseases and their treatments. Now that we have a major new piece in the puzzle, the effort has to be linking this finding to other pieces until the picture of HCV resolution is sufficiently clear to improve, and perhaps personalize, HCV treatment and prevention worldwide.
Acknowledgments
The authors thank Stuart Ray for helpful comments on the editorial and figure. Dr. Thomas and Dr. Thio participate with Dr Rauch et al in a multicenter consortium on genetics of HCV infection.
Funding
Supported in part by U.S. Public Health Service grant R01013324.
This is a commentary on article Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J, Colombo S, Cerny A, Dufour JF, Furrer H, Günthard HF, Heim M, Hirschel B, Malinverni R, Moradpour D, Müllhaupt B, Witteck A, Beckmann JS, Berg T, Bergmann S, Negro F, Telenti A, Bochud PY; Swiss Hepatitis C Cohort Study; Swiss HIV Cohort Study. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138(4):1338-45.
Footnotes
Conflicts of interest
The authors declare no conflicts.
References
- 1.Jessner W, Gschwantler M, Steindl-Munda P, et al. Primary interferon resistance and treatment response in chronic hepatitis C infection: a pilot study. Lancet. 2001;358:1241–1242. doi: 10.1016/S0140-6736(01)06356-5. [DOI] [PubMed] [Google Scholar]
- 2.Veldt BJ, Heathcote EJ, Wedemeyer H, et al. Sustained virologic response and clinical outcomes in patients with chronic hepatitis C and advanced fibrosis. Ann Intern Med. 2007;147:677–684. doi: 10.7326/0003-4819-147-10-200711200-00003. [DOI] [PubMed] [Google Scholar]
- 3.Thomas DL, Astemborski J, Rai RM, et al. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284:450–456. doi: 10.1001/jama.284.4.450. [DOI] [PubMed] [Google Scholar]
- 4.Kenny-Walsh E. Clinical outcomes after hepatitis C infection from contaminated anti-D immune globulin. Irish Hepatology Research Group. N Engl J Med. 1999;340:1228–1233. doi: 10.1056/NEJM199904223401602. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Torres M, Jeffers LJ, Sheikh MY, et al. Peginterferon alfa-2a and ribavirin in Latino and non-Latino whites with hepatitis C. N Engl J Med. 2009;360:257–267. doi: 10.1056/NEJMoa0805062. [DOI] [PubMed] [Google Scholar]
- 6.Jeffers LJ, Cassidy W, Howell CD, et al. Peginterferon alfa-2a (40 kd) and ribavirin for black American patients with chronic HCV genotype 1. Hepatology. 2004;39:1702–1708. doi: 10.1002/hep.20212. [DOI] [PubMed] [Google Scholar]
- 7.Hoofnagle JH, Wahed AS, Brown RS, Jr, et al. Early changes in hepatitis C virus (HCV) levels in response to peginterferon and ribavirin treatment in patients with chronic HCV genotype 1 infection. J Infect Dis. 2009;199:1112–1120. doi: 10.1086/597384. [DOI] [PubMed] [Google Scholar]
- 8.Thio CL, Gao X, Goedert JJ, et al. HLA-Cw*04 and hepatitis C virus persistence. J Virol. 2002;76:4792–4797. doi: 10.1128/JVI.76.10.4792-4797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thio CL, Thomas DL, Goedert JJ, et al. Racial differences in HLA class II associations with hepatitis C virus outcomes. J Infect Dis. 2001;184:16–21. doi: 10.1086/321005. [DOI] [PubMed] [Google Scholar]
- 10.Khakoo SI, Thio CL, Martin MP, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 11.Thursz M, Yallop R, Goldin R, et al. Influence of MHC class II genotype on outcome of infection with hepatitis C virus. The HENCORE group. Hepatitis C European Network for Cooperative Research. Lancet. 1999;354:2119–2124. doi: 10.1016/s0140-6736(99)91443-5. [DOI] [PubMed] [Google Scholar]
- 12.Thio CL, Goedert JJ, Mosbruger T, et al. An analysis of tumor necrosis factor alpha gene polymorphisms and haplotypes with natural clearance of hepatitis C virus infection. Genes Immun. 2004;5:294–300. doi: 10.1038/sj.gene.6364072. [DOI] [PubMed] [Google Scholar]
- 13.An P, Thio CL, Kirk GD, et al. Regulatory polymorphisms in the interleukin-18 promoter are associated with hepatitis C virus clearance. J Infect Dis. 2008;198:1159–1165. doi: 10.1086/592047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su X, Yee LJ, Im K, et al. Association of single nucleotide polymorphisms in interferon signaling pathway genes and interferon-stimulated genes with the response to interferon therapy for chronic hepatitis C. J Hepatol. 2008;49:184–191. doi: 10.1016/j.jhep.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welzel TM, Morgan TR, Bonkovsky HL, et al. Variants in interferon-alpha pathway genes and response to pegylated interferon-Alpha2a plus ribavirin for treatment of chronic hepatitis C virus infection in the hepatitis C antiviral long-term treatment against cirrhosis trial. Hepatology. 2009;49:1847–1858. doi: 10.1002/hep.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yee LJ, Tang J, Gibson AW, et al. Interleukin 10 polymorphisms as predictors of sustained response in antiviral therapy for chronic hepatitis C infection. Hepatology. 2001;33:708–712. doi: 10.1053/jhep.2001.22347. [DOI] [PubMed] [Google Scholar]
- 17.Mosbruger T, Duggal P, Goedert JJ, et al. Large-scale candidate gene analysis of spontaneous hepatitis C virus clearance. JID. doi: 10.1086/651606. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 19.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rauch A, Kutalik Z, Descombes P, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 21.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 23.Ge D, Fellay J, Thompson AJ, Simon JS, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 24.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibbs RA, Belmont JW, Hardenbol P, et al. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 26.Larkin J, Jin L, Farmen M, et al. Synergistic antiviral activity of human interferon combinations in the hepatitis C virus replicon system. J Interferon Cytokine Res. 2003;23:247–257. doi: 10.1089/107999003321829962. [DOI] [PubMed] [Google Scholar]
- 27.Marcello T, Grakoui A, Barba-Spaeth G, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 28.Gad HH, Dellgren C, Hamming OJ, et al. Interferon-lambda is functionally an interferon but structurally related to the interleukin-10 family. J Biol Chem. 2009;284:20869–20875. doi: 10.1074/jbc.M109.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, Sun J, Meng L, et al. ISG15, a ubiquitin-like interferon stimulated gene, promotes Hepatitis C Virus production in vitro: Implications for chronic infection and response to treatment. J Gen Virol. 2010;91:382–388. doi: 10.1099/vir.0.015388-0. [DOI] [PubMed] [Google Scholar]
- 31.Jordan WJ, Eskdale J, Srinivas S, et al. Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun. 2007;8:254–261. doi: 10.1038/sj.gene.6364382. [DOI] [PubMed] [Google Scholar]