Abstract
Sand fly and Leishmania are one of the best studied vector-parasite models. Much is known about the development of these parasites within the sand fly, and how transmission to a suitable vertebrate host takes place. Various molecules secreted by the vector assist the establishment of the infection in a vertebrate, and changes to the vector are promoted by the parasites in order to facilitate or enhance transmission. Despite a generally accepted view that sand flies and Leishmania are also one of the oldest vector-pathogen pairs known, such long history has not been translated into a harmonic relationship. Leishmania are faced with many barriers to the establishment of a successful infection within the sand fly vector, and specific associations have been developed which are thought to represent aspects of a co-evolution between the parasite and its vectors. In this review, we highlight the journey taken by Leishmania during its development within the vector, and describe the issues associated with the natural barriers encountered by the parasite. Recent data revealed sexual replication of Leishmania within the sand fly, but it is yet unknown if such reproduction affects disease outcome. New approaches targeting sand fly molecules to prevent parasite transmission are being sought, and various techniques related to genetic manipulation of sand flies are being utilized.
INTRODUCTION
Sand flies (Diptera: Psychodidae: Phlebotominae) are the main vectors of leishmaniasis, a multi-spectrum disease ranging from self-healing skin lesions to fatal visceral illness. Approximately 40 species of Leishmania have been described [1], and different species are associated with distinct disease outcomes. In parallel, over 900 species and subspecies of sand flies have been identified to date (from a recent compilation of all sand fly species, E. Galati personal communication), but only a limited number have been proven or incriminated as vectors of Leishmania [1]. A specific relationship exists between sand flies and Leishmania such that, in nature, only certain species of sand flies are able to transmit certain species of Leishmania [2]. This species-specificity is driven by several molecular factors that allow the parasite to infect, survive and multiply within the midgut of the sand fly and be transmitted to a suitable vertebrate host during a blood meal. Some sand fly species are considered permissive (or non specific) as they are able to harbor experimental infections of several Leishmania species (e.g. Lutzomyia longipalpis and Leishmania (L.) infantum chagasi or Leishmania (L.) mexicana); other sand fly species are considered restrictive (or specific) as they only can be infected with the Leishmania species that they carry in nature (e.g. Phlebotomus papatasi and Leishmania major). However, the precise interactions that lead to this vectorial capacity in sand flies, whether for permissive or restrictive vectors, remain to be fully elucidated.
During its life within the sand fly vector, Leishmania undergo a complex developmental process, mainly restricted to the midgut, in which the parasites must overcome several documented difficulties. Both parasite and vector achieve a balance which involves significant interplay. In one end, for its survival, Leishmania strives to manipulate aspects of the physiology of the sand fly, interfering with digestive proteases [3–5], secreting a myoinhibitory peptide that arrests hindgut peristalsis [6] and causing significant damage to the stomodeal valve (or cardia) of the fly [7, 8].
In contrast, sand flies probably recognize the presence of Leishmania and likely mount an immune response to the infection [9]. In L. longipalpis, defensin, glycine-rich proteins (GRP), which are transcripts associated with the innate immunity of insects, are up-regulated following a blood meal [9]. Serine protease inhibitors (serpins) also are up-regulated following a blood meal and possibly also by Leishmania [9], while digestive enzymes may be regulated at the level of transcription and/or activity by the parasite [10, 11]. Sand flies also seem to induce programmed cell death (apoptosis) of midgut cells following infection [9, 12]. Apoptosis is an innate defense mechanism in insects and known to be used by mosquitoes to eliminate Plasmodium-infected midgut cells [13]. Although Leishmania do not invade the midgut cells, close contact between the parasites and these cells i.e., adhesion to epithelial cells through the parasite surface, is well documented [14]. Thus, despite their long evolutionary history (fossil records indicate the presence of flagellates, possibly trypanosomatids, within sand flies in the Early Cretaceous [15]) the relationship between Leishmania and sand flies can be considered an active and intense evolutionary arms-race. In this review we highlight some of the most important events during the development of Leishmania in a suitable sand fly vector, and discuss issues associated with Leishmania-sand fly specificity, metacyclogenesis, and sand fly midgut responses to Leishmania. Additional themes related to current and future strategies to prevent transmission of Leishmania parasites targeting the vector are also discussed.
THE INFECTION PROCESS
The infection of sand flies with Leishmania starts upon the release of amastigotes from infected macrophages ingested by the fly during the blood meal, and culminates with the injection of metacyclic promastigotes into a vertebrate host during the fly’s next attempt to blood feed. The approximate 4–5 days that will take Leishmania to develop from amastigotes to infective metacyclics are met with many formidable barriers that the parasite must overcome in order to survive and establish an infection in the sand fly (Figure 1). Digestive proteases pose the first and one of the most significant barriers to the parasite survival. Within the first 6–12h following the infection, most Leishmania are killed, possibly by the action of such proteases. Parasites in the transitional stage between amastigotes and promastigotes (procyclic) are most vulnerable to this proteolytic attack, and become less susceptible as they fully mature into the next stage of development, the procyclic promastigote. This was demonstrated by assessing L. major survival after P. papatasi artificial feeding on blood containing exogenous chitinase [17], which led to the absence of the peritrophic matrix (PM), and prevented parasites from colonizing the midgut. A faster diffusion of digestive enzymes killing the Leishmania before they were able to transform into the more resistant promastigote forms was likely the reason. This was confirmed by the addition of protease inhibitors to the blood meal, which allowed the parasites to survive in the midgut [17]. Further support for the role of digestive enzymes in Leishmania killing was obtained using P. papatasi-L. donovani (a non-natural parasite-sand fly pair) [3]. In this case, only 5% of parasites survived within the midgut when whole blood was used in artificial feeding, but this number increased to 70% when blood was combined with trypsin inhibitors or when the insects were fed erythrocytes plus saline.
Figure 1.
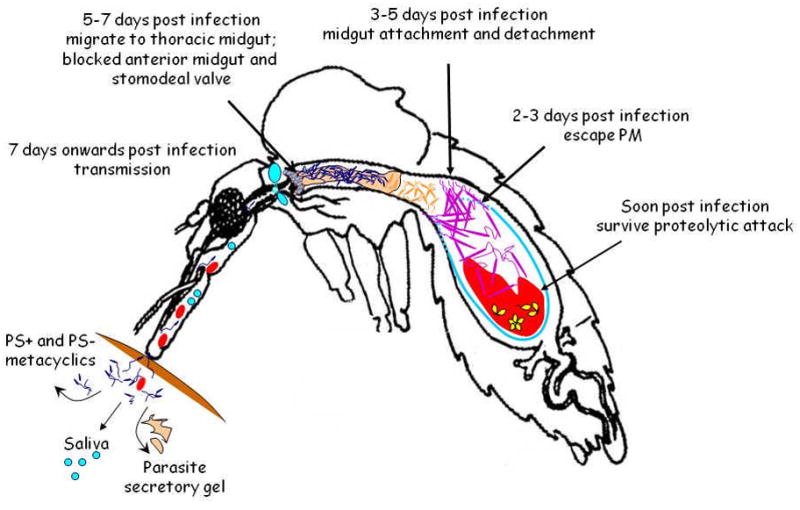
Leishmania life cycle within the sand fly vector midgut (modified from [16]). Barriers faced by Leishmania during its development are illustrated. The first barrier is mounted by a proteolytic attack by digestive proteases secreted immediately following a blood meal. A second barrier is posed by the peritrophic matrix (PM) from which parasites must escape, and believed to be accomplished between days 2 and 3 post infection (PI). The third barrier is the required midgut attachment to prevent excretion of parasites with the remnants of the blood meal, which in general occurs 3–5 days PI. Finally, parasites must detach from the midgut and migrate towards the thoracic midgut. Secretion of parasite secretory gel (PSG) together with a destruction of the stomodeal valve produces the “blocked” sand fly. Blocked sand flies are unable to complete a full blood meal and thus attempt to feed with greater frequency, increasing the chance of parasite transmission. Metacyclics either expressing (PS+) or not expressing (PS−) phosphatidylserine are found within the sand fly and are probably injected in the mammalian host. Both forms are essential for lesion development (see text).
In Leishmania, protection against proteolytic attack has been attributed to phosphoglycan (PG)-containing molecules, either released or surface-bound (reviewed in [2]), possibly acting through the inhibition of the proteolytic enzymes released in the sand fly midgut following a blood meal. Additionally, secreted PG-containing products, by virtue of their negative charge, might protect the promastigote by acting as a transient barrier against digestive enzymes in the vicinity of the parasite [18], and the surface LPG might further protect the cell from the digestive proteases. Nevertheless, these hypotheses remain to be tested.
Enzymes such as trypsins and chymotrypsins are responsible for the bulk of the blood digestion in sand flies. Considering the importance of these enzymes in digestion and parasite survival, trypsin and chymotrypsin-like expression and activity were investigated during sand fly infection with Leishmania [3–5]. Both the type of meal ingested and the presence of Leishmania were linked to distinct levels of proteolytic enzymes in P. papatasi [3, 5], and the authors suggested that a specific component of the trypsin-like activity within the P. papatasi gut prevents L. donovani from surviving in this vector. In contrast, the ability of L. major to survive and thrive was correlated with the modulation of such component [5]. Accordingly, the use of trypsin inhibitor enabled L. donovani to successfully infect P. papatasi [3, 5].
Similar experiments [11] showed significant decrease in activity of alkaline protease, trypsin and aminopeptidase in P. papatasi infected by L. major. However, in the non-natural P. langeroni-L. major pair the situation was somewhat different, with an apparent delay of the (blood) digestion process, and persistence of high levels of digestive enzymes. Extending the digestion period might bring some advantage for the survival of the parasites. Serine protease inhibitors (SPI) with inhibitory effect over vertebrate macrophage serine proteases, such as neutrophil elastase, trypsin and chymotrypsin were found in L. major [19]. The possibility raised by these authors, of SPI having an effect on insect midgut proteases, remains to be demonstrated.
For L. longipalpis, there is less information on the correlation of parasite presence and midgut enzymatic activity, although new data suggest a reduced trypsin activity related to the presence of L. i. chagasi ([20]). Also, inhibition of trypsin expression in L. longipalpis through RNAi caused an increased survival of Leishmania in the vector [21].
Recent studies have relied on assessment of transcription profiles of digestive proteases in sand flies. In L. longipalpis, the transcription pattern of two trypsin genes was determined. One (Lltryp2) showed a constitutive transcription pattern, only marginally reduced when blood ingestion occurred, while the other (Lltryp1) was induced by blood intake, with a peak of transcription at 12h to 24h after blood feeding. Although differences were not observed (by RT-PCR) in the expression profiles of both transcripts at 72h after a blood meal containing L. i. chagasi [22], Lltryp2 expression differ between infected and non infected flies 5 to 7 days post blood digestion by comparing transcripts from cDNA libraries [23]. Modulation of expression before and after completion of the blood digestion is presently being investigated. For P. papatasi, proteases have been characterized [24] but studies to show the effect of infective blood meals on the transcription of these enzymes are yet to be performed.
Modulation of transcription of digestive enzymes genes was reported through extensive EST sequencing of libraries constructed from blood fed and Leishmania infected sand flies. In cDNA libraries from L. i. chagasi-infected L. longipalpis, transcripts of Lltryp1, a blood-modulated trypsin gene, were less abundant while transcripts for a constitutive trypsin (Lltryp2) were more abundant in relation to libraries from uninfected flies [23]. In P. papatasi, a down regulation was observed for the chymotrypsin transcript PpChym2, a gene expressed only following a blood meal, in a cDNA library constructed from L. major infected flies (Table 1 and [10]). Increased expression for the trypsin PpTryp1 also was detected in the same library (Table 1 and [10]).
Table 1.
P. papatasi selected midgut transcript abundance.
| Transcript name | Putative function | Blood-fed P. papatasi | L. major-infected P. papatasi | P value |
|---|---|---|---|---|
|
| ||||
| PpMVP1 | Microvilli protein | 134 | 70 | 5.8×10−7 |
| PpMVP2 | Microvilli protein | 60 | 42 | 4.1×10−2 |
| PpPer1 | Peritrophin | 54 | 16 | 1.7×10−6 |
| PpPer2 | Peritrophin | 45 | 35 | 1.8×10−2 |
| PpFLC | Ferritin light chain | 18 | 3 | 7.1×10−4 |
| PpChym2 | Chymotrypsin | 36 | 8 | 1.1×10−5 |
| PpTryp1 | Trypsin | 10 | 82 | 1.0×10−13 |
Interestingly, the sand fly peritrophic matrix (PM) which constitutes the second barrier to Leishmania development (see below), also serves to protect the parasites from an onslaught of digestive proteases [25]. Leishmania that survive the proteolytic attack must exit the endoperitrophic space to continue their development. At the end of blood digestion, the PM is probably degraded by a midgut-secreted chitinase, as suggested by data demonstrating that the transcription of chitinase genes is modulated by blood ingestion both in L. longipalpis [26] and P. papatasi [27]. Chitinases, both from the insect and the parasite, seem to be important molecules involved in many aspects of the sand fly-Leishmania interaction. Escaping from the endoperitrophic space, a necessary step for Leishmania attachment to the sand fly midgut epithelia was shown to be aided by a parasite secreted chitinase. Histological sections of infected flies provided evidence for the degradation of the chitin layer in the anterior region of the PM that allowed the forward migration of a large number of parasites [28], which was attributed to the parasite chitinase activity. Later work identified genes coding for secreted chitinases in many Leishmania species [29, 30]. One such gene was identified and overexpressed in L. mexicana [31] used to infect L. longipalpis. Overexpression of this L. mexicana chitinase within the midgut of the sand fly allowed the parasites to escape from the PM and arrive at the stomodeal valve sooner than wild type parasites [32]. It also led to an increased number of parasites per fly. Nevertheless, the fact that a significant chitinolytic activity was later detected within sand fly midguts following a blood meal [27] also suggests that Leishmania may take advantage of this natural physiological condition of sand flies in order to trespass the PM. The coincidence of the timing of escape with high levels of chitinolytic activity in the sand fly gut is also relevant. Thus, it is possible that the parasite escape from the PM is facilitated by the effects of the two chitinases (from the Leishmania and from the sand fly) acting upon the chitin fibers of the PM. Such action may be synergistic, but formal demonstration of such an effect is still required.
Following the crossing of the PM, the next step for Leishmania is to attach to the midgut epithelia of the sand fly when the blood meal remains are passed to the hindgut at the completion of the digestive process. Attachment can be promoted by LPG, as in the case of restrictive vectors such as P. papatasi, or via a LPG-independent mechanism [33], discussed below.
As the last step in the development of the parasites and prior to the sand fly attempting to feed on another host, metacyclic Leishmania migrate to the thoracic midgut area of the stomodeal valve. According to observations [28], chitinases secreted by Leishmania also attack the lining of this valve, which contains chitin, irreparably damaging the valve impeding its proper function.
Additionally, the parasite secretory gel (PSG) forms a plug that prevents or significantly reduces the influx of blood into the midgut [34, 35]. Combined, the PSG plug and the lack of a functioning stomodeal valve create a mechanism that facilitates expelling (or regurgitation) of parasites located in front of or within the plug onto the skin of the vertebrate host. It was suggested that another round of parasite replication takes place following the parasite migration towards the stomodeal valve area [36]. In this case, leptomonad forms of Leishmania multiply and develop into more metacyclics, increasing the number of infectious forms behind the PSG plug, and enhancing the plug’s effect on the blocked fly.
METACYCLOGENESIS
Leishmania undergoes a differentiation process within the sand fly vector from non-infective to infective promastigotes, a phenomenon known as metacyclogenesis. Amastigotes taken up by the sand fly during a blood meal transform into the procyclic promastigotes, a motile form that multiplies in the blood contained within the PM. Procyclics then develop into nectomonads, a large and slender form that crosses the PM, adheres to the gut epithelia and migrates towards the anterior gut. Nectomonads transform into leptomonads (smaller and dividing forms), that secrete PSG. At the stomodeal valve, haptomonads (precursor unknown), small leaf-like forms, are found adhered to the epithelia. Finally, metacyclics (short and slender cell body, with a flagellum twice the size of the body that confers its characteristic rapid motility) differentiate from leptomonads [16, 37–39].
Metacyclogenesis, occurring either in the sand fly gut or in sterile culture, is associated with morphological alterations, and also with regulated biochemical changes. One of the best described modifications occurs at the level of the lipophosphoglycan (LPG), an abundant glycoconjugate expressed on the promastigote surface membrane, which plays key roles in the host–parasite interactions. LPG is expressed by all Leishmania species and is composed of four domains: (a) a 1-O-alkyl-2-lysophosphatidylinositol lipid anchor linked to (b) a heptasaccharide core; (c) a long phosphoglycan polymer of Gal(β1,4)Man(α1)-PO4 repeating units; and (d) a cap oligosaccharide structure. While the lipid anchor and saccharide core are conserved between species, there are intra- and interspecies polymorphisms in the disaccharide repeating units and in the cap structure [2, 39]. Variations in the LPG side chains linked to the Gal residues vary from almost completely substituted by a variety of saccharides as in L. major and L. tropica, to un-substituted as in L. donovani from Sudan and L. i. chagasi, or partially substituted as in L. mexicana and L. donovani from India (reviewed in [39]).
Metacyclics can be purified from in vitro stationary culture or from sand fly gut by specific lectins and/or monoclonal antibodies according to their LPG composition, or by a LPG-independent Ficoll gradient [40, 41]. These forms can be also detected in a mixed population by flow cytometry [42].
In the best characterized metacyclogenesis model, the L. major Friedlin strain, LPG undergoes several molecular modifications initially detected using peanut lectin and stage-specific monoclonal antibodies [43, 44]. Metacyclic LPG increases its length by 2 to 3 fold due to an increment in the number of phosphorylated saccharide repeated units; it also changes its side-chain terminal carbohydrates composition by capping the terminal βgalactose with arabinose residues [45]. Interestingly, the galactose capping allows the release of parasites tethered to the midgut by the LPG interaction with a P. papatasi galectin [46]. Also, the size increment of metacyclic LPG correlates with an increase in the thickness of the L. major surface glycocalyx [47]. Remarkable heterogeneity has been reported in the nature of carbohydrate substitutions in LPG of L. major from different countries, probably reflecting the pattern of galactosyl transferases expression [39, 48].
A slightly different mechanism was described for LPG alterations during metacyclogenesis of the Sudanese L. donovani, in which a similar elongation in the phosphoglycan chain of metacyclic LPG increases the thickness of the metacyclic glycocalyx. However, procyclic and metacyclic LPGs are structurally similar, expressing the same sugars. Clustering and folding of the terminal capping carbohydrates of the elongated L. donovani LPG may be responsible for the loss of metacyclic binding to the insect midgut [49]. This same folding masks sugars recognized by lectins, allowing negative selection of metacyclic forms [49]. In procyclic LPG from an Indian strain of L. donovani, side-chains and cap structures consist of β1,3-linked glucose residues, which are down regulated in the elongated chain of the metacyclic LPG resulting in failure of the metacyclics to adhere to P. argentipes midgut [50]. Likewise, metacyclogenesis of L. i. chagasi, the agent of visceral leishmaniasis in the New World, is also associated with lack of β1,3-linked glucose residues resulting in loss of metacyclic’s ability to bind L. longipalpis midgut [51].
It was recently demonstrated that L. braziliensis LPG elongates during metacyclogenesis, and that procyclic LPG lacks side chain sugar substitutions [52]. However, in contrast to LPG of other Leishmania species, metacyclics of L. braziliensis make less LPG and add β1,3-glucose residues that branch off the disaccharide-phosphate repeat units of LPG. Metacyclics of L. braziliensis are selectively agglutinated by the mannose specific lentil lectin [53], and are not recognized by Bauhinia purpurea lectin, which has affinity by β-Gal(1,3)-GalNAc residues [54].
These inter- and intra-specific structural polymorphisms in the LPG molecule are probably responsible for the sand fly vectorial competence for different Leishmania species, mediating attachment to the midgut epithelium, and, in this way, avoiding excretion of the parasite with the remnants of the digested blood meal [16, 39]. Evidence supporting this role was reported in studies in vitro using explanted midguts, demonstrating that procyclic LPG binds to the epithelia and inhibits the attachment of procyclic promastigotes [46, 55, 56]. These studies were later confirmed with the use of mutants deficient in LPG expression, which fail both to adhere to explanted midguts in vitro, as well as to persist in vivo after the blood meal elimination [57]. In a seminal work, a galectin expressed in the midgut of P. papatasi and P. duboscqi (two restrictive sand flies) was identified as a receptor for L. major procyclic LPG, supporting the vectorial capacity idea [46].
Although LPG expression explains some of the restrictive vectorial capacity (such as in the L. major-P. papatasi pair), LPG-independent interactions were reported associated with permissive sand fly species, including L. longipalpis in the New World and P. arabicus in the Old World [33, 58]. One interesting possibility is that permissive sand fly species express more than one midgut receptor able to bind different glycoconjugates from the parasite cell surface.
Interestingly, phosphoglycan repeat units similar to those from LPG are present in other promastigote molecules, such as proteophosphoglycan and acid phosphatase, but the role of these molecules has not been investigated during metacyclogenesis.
Although the modifications that occur in the LPG of some Leishmania species during metacyclogenesis are well described, the signals that trigger these changes are unknown. Similarly, the role each morphological form of the parasite plays in the development of metacyclics is puzzling. LPG and other morphological changes occur upon parasite’s sensing and transduction of stimuli from the surrounding microenvironment. Thus, signals for promastigote differentiation include acidic pH [59], decreased levels of tetrahydrobiopterin [60], absence of hemoglobin [61] or oxygen [62], and exposure to sand fly saliva [63]. It is possible that some or all of these stimuli act in concert to promote metacyclogenesis within the vector. In vitro, metacyclogenesis is associated with autophagy, since a mutant that lacks a functional autophagic pathway, is unable to differentiate into metacyclic [64]. Indeed, nutritional starvation and high population density seem to occur in the infected sand flies, and these two conditions are strong inducers of autophagy [64]. Since autophagic and apoptotic cell death mechanisms can be redundant, it is interesting to note that phosphatidylserine (PS), an apoptotic marker, is expressed by metacyclics during their development within the sand fly [65]. Apoptotic, annexin V-positive metacyclics increase in the sand fly gut over time following infection and tunnel-positive parasites are seen associated with the stomodeal valve, suggesting that PS exposure is part of the metacyclic differentiation process. Interestingly, mammalian host infection occurs only when the metacyclic population is composed by PS-positive and PS-negative forms (Figure 1), and the former is essential for the infectivity of the latter [65, 66].
During metacyclogenesis, the parasite’s cell body grows to its maximum length (reached during the nectomonad stage), and then shrinks to the slender, shorter metacyclic. In contrast, the flagellum positioned in the anterior extremity of the parasite grows continually, reaching its maximum length in the metacyclic stage. Flagellum movement propels the parasite forward, and is the first parasite structure to contact the sand fly midgut epithelia. The importance of the flagellum for parasite adhesion to the midgut was suggested because an L. amazonensis mutant with a short flagellum is excreted from L. longipalpis together with the undigested blood meal [67]. Elimination of this mutant might be due to its inability to move and bind to midgut receptors after crossing the PM. However, considering the extremely short distance between the PM and midgut epithelium, it is unclear why these mutants are unable to reach and bind to the epithelial cell receptors after escaping the PM. Since this mutant expresses normal levels of LPG, the LPG-midgut receptor interaction is probably not sufficient to hold the promastigotes and, thus, prevent its excretion. In Leishmania promastigotes, LPG is expressed throughout the flagellar and cell body membranes [68]. Nevertheless, it seems that flagellar insertion in the epithelial microvilli is also important to avoid parasite excretion.
Recent results show that the flagellum is critical not only for parasite motility, but also for flagellar pocket organization, exo- and endocytosis, and cell size regulation and division [69]. The idea of a flagellum functioning as a sensory organ was proposed based on the flagellar localization and function of proteins such as, kinesin, thymidine kinase and mitogen-activated protein (MAP) kinase [69]. Indeed, since promastigotes move from the midgut towards the anterior end of the sand fly during metacyclogenesis and the flagellum in the anterior portion of the parasite, the hypothesis that the flagellum touches, senses and is responsible for signal transduction is very appealing.
TRANSMISSION OF LEISHMANIA TO THE MAMMALIAN HOST
The number of metacyclics inoculated during the sand fly bite was first estimated to range from 103 to 104 using an artificial feeding system [58, 70]. With a natural transmission model using P. duboscqi and L. major (naturally occurring sand fly-Leishmania pair) new data suggest a much wider range from 10 to 100,000 parasites deposited per sand fly bite into mouse ears [71]. Here, high-dose parasite inocula correlated with larger, more severe lesions, which developed earlier, whereas low-dose inocula produced smaller lesions that developed later. These results suggest that parasite inoculation dose can be a potential factor determining the clinical outcome of Leishmania infection [71].
Leishmania metacyclics are inoculated into the skin together with the sand fly saliva (Figure 1), and in a pioneering work, it was demonstrated that saliva, besides circumventing the host haemostatic system, also enhanced Leishmania infection [72]. Thus, suggesting for the first time that immunization with saliva could protect against parasite infection. This possibility has been proven feasible in subsequent works [73–78]. Besides saliva, metacyclics are also inoculated with the promastigote secretory gel (PSG) produced by the parasites (Figure 1). PSG obstructs the stomodeal valve facilitating metacyclic transmission, and also exacerbates the disease in the mammalian host [58]. Considering the complexity of metacyclogenesis and Leishmania transmission, further studies are needed to understand how saliva, PSG, metacyclic phosphatidylserine expression, and/or inoculums load contribute for disease development. Most likely, a redundancy of all these factors is relevant for the establishment of leishmaniasis.
GENETIC EXCHANGE IN LEISHMANIA
One of the most significant recent findings regarding Leishmania development was the demonstration of chromosomal exchange in L. major during development within the sand fly midgut. The ability to obtain sexual crosses of Leishmania will provide a significant step forward on the genetics of these parasites, and is an important tool to identify relevant genes in the parasite that may impact pathogenesis. Nevertheless, important questions as to when during parasite development within the sand fly midgut such mating between Leishmania occurs, and why were these genetic hybrids only detected within the sand flies and not in the vertebrate remain unanswered.
Previous studies indicated the presence of L. major-L. infantum hybrids in P. papatasi [79, 80], suggesting that hybrids (and sex) are fairly common within Leishmania parasites. This being the case, perhaps the presence of hybrid forms would in part account for some of the different outcomes observed for the disease. These novel findings regarding how Leishmania can exchange chromosomes even between distinct species should bring renewed interest regarding field samples of infected sand flies from endemic areas. Beyond the realization that at least in L. major chromosome exchange does occur, the formation of hybrids through genetic crosses can be used to identify parasite genes important for development within the sand fly vector. These studies will likely be facilitated by our current knowledge of this parasite’s genome.
COMPARATIVE TRANSCRIPTOME ANALYSES AND FUNCTIONAL GENOMICS IN SAND FLIES
Studies on differential expression in sand fly midguts were first performed by comparing L. longipalpis midguts from flies that were either sugar fed, blood fed or infected with L. i. chagasi [81]. These studies led to the identification and characterization of several transcripts that are up-regulated during blood feeding and possibly linked to parasite presence. More recently, greater throughput transcriptome analyses of sand flies have produced relevant data towards the understanding of the interplay between the sand fly vector and the infecting Leishmania [9, 10, 23, 82, 83]. Among such findings were the characterization of several molecules, including some associated with specific sand fly-Leishmania pairs, such as PpGalec [46]. Moreover, these analyses also provided significant clues on the gene expression within the sand fly midgut. For instance, in P. papatasi, transcripts coding for midgut proteins believed to participate in some aspect of L. major development, and only expressed following a blood meal, were identified [10]. Subsequent comparative transcriptome analyses of sugar fed, blood fed and L. i. chagasi-infected L. longipalpis led to similar conclusions regarding midgut transcripts regulated by blood feeding [9, 23]. Among the most abundant midgut transcripts identified in the studies done with P. papatasi and L. longipalpis were transcripts coding for microvilli associated proteins, peritrophins, chymotrypsin, trypsin and others.
Table 1 (modified from [10]) summarizes the data obtained through comparative transcriptome analyses of P. papatasi cDNA libraries established from midguts dissected from flies that artificially fed on blood or on blood infected with L. major parasites (4×106 amastigotes/ml). Columns 1 and 2 represent the transcripts identified and their putative function based on sequence data analyses. Numbers in columns 3 and 4 indicate the number of transcripts for each of the contigs listed found in the two cDNA libraries analyzed. P values were determined for the differences found in transcript abundance per treatment (non-infected vs. L. major-infected). P. papatasi sand flies were artificially fed on non-infected (column 3) or L. major-infected (column 4) mouse blood.
Many transcripts identified from such studies appear to be regulated during the infection with Leishmania (Tables 1 and 2), and thus may be directly involved in parasite development. More importantly, these analyses also suggested that similar transcripts (orthologs or paralogs) from these two distantly related sand fly species were regulated in a similar fashion by the parasites used to infect them, possibly indicating similar strategies used by Leishmania to overcome barriers of defense mechanisms put forth by the sand flies. The data collected from the comparative transcriptome analyses of sand flies is now being applied towards reverse genetic studies of these vectors.
Table 2.
L. longipalpis selected midgut transcript abundance.
| Transcript name | Putative function | Blood-fed L. longipalpis | L. chagasi-infected L. longipalpis | P value |
|---|---|---|---|---|
| LuloMVP1 | Microvilli protein | 109 | 55 | 2.0×10−4 |
| LuloMVP2 | Microvilli protein | 87 | 40 | 2.0×10−4 |
| LuloMVP3 | Microvilli protein | 24 | 7 | 4.9×10−3 |
| LuloMVP4 | Microvilli protein | 60 | 27 | 1.6×10−3 |
| LuloPer1 | Peritrophin | 6 | 22 | 1.0×10−3 |
| Lltryp2 | Trypsin | 6 | 15 | 2.9×10−2 |
| LuloChym1A | Chymotrypsin | 51 | 22 | 2.4×10−3 |
| LuloCpepA1 | Carboxypepti dase | 14 | 3 | 1.3×10−2 |
Studies on sand fly-Leishmania interaction are beginning a new chapter with significant insights achieved through the application of various molecular techniques to identify and characterize sand fly molecules possibly involved in Leishmania development. Functional (reverse genomics) studies on sand flies utilizing RNAi are beginning to take hold. From studies performed using sand fly cell lines [84], as well as from the identification of cDNA transcripts [9], it is clear that the RNAi machinery is present and functional in sand flies. Recently, xanthine dehydrogenase (XDH) was knocked down in L. longipalpis [85] causing a 40% reduction of XDH transcripts and a reduction in the levels of urate. The dsRNA targeting XDH also led to a reduction of the life span of the flies. Also, treatment of adult L. longipalpis with dsRNA for trypsin caused a decreased in the expression and activity of this digestive enzyme and increased the survival of L. mexicana [21].
RNAi effects may vary depending on the targeted transcript. Sand flies RNAi silencing is achievable whether transcripts are expressed in sugar fed flies, or if the transcripts are only expressed after blood feeding (MRO unpublished observations). The ability to obtain viable sand flies with targeted transcript reduction (and reduced protein level) will provide an important tool to address questions associated with the physiology of sand flies, prior and after a blood meal. Such approaches will also contribute towards assessing the role of sand fly molecules in Leishmania development within the vector.
TRANSLATIONAL RESEARCH AND FUTURE APPLICATIONS
As with other disease vectors, sand fly molecules that may directly or indirectly interfere with Leishmania development are being screened for their potential application as transmission blocking vaccines (TBV). Currently, several P. papatasi midgut molecules that are modulated by or associated with Leishmania development within the sand fly vector are under investigation by different groups. TBV against Leishmania was first proposed for PpGalec, a L. major LPG receptor expressed in the midgut of P. papatasi [46]. Blockage of PpGalec using specific antisera led to a 10 fold reduction of L. major in the gut of P. papatasi. Thus, targeting sand fly molecules that are directly or indirectly related to Leishmania within the sand fly vector may lead to the disruption of parasite development preventing transmission of infective metacyclics, or to the elimination of parasites during sand fly defecation after the blood meal is digested.
Recently, Leishmune® a vaccine against L. i. chagasi based on the L. donovani fucose-mannose ligand (FML) in combination with saponin (adjuvant) was shown to prevent infection of sand flies, thus functioning as a TBV [86]. Although the surface molecule FML was isolated from L. donovani, Leishmune® exhibited transmission blocking activity in the New World sand fly L. longipalpis when infected with L. i. chagasi [86], suggesting that cross-species effects from TBVs (either pathogen-based or insect-based) are possible. Concurrent with this idea, mouse antisera raised against the P. papatasi midgut chitinase PpChit1 reduced midgut chitinolytic activity in vitro from this vector, but also, from P. duboscqi and P. argentipes [27]. Moreover, the antibodies produced in dogs following Leishmune® injection reduced L. longipalpis infectivity by 79.3% and parasite load by 74.3% even after 12 months of immunization [86], pointing to the possible long term effects of TBVs.
The use of sand fly midgut symbionts to express molecules targeting Leishmania is being considered. The use of symbiotic organisms such as bacteria from disease vectors to express molecules toxic to pathogens they transmit are collectively referred to as paratransgenesis, and was first demonstrated in triatomines [87]. For sand flies, midgut bacterial symbionts have been identified in New and Old World vectors [88–90], and the data suggest that the adult sand fly midgut microbiota vary according to the substrate in which the larvae develop [88]. Moreover, insect midgut microbiota may also directly influence how a disease vector respond to infection with a pathogen [91]. Whether sand fly symbionts affect Leishmania development is not clear. Other symbiotic bacteria such as Wolbachia also have been selected based on their capacity to reduce the lifespan of disease vectors, thus reducing vector competence without the use of molecular genetics techniques [92, 93]. Potentially, such Wolbachia strains may also be used on sand flies to reduce disease transmission.
Regarding direct genetic manipulation (or vector transgenesis) of sand flies, to date no genetically modified sand fly line is available. Towards this, heterologous promoters have been identified using sand fly cell lines [94], and with the significant advancement of our understanding of gene expression and the characterization of many sand fly genes, some of which presenting tissue specific expression, transgenesis may soon be realized for these vectors. Many underlying aspects of sand fly genome organization and regulation of gene expression are expected to be identified through the ongoing genome projects of two sand fly species, P. papatasi and L. longipalpis [95]. With such information at hand, it will be possible to utilize transformation tools, such as those provided by the use of alphaviruses [96–98] to genetically transform sand flies. Already, microinjection of adult flies with dsRNA for targeted mRNA silencing via RNAi is commonplace in several laboratories. With specific training, these techniques can easily be transferred to other laboratories, in a fashion similar to that employed for mosquito researchers. Possibly, stable transgenic sand flies will also become available in the near future, and the contribution transgenic sand fly lines would have on the sand fly research community can be significant. Many theories related to vectorial capacity and to Leishmania-sand fly interaction can be tested, bringing sand fly basic research to a new level of investigation.
Acknowledgments
MR-O is funded by grants from the NIAID/NIH 7RO1AI074691 and by the Kansas Research Experimental Station (KAES contribution number 10-286-J). EMS is supported by CNPq and FAPERJ and YMTC by CNPq, FAPERJ and IOC-Fiocruz.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Killick-Kendrick R. Phlebotomine vectors of the leishmaniases: a review. Med Vet Entomol. 1990;4:1–24. doi: 10.1111/j.1365-2915.1990.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 2.Sacks D, Kamhawi S. Molecular aspects of parasite-vector and vector-host interactions in leishmaniasis. Annu Rev Microbiol. 2001;55:453–83. doi: 10.1146/annurev.micro.55.1.453. [DOI] [PubMed] [Google Scholar]
- 3.Schlein Y, Jacobson RL. Resistance of Phlebotomus papatasi to infection with Leishmania donovani is modulated by components of the infective bloodmeal. Parasitology. 1998;117:467–73. doi: 10.1017/s0031182098003321. [DOI] [PubMed] [Google Scholar]
- 4.Schlein Y, Warburg A, Schnur LF, Shlomai J. Vector compatibility of Phlebotomus papatasi dependent on differentially induced digestion. Acta Trop. 1983;40:65–70. [PubMed] [Google Scholar]
- 5.Borovsky D, Schlein Y. Trypsin and chymotrypsin-like enzymes of the sandfly Phlebotomus papatasi infected with Leishmania and their possible role in vector competence. Med Vet Entomol. 1987;1:235–42. doi: 10.1111/j.1365-2915.1987.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 6.Vaidyanathan R. Isolation of a myoinhibitory peptide from Leishmania major (Kinetoplastida: Trypanosomatidae) and its function in the vector sand fly Phlebotomus papatasi (Diptera: Psychodidae) J Med Entomol. 2005;42:142–52. doi: 10.1093/jmedent/42.2.142. [DOI] [PubMed] [Google Scholar]
- 7.Schlein Y, Jacobson RL, Messer G. Leishmania infections damage the feeding mechanism of the sandfly vector and implement parasite transmission by bite. Proc Natl Acad Sci U S A. 1992;89:9944–8. doi: 10.1073/pnas.89.20.9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volf P, Hajmova M, Sadlova J, Votypka J. Blocked stomodeal valve of the insect vector: similar mechanism of transmission in two trypanosomatid models. Int J Parasitol. 2004;34:1221–7. doi: 10.1016/j.ijpara.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Pitaluga AN, Beteille V, Lobo AR, Ortigao-Farias JR, Davila AM, Souza AA, Ramalho-Ortigao JM, Traub-Cseko YM. EST sequencing of blood-fed and Leishmania-infected midgut of Lutzomyia longipalpis, the principal visceral leishmaniasis vector in the Americas. Mol Genet Genomics. 2009;282:307–17. doi: 10.1007/s00438-009-0466-2. [DOI] [PubMed] [Google Scholar]
- 10.Ramalho-Ortigao M, Jochim RC, Anderson JM, Lawyer PG, Pham VM, Kamhawi S, Valenzuela JG. Exploring the midgut transcriptome of Phlebotomus papatasi: comparative analysis of expression profiles of sugar-fed, blood-fed and Leishmania major-infected sandflies. BMC Genomics. 2007;8:300. doi: 10.1186/1471-2164-8-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dillon RJ, Lane RP. Influence of Leishmania infection on blood-meal digestion in the sandflies Phlebotomus papatasi and P. langeroni. Parasitol Res. 1993;79:492–6. doi: 10.1007/BF00931590. [DOI] [PubMed] [Google Scholar]
- 12.Boulanger N, Lowenberger C, Volf P, Ursic R, Sigutova L, Sabatier L, Svobodova M, Beverley SM, Spath G, Brun R, Pesson B, Bulet P. Characterization of a defensin from the sand fly Phlebotomus duboscqi induced by challenge with bacteria or the protozoan parasite Leishmania major. Infect Immun. 2004;72:7140–6. doi: 10.1128/IAI.72.12.7140-7146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurd H, Carter V. The role of programmed cell death in Plasmodium-mosquito interactions. Int J Parasitol. 2004;34:1459–72. doi: 10.1016/j.ijpara.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Killick Kendrick R, Rioux JA. Intravectorial cycle of Leishmania in sandflies. Ann Parasitol Hum Comp. 1991;66 (Suppl 1):71–4. [PubMed] [Google Scholar]
- 15.Poinar GJ. Early Cretaceous trypanosomatids associated with fossil sand fly larvae in Burmese amber. Mem Inst Oswaldo Cruz. 2007;102:635–7. doi: 10.1590/s0074-02762007005000070. [DOI] [PubMed] [Google Scholar]
- 16.Kamhawi S. Phlebotomine sand flies and Leishmania parasites: friends or foes? Trends Parasitol. 2006;22:439–45. doi: 10.1016/j.pt.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Pimenta PF, Modi GB, Pereira ST, Shahabuddin M, Sacks DL. A novel role for the peritrophic matrix in protecting Leishmania from the hydrolytic activities of the sand fly midgut. Parasitology. 1997;115:359–69. doi: 10.1017/s0031182097001510. [DOI] [PubMed] [Google Scholar]
- 18.Sacks D. Leishmania-sand fly interactions controlling species-specific vector competence. Cell Microbiol. 2001;3:189–196. doi: 10.1046/j.1462-5822.2001.00115.x. [DOI] [PubMed] [Google Scholar]
- 19.Eschenlauer S, Faria M, Morrison L, Bland N, Ribeiro-Gomes F, DosReis G, Coombs G, Lima A, Mottram J. Influence of parasite encoded inhibitors of serine peptidases in early infection of macrophages with Leishmania major. Cell Microbiol. 2009;11:106–120. doi: 10.1111/j.1462-5822.2008.01243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Telleria EL, de Araújo AP, Secundino NF, d'Avila-Levy CM, Traub-Csekö YM. Trypsin-like serine proteases in Lutzomyia longipalpis--expression, activity and possible modulation by Leishmania infantum chagasi. PLoS One. 2010;5:e10697. doi: 10.1371/journal.pone.0010697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sant'anna MR, Diaz-Albiter H, Mubaraki M, Dillon RJ, Bates PA. Inhibition of trypsin expression in Lutzomyia longipalpis using RNAi enhances the survival of Leishmania. Parasit Vectors. 2009;2:62. doi: 10.1186/1756-3305-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Telleria E, Pitaluga A, Ortigão-Farias J, de Araújo A, Ramalho-Ortigão J, Traub-Cseko Y. Constitutive and blood meal-induced trypsin genes in Lutzomyia longipalpis. Arch Insect Biochem Physiol. 2007;66:53–63. doi: 10.1002/arch.20198. [DOI] [PubMed] [Google Scholar]
- 23.Jochim RC, Teixeira CR, Laughinghouse A, Mu J, Oliveira F, Gomes RB, Elnaiem DE, Valenzuela JG. The midgut transcriptome of Lutzomyia longipalpis: comparative analysis of cDNA libraries from sugar-fed, blood-fed, post-digested and Leishmania infantum chagasi-infected sand flies. BMC Genomics. 2008;9:15. doi: 10.1186/1471-2164-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramalho-Ortigao JM, Kamhawi S, Rowton ED, Ribeiro JM, Valenzuela JG. Cloning and characterization of trypsin- and chymotrypsin-like proteases from the midgut of the sand fly vector Phlebotomus papatasi. Insect Biochem Mol Biol. 2003;33:163–71. doi: 10.1016/s0965-1748(02)00187-x. [DOI] [PubMed] [Google Scholar]
- 25.Pimenta PF, Modi GB, Pereira ST, Shahabuddin M, Sacks DL. A novel role for the peritrophic matrix in protecting Leishmania from the hydrolytic activities of the sand fly midgut. Parasitology. 1997;115 ( Pt 4):359–69. doi: 10.1017/s0031182097001510. [DOI] [PubMed] [Google Scholar]
- 26.Ramalho-Ortigao JM, Traub-Cseko YM. Molecular characterization of LlChit1, a midgut chitinase cDNA from the leishmaniasis vector Lutzomyia longipalpis. Insect Biochem Mol Biol. 2003;33:279–87. doi: 10.1016/s0965-1748(02)00209-6. [DOI] [PubMed] [Google Scholar]
- 27.Ramalho-Ortigao JM, Kamhawi S, Joshi MB, Reynoso D, Lawyer PG, Dwyer DM, Sacks DL, Valenzuela JG. Characterization of a blood activated chitinolytic system in the midgut of the sand fly vectors Lutzomyia longipalpis and Phlebotomus papatasi. Insect Mol Biol. 2005;14:703–12. doi: 10.1111/j.1365-2583.2005.00601.x. [DOI] [PubMed] [Google Scholar]
- 28.Schlein Y, Jacobson RL, Shlomai J. Chitinase secreted by Leishmania functions in the sandfly vector. Proc R Soc Lond B Biol Sci. 1991;245:121–6. doi: 10.1098/rspb.1991.0097. [DOI] [PubMed] [Google Scholar]
- 29.Shakarian A, Dwyer D. The LdCht1 gene encodes the secretory chitinase of the human pathogen Leishmania donovani. Gene. 1998;208:315–22. doi: 10.1016/s0378-1119(98)00011-0. [DOI] [PubMed] [Google Scholar]
- 30.Shakarian A, Dwyer D. Pathogenic Leishmania secrete antigenically related chitinases which are encoded by a highly conserved gene locus. Exp Parasitol. 2000;94:238–42. doi: 10.1006/expr.2000.4493. [DOI] [PubMed] [Google Scholar]
- 31.Joshi M, Rogers M, Shakarian A, Yamage M, Al-Harthi S, Bates P, Dwyer D. Molecular characterization, expression, and in vivo analysis of LmexCht1: the chitinase of the human pathogen, Leishmania mexicana. J Biol Chem. 2005;280:3847–61. doi: 10.1074/jbc.M412299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers ME, Hajmova M, Joshi MB, Sadlova J, Dwyer DM, Volf P, Bates PA. Leishmania chitinase facilitates colonization of sand fly vectors and enhances transmission to mice. Cell Microbiol. 2008;10:1363–72. doi: 10.1111/j.1462-5822.2008.01132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myskova J, Svobodova M, Beverley S, Volf P. A lipophosphoglycan-independent development of Leishmania in permissive sand flies. Microbes Infect. 2007;9:317–24. doi: 10.1016/j.micinf.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers ME, Chance ML, Bates PA. The role of promastigote secretory gel in the origin and transmission of the infective stage of Leishmania mexicana by the sandfly Lutzomyia longipalpis. Parasitology. 2002;124:495–507. doi: 10.1017/s0031182002001439. [DOI] [PubMed] [Google Scholar]
- 35.Stierhof Y, Bates P, Jacobson R, Rogers M, Schlein Y, Handman E, Ilg T. Filamentous proteophosphoglycan secreted by Leishmania promastigotes forms gel-like three-dimensional networks that obstruct the digestive tract of infected sandfly vectors. Eur J Cell Biol. 1999;78:675–89. doi: 10.1016/S0171-9335(99)80036-3. [DOI] [PubMed] [Google Scholar]
- 36.Bates PA, Rogers ME. New insights into the developmental biology and transmission mechanisms of Leishmania. Curr Mol Med. 2004;4:601–9. doi: 10.2174/1566524043360285. [DOI] [PubMed] [Google Scholar]
- 37.Saraiva EM, Pimenta PF, Brodin TN, Rowton E, Modi GB, Sacks DL. Changes in lipophosphoglycan and gene expression associated with the development of Leishmania major in Phlebotomus papatasi. Parasitology. 1995;111:275–87. doi: 10.1017/s003118200008183x. [DOI] [PubMed] [Google Scholar]
- 38.Bates PA. Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int J Parasitol. 2007;37:1097–106. doi: 10.1016/j.ijpara.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sacks D, Lawyer P, Kamhawi S. The biology of Leishmania-sandfly interactions. In: Myler P, Fasel N, editors. Leishmania after the genome. Caister Academic Press; Norfolk, UK: 2008. p. 205. [Google Scholar]
- 40.Späth G, Beverley S. A lipophosphoglycan-independent method for isolation of infective Leishmania metacyclic promastigotes by density gradient centrifugation. Exp Parasitol. 2001;99:97–103. doi: 10.1006/expr.2001.4656. [DOI] [PubMed] [Google Scholar]
- 41.Yao C, Chen Y, Sudan B, Donelson J, Wilson M. Leishmania chagasi: homogenous metacyclic promastigotes isolated by buoyant density are highly virulent in a mouse model. Exp Parasitol. 2008;118:129–33. doi: 10.1016/j.exppara.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saraiva E, Pinto-da-Silva L, Wanderley J, Bonomo A, Barcinski M, Moreira M. Flow cytometric assessment of Leishmania spp metacyclic differentiation: validation by morphological features and specific markers. Exp Parasitol. 2005;110:39–47. doi: 10.1016/j.exppara.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Sacks DL, da Silva RP. The generation of infective stage Leishmania major promastigotes is associated with the cell-surface expression and release of a developmentally regulated glycolipid. J Immunol. 1987;139:3099–106. [PubMed] [Google Scholar]
- 44.Sacks DL, Hieny S, Sher A. Identification of cell surface carbohydrate and antigenic changes between noninfective and infective developmental stages of Leishmania major promastigotes. J Immunol. 1985;135:564–9. [PubMed] [Google Scholar]
- 45.McConville MJ, Turco SJ, Ferguson MA, Sacks DL. Developmental modification of lipophosphoglycan during the differentiation of Leishmania major promastigotes to an infectious stage. Embo J. 1992;11:3593–600. doi: 10.1002/j.1460-2075.1992.tb05443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamhawi S, Ramalho-Ortigao M, Pham VM, Kumar S, Lawyer PG, Turco SJ, Barillas-Mury C, Sacks DL, Valenzuela JG. A role for insect galectins in parasite survival. Cell. 2004;119:329–41. doi: 10.1016/j.cell.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Pimenta PF, Saraiva EM, Sacks DL. The comparative fine structure and surface glycoconjugate expression of three life stages of Leishmania major. Exp Parasitol. 1991;72:191–204. doi: 10.1016/0014-4894(91)90137-l. [DOI] [PubMed] [Google Scholar]
- 48.Dobson D, Mengeling B, Cilmi S, Hickerson S, Turco S, Beverley S. Identification of genes encoding arabinosyltransferases (SCA) mediating developmental modifications of lipophosphoglycan required for sand fly transmission of Leishmania major. J Biol Chem. 2003;278:28840–8. doi: 10.1074/jbc.M302728200. [DOI] [PubMed] [Google Scholar]
- 49.Sacks DL, Pimenta PF, McConville MJ, Schneider P, Turco SJ. Stage-specific binding of Leishmania donovani to the sand fly vector midgut is regulated by conformational changes in the abundant surface lipophosphoglycan. J Exp Med. 1995;181:685–97. doi: 10.1084/jem.181.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahoney AB, Sacks DL, Saraiva E, Modi G, Turco SJ. Intra-species and stage-specific polymorphisms in lipophosphoglycan structure control Leishmania donovani-sand fly interactions. Biochemistry. 1999;38:9813–23. doi: 10.1021/bi990741g. [DOI] [PubMed] [Google Scholar]
- 51.Soares R, Macedo M, Ropert C, Gontijo N, Almeida I, Gazzinelli R, Pimenta P, Turco S. Leishmania chagasi: lipophosphoglycan characterization and binding to the midgut of the sand fly vector Lutzomyia longipalpis. Mol Biochem Parasitol. 2002;121:213–24. doi: 10.1016/s0166-6851(02)00033-6. [DOI] [PubMed] [Google Scholar]
- 52.Soares R, Cardoso T, Barron T, Araújo M, Pimenta P, Turco S. Leishmania braziliensis: a novel mechanism in the lipophosphoglycan regulation during metacyclogenesis. Int J Parasitol. 2005;35:245–53. doi: 10.1016/j.ijpara.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Almeida M, Cuba C, de Sa C, Pharoah M, Howard K, Miles M. Metacyclogenesis of Leishmania (Viannia) braziliensis in vitro: evidence that lentil lectin is a marker of complement resistance and enhanced infectivity. Trans R Soc Trop Med Hyg. 1993;87:325–9. doi: 10.1016/0035-9203(93)90150-o. [DOI] [PubMed] [Google Scholar]
- 54.Pinto-da-Silva L, Camurate M, Costa K, Oliveira S, da Cunha-e-Silva N, Saraiva E. Leishmania (Viannia) braziliensis metacyclic promastigotes purified using Bauhinia purpurea lectin are complement resistant and highly infective for macrophages in vitro and hamsters in vivo. Int J Parasitol. 2002;32:1371–7. doi: 10.1016/s0020-7519(02)00137-6. [DOI] [PubMed] [Google Scholar]
- 55.Pimenta PF, Saraiva EM, Rowton E, Modi GB, Garraway LA, Beverley SM, Turco SJ, Sacks DL. Evidence that the vectorial competence of phlebotomine sand flies for different species of Leishmania is controlled by structural polymorphisms in the surface lipophosphoglycan. Proc Natl Acad Sci U S A. 1994;91:9155–6. doi: 10.1073/pnas.91.19.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pimenta PF, Turco SJ, McConville MJ, Lawyer PG, Perkins PV, Sacks DL. Stage-specific adhesion of Leishmania promastigotes to the sandfly midgut. Science. 1992;256:1812–5. doi: 10.1126/science.1615326. [DOI] [PubMed] [Google Scholar]
- 57.Sacks DL, Modi G, Rowton E, Spath G, Epstein L, Turco SJ, Beverley SM. The role of phosphoglycans in Leishmania-sand fly interactions. Proc Natl Acad Sci U S A. 2000;97:406–11. doi: 10.1073/pnas.97.1.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogers M, Ilg T, Nikolaev A, Ferguson M, Bates P. Transmission of cutaneous leishmaniasis by sand flies is enhanced by regurgitation of fPPG. Nature. 2004;430:463–7. doi: 10.1038/nature02675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zakai H, Chance M, Bates P. In vitro stimulation of metacyclogenesis in Leishmania braziliensis, L. donovani, L. major and L. mexicana. Parasitol Res. 1998;116:305–9. doi: 10.1017/s0031182097002382. [DOI] [PubMed] [Google Scholar]
- 60.Cunningham ML, Titus RG, Turco SJ, Beverley SM. Regulation of differentiation to the infective stage of the protozoan parasite Leishmania major by tetrahydrobiopterin. Science. 2001;292:285–7. doi: 10.1126/science.1057740. [DOI] [PubMed] [Google Scholar]
- 61.Schlein Y, Jacobson RL. Haemoglobin inhibits the development of infective promastigotes and chitinase secretion in Leishmania major cultures. Parasitology. 1994;109:23–8. doi: 10.1017/s0031182000077726. [DOI] [PubMed] [Google Scholar]
- 62.Méndez S, Fernández-Pérez F, de la Fuente C, Cuquerella M, Gómez-Muñoz M, Alunda J. Partial anaerobiosis induces infectivity of Leishmania infantum promastigotes. Parasitol Res. 1999;85:507–9. doi: 10.1007/s004360050587. [DOI] [PubMed] [Google Scholar]
- 63.Charlab R, Tesh RB, Rowton ED, Ribeiro JM. Leishmania amazonensis: sensitivity of different promastigote morphotypes to salivary gland homogenates of the sand fly Lutzomyia longipalpis. Exp Parasitol. 1995;80:167–75. doi: 10.1006/expr.1995.1021. [DOI] [PubMed] [Google Scholar]
- 64.Besteiro S, Williams R, Morrison L, Coombs G, Mottram J. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J Biol Chem. 2006;281:11384–96. doi: 10.1074/jbc.M512307200. [DOI] [PubMed] [Google Scholar]
- 65.Wanderley J, Pinto da Silva L, Deolindo P, Soong L, Borges V, Prates D, de Souza A, Barral A, Balanco J, do Nascimento M, Saraiva E, Barcinski M. Cooperation between apoptotic and viable metacyclics enhances the pathogenesis of Leishmaniasis. PLoS One. 2009;29:e5733. doi: 10.1371/journal.pone.0005733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Zandbergen G, Bollinger A, Wenzel A, Kamhawi S, Voll R, Klinger M, Müller A, Hölscher C, Herrmann M, Sacks D, Solbach W, Laskay T. Leishmania disease development depends on the presence of apoptotic promastigotes in the virulent inoculum. Proc Natl Acad Sci U S A. 2006;103:13837–42. doi: 10.1073/pnas.0600843103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cuvillier A, Miranda J, Ambit A, Barral A, Merlin G. Abortive infection of Lutzomyia longipalpis insect vectors by aflagellated LdARL-3A-Q70L overexpressing Leishmania amazonensis parasites. Cell Microbiol. 2003;5:717–28. doi: 10.1046/j.1462-5822.2003.00316.x. [DOI] [PubMed] [Google Scholar]
- 68.Chaves C, Soares D, Da Silva R, Saraiva E. Characterization of the species- and stage-specificity of two monoclonal antibodies against Leishmania amazonensis. Exp Parasitol. 2003;103:152–9. doi: 10.1016/s0014-4894(03)00098-5. [DOI] [PubMed] [Google Scholar]
- 69.Rotureau B, Morales M, Bastin P, Späth G. The flagellum-MAP kinase connection in Trypanosomatids: a key sensory role in parasite signaling and development? Cell Microbiol. 2009 doi: 10.1111/j.1462-5822.2009.01295.x. [DOI] [PubMed] [Google Scholar]
- 70.Warburg A, Schlein Y. The effect of post-bloodmeal nutrition of Phlebotomus papatasi on the transmission of Leishmania major. Am J Trop Med Hyg. 1986;35:926–30. doi: 10.4269/ajtmh.1986.35.926. [DOI] [PubMed] [Google Scholar]
- 71.Kimblin N, Peters N, Debrabant A, Secundino N, Egen J, Lawyer P, Fay M, Kamhawi S, Sacks D. Quantification of the infectious dose of Leishmania major transmitted to the skin by single sand flies. Proc Natl Acad Sci U S A. 2008;105:10125–30. doi: 10.1073/pnas.0802331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Titus RG, Ribeiro JM. Salivary gland lysates from the sand fly Lutzomyia longipalpis enhance Leishmania infectivity. Science. 1988;239:1306–8. doi: 10.1126/science.3344436. [DOI] [PubMed] [Google Scholar]
- 73.Belkaid Y, Kamhawi S, Modi G, Valenzuela J, Noben-Trauth N, Rowton E, Ribeiro J, Sacks DL. Development of a natural model of cutaneous leishmaniasis: powerful effects of vector saliva and saliva preexposure on the long-term outcome of Leishmania major infection in the mouse ear dermis. J Exp Med. 1998;188:1941–53. doi: 10.1084/jem.188.10.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kamhawi S. The biological and immunomodulatory properties of sand fly saliva and its role in the establishment of Leishmania infections. Microbes Infect. 2000;2:1–9. doi: 10.1016/s1286-4579(00)01331-9. [DOI] [PubMed] [Google Scholar]
- 75.Valenzuela JG, Belkaid Y, Garfield MK, Mendez S, Kamhawi S, Rowton ED, Sacks DL, Ribeiro JMC. Toward a defined anti-Leishmania vaccine targeting vector antigens: Characterization of a protective salivary protein. J. Exp. Med. 2001;194:331–342. doi: 10.1084/jem.194.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oliveira F, Lawyer P, Kamhawi S, Valenzuela J. Immunity to Distinct Sand Fly Salivary Proteins Primes the Anti-Leishmania Immune Response towards Protection or Exacerbation of Disease. PLoS Negl Trop Dis. 2008;2:e226. doi: 10.1371/journal.pntd.0000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gomes R, Teixeira C, Teixeira M, Oliveira F, Menezes M, Silva C, de Oliveira C, Miranda J, Elnaiem D, Kamhawi S, Valenzuela J, Brodskyn C. Immunity to a salivary protein of a sand fly vector protects against the fatal outcome of visceral leishmaniasis in a hamster model. Proc Natl Acad Sci U S A. 2008;105:7845–50. doi: 10.1073/pnas.0712153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Collin N, Gomes R, Teixeira C, Cheng L, Laughinghouse A, Ward J, Elnaiem D, Fischer L, Valenzuela J, Kamhawi S. Sand fly salivary proteins induce strong cellular immunity in a natural reservoir of visceral leishmaniasis with adverse consequences for Leishmania. PLoS Pathog. 2009;5:e1000441. doi: 10.1371/journal.ppat.1000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Volf P, Benkova I, Myskova J, Sadlova J, Campino L, Ravel C. Increased transmission potential of Leishmania major/Leishmania infantum hybrids. Int J Parasitol. 2007;37:589–93. doi: 10.1016/j.ijpara.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ravel C, Cortes S, Pratlong F, Morio F, Dedet JP, Campino L. First report of genetic hybrids between two very divergent Leishmania species: Leishmania infantum and Leishmania major. Int J Parasitol. 2006;36:1383–8. doi: 10.1016/j.ijpara.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 81.Ramalho-Ortigao JM, Temporal P, de Oliveira SM, Barbosa AF, Vilela ML, Rangel EF, Brazil RP, Traub-Cseko YM. Characterization of constitutive and putative differentially expressed mRNAs by means of expressed sequence tags, differential display reverse transcriptase-PCR and randomly amplified polymorphic DNA-PCR from the sand fly vector Lutzomyia longipalpis. Mem Inst Oswaldo Cruz. 2001;96:105–11. doi: 10.1590/s0074-02762001000100012. [DOI] [PubMed] [Google Scholar]
- 82.Dillon RJ, Ivens AC, Churcher C, Holroyd N, Quail MA, Rogers ME, Soares MB, Bonaldo MF, Casavant TL, Lehane MJ, Bates PA. Analysis of ESTs from Lutzomyia longipalpis sand flies and their contribution toward understanding the insect-parasite relationship. Genomics. 2006;88:831–40. doi: 10.1016/j.ygeno.2006.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oliveira F, Jochim R, Valenzuela J, Kamhawi S. Sand flies, Leishmania, and transcriptome-borne solutions. Parasitol Int. 2009;58:1–5. doi: 10.1016/j.parint.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pitaluga AN, Mason PW, Traub-Cseko YM. Non-specific antiviral response detected in RNA-treated cultured cells of the sandfly, Lutzomyia longipalpis. Dev Comp Immunol. 2008;32:191–7. doi: 10.1016/j.dci.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 85.Sant'Anna MR, Alexander B, Bates PA, Dillon RJ. Gene silencing in phlebotomine sand flies: Xanthine dehydrogenase knock down by dsRNA microinjections. Insect Biochem Mol Biol. 2008;38:652–60. doi: 10.1016/j.ibmb.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saraiva EM, de Figueiredo Barbosa A, Santos FN, Borja-Cabrera GP, Nico D, Souza LO, de Oliveira Mendes-Aguiar C, de Souza EP, Fampa P, Parra LE, Menz I, Dias JG, Jr, de Oliveira SM, Palatnik-de-Sousa CB. The FML-vaccine (Leishmune) against canine visceral leishmaniasis: a transmission blocking vaccine. Vaccine. 2006;24:2423–31. doi: 10.1016/j.vaccine.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 87.Durvasula RV, Gumbs A, Panackal A, Kruglov O, Aksoy S, Merrifield RB, Richards FF, Beard CB. Prevention of insect-borne disease: an approach using transgenic symbiotic bacteria. Proc Natl Acad Sci U S A. 1997;94:3274–8. doi: 10.1073/pnas.94.7.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hillesland H, Read A, Subhadra B, Hurwitz I, McKelvey R, Ghosh K, Das P, Durvasula R. Identification of aerobic gut bacteria from the kala azar vector, Phlebotomus argentipes: a platform for potential paratransgenic manipulation of sand flies. Am J Trop Med Hyg. 2008;79:881–6. [PubMed] [Google Scholar]
- 89.Gouveia C, Asensi MD, Zahner V, Rangel EF, Oliveira SM. Study on the bacterial midgut microbiota associated to different Brazilian populations of Lutzomyia longipalpis (Lutz & Neiva) (Diptera: Psychodidae) Neotrop Entomol. 2008;37:597–601. doi: 10.1590/s1519-566x2008000500016. [DOI] [PubMed] [Google Scholar]
- 90.Dillon RJ, el Kordy E, Shehata M, Lane RP. The prevalence of a microbiota in the digestive tract of Phlebotomus papatasi. Ann Trop Med Parasitol. 1996;90:669–73. doi: 10.1080/00034983.1996.11813102. [DOI] [PubMed] [Google Scholar]
- 91.Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McMeniman CJ, Lane RV, Cass BN, Fong AW, Sidhu M, Wang YF, O'Neill SL. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science. 2009;323:141–4. doi: 10.1126/science.1165326. [DOI] [PubMed] [Google Scholar]
- 93.Turley AP, Moreira LA, O'Neill SL, McGraw EA. Wolbachia Infection Reduces Blood-Feeding Success in the Dengue Fever Mosquito, Aedes aegypti. PLoS Negl Trop Dis. 2009;3:e516. doi: 10.1371/journal.pntd.0000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saraiva E, Fampa P, Cedeno V, Bergoin M, Mialhe E, Miller L. Expression of heterologous promoters in Lutzomyia longipalpis and Phlebotomus papatasi (Diptera: Psychodidae) cell lines. J Med Entomol. 2000;37:802–806. doi: 10.1603/0022-2585-37.6.802. [DOI] [PubMed] [Google Scholar]
- 95.McDowell MA, Collins F, Ramalho-Ortigao M, Valenzuela JG, Kamhawi S, Dillon R, Bates P, Lehane M. Proposal for Sequencing the Genome of the Sand Flies, Lutzomyia longipalpis and Phlebotomus papatasi. http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/SandFliesSeq.pdf.
- 96.Campbell CL, Keene KM, Brackney DE, Olson KE, Blair CD, Wilusz J, Foy BD. Aedes aegypti uses RNA interference in defense against Sindbis virus infection. BMC Microbiol. 2008;8:47. doi: 10.1186/1471-2180-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Travanty EA, Adelman ZN, Franz AW, Keene KM, Beaty BJ, Blair CD, James AA, Olson KE. Using RNA interference to develop dengue virus resistance in genetically modified Aedes aegypti. Insect Biochem Mol Biol. 2004;34:607–13. doi: 10.1016/j.ibmb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 98.Sanchez-Vargas I, Travanty EA, Keene KM, Franz AW, Beaty BJ, Blair CD, Olson KE. RNA interference, arthropod-borne viruses, and mosquitoes. Virus Res. 2004;102:65–74. doi: 10.1016/j.virusres.2004.01.017. [DOI] [PubMed] [Google Scholar]


