Abstract
OBJECTIVES
The Lynch syndrome (LS) is an inherited cancer syndrome showing a preponderance of colorectal cancer (CRC) in context with endometrial cancer and several other extracolonic cancers, which is due to pathogenic mutations in the mismatch repair (MMR) genes, MLH1, MSH2, MSH6, and PMS2. Some families were found to show a LS phenotype without an identified MMR mutation, although there was microsatellite instability and absence of MSH2 expression by immunohistochemistry. Studies of a subset of these families found a deletion at the 3′ end of the epithelial cell adhesion molecule (EPCAM) gene, causing transcription read-through resulting in silencing of MSH2 through hypermethylation of its promoter. The tumor spectrum of such families appears to differ from classical LS.
METHODS
Our study of two large families (USA Family R and Dutch Family A) with an EPCAM deletion was carried out using each institution’s standard family study protocol. DNA was extracted from peripheral blood and EPCAM deletion analysis was performed.
RESULTS
Both families were found to harbor the same deletion at the 3′ end of EPCAM. Analysis showed that the deletion originated from a common ancestor. Family R and Family A members showed segregation of CRC with the presence of this EPCAM mutation. Compared with classic LS, there were almost no extracolonic cancers.
CONCLUSIONS
Members of Family R and Family A, all with the same EPCAM deletion, predominantly presented with CRC but no LS-associated endometrial cancer, confirming findings seen in other, smaller, LS families with EPCAM mutations. In these EPCAM mutation carriers, cancer surveillance should be focused on CRC.
INTRODUCTION
The Lynch syndrome (LS) predisposes to colon cancer and several extracolonic cancers, the most common of which is endometrial cancer. These are caused by mutations in the DNA mismatch repair (MMR) genes MLH1, MSH2, MSH6, and PMS2. A subset of families with phenotypes consonant with LS, including microsatellite instability and abnormal MMR immunohistochemistry have been found to harbor a deletion at the 3′ end of the gene encoding epithelial cell adhesion molecule (EPCAM; previously referred to as TACSTD1) (1–6). The tumors with EPCAM mutations showed high-level microsatellite instability; immunohistochemistry tests disclosed a loss of MSH2 protein, with no identifiable mutation in the MSH2gene (1,3). EPCAM mutations were found to cosegregate with the LS phenotype in a considerable fraction (19%) of LS families that lacked MLH1/MSH2 mutations (3) and to account for at least 2.3% of explained MSH2-deficient families (5). EPCAM is located immediately 5′ of MSH2 and deletion of its termination signal allows transcription to follow through across the MSH2 promoter leading to promoter hypermethylation and gene silencing. In EPCAM deletion-positive subjects, the risk of developing colorectal cancer (CRC) was shown to be comparable to that of carriers of a MSH2 mutation. However, their risk of developing endometrial cancer was lower than in MSH2 mutation carriers, and appeared to be restricted to carriers of deletions that extend close to the MSH2 gene (7).
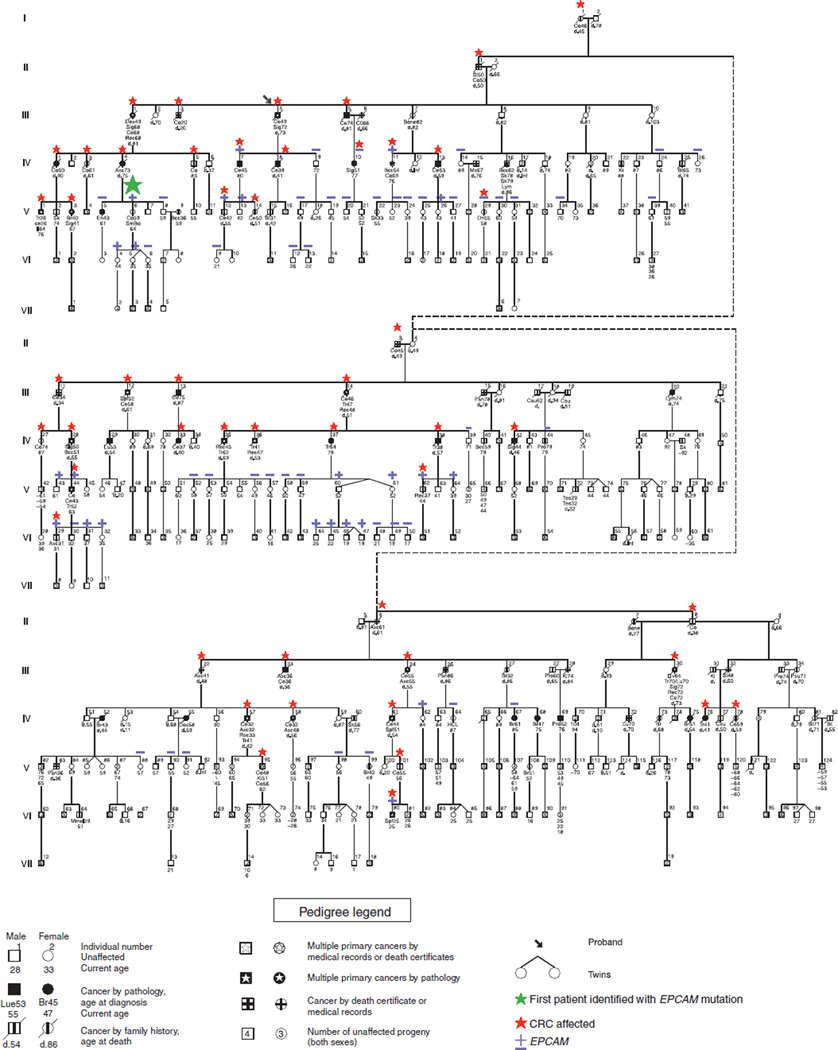
Family R (Figure 1) is a large LS family that was referred to the Creighton research group 35 years ago by the proband’s surgeon, who knew that CRC was highly prevalent throughout five generations of the family. Study of Family R contributed to the template for the classic description of LS and it was the fourth hereditary CRC family studied by Dr Lynch. Family R was first reported at a conference in 1976 (8), and further updated in 1977 (9). Repeated comprehensive DNA testing by several groups failed to find a mutation in a MMR gene until EPCAM deletions were reported to cause LS (1).
Figure 1.
Pedigree of Family R. Numbers below the symbols refer to the age in years of last clinical follow-up.
In this study we show that Family R carries the same EPCAM mutation as Dutch Family A, the first family in which a 3′ end EPCAM deletion was recognized as the cause of MSH2-deficient tumors due to hypermethylation of the MSH2 promoter (1). These two families are the largest common ancestor pedigrees reported to date. Analysis of their tumor spectrum is used to determine the risk of extracolonic tumors in carriers of this specific EPCAM mutation.
METHODS
Subjects
This study was approved by the Institutional Review Boards of the institutions involved.
USA Family R
Contact was continually maintained with Family R through telephone calls, correspondence, and personal contact through several educational family information services (FISs) (10). The FIS is comprised of a gathering of as many family members as wish to attend the meeting so that they can benefit from an informal educational coverage of the “family disease”, including its diagnosis, the significance of a deleterious germline mutation, and its phenotypic features. It then deals with the patient’s risk of developing CRC with focus upon targeted screening recommendations based upon a patient’s position in the pedigree. The proband (Figure 1, III-4) and several other family members were instrumental in gathering information from and distributing information to the family. Genetic counseling was based upon the compelling evidence for a Mendelian autosomal dominant pattern of CRC throughout the family in concert with its striking clinical and pathology features. Given these findings, first-degree relatives of individuals who developed CRC were strongly encouraged to undergo colonoscopy every other year starting at age 20–25 and then annually beginning at age 40 (8,11–14). At the time of the FIS, the initial finding of a lack of endometrial cancer risk had not yet been confirmed and patients were recommended standard LS endometrial cancer surveillance and consideration of prophylactic hysterectomy.
Following the discovery of MMR mutations in LS families in the mid-1990s (15–20), we began DNA testing on the most genetically informative members of Family R, namely those with early onset CRC and/or metachronous CRC; however, we were unable to identify a MMR gene mutation.
In October 2009, the EPCAM mutation c.859-1462_* 1999del was identified in a member of Family R (Figure 1, V-6). Shortly afterward, we held another FIS wherein > 70 Family R members attended in order to learn about this new finding and how it might impact them. Family members for whom we did not have a stored sample were invited to provide blood for testing. Forty-seven DNA samples were tested for the EPCAM mutation. Another FIS was held in March 2010 in order to provide individuals with their DNA test results in private genetic counseling sessions. This FIS also gave other family members an opportunity to provide a DNA sample for testing. All living relatives found it acceptable to include their genotyping information in the pedigree.
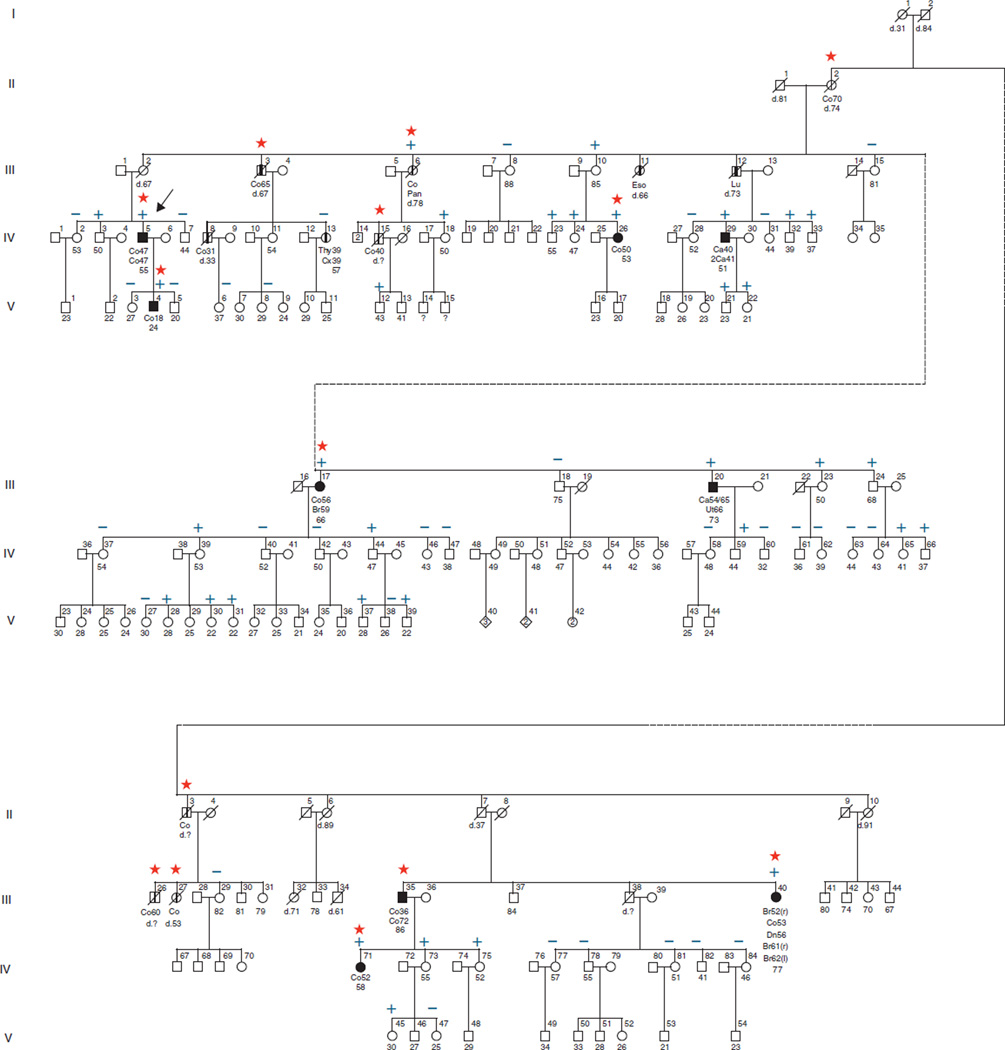
Dutch Family A
The proband of the Dutch Family A (Figure 2) visited the outpatient clinic for family cancer of the Radboud University Nijmegen Medical Centre, because he developed two colorectal carcinomas at age 47. He was the first patient in whom a deletion of the 3′ end of EPCAM was observed (1). This deletion was also detected in his son, who visited our outpatient clinic because he developed an MSH2-deficient CRC at age 18. This deletion was also present in three more distantly related family members affected with CRC, who had been contacted by the proband and underwent genetic counseling and testing for this mutation. After the pathogenicity of the deletion was proven, presymptomatic testing was offered to other family members using routine procedures by which the information is disseminated by contact persons within the family. Individuals in the pedigree were de-identified to protect their privacy without loss of the scientific information.
Figure 2.
Pedigree of Family A. The clinical data of the relatives that have not been tested are less reliable than those of the tested subjects and the obligate carriers. For pedigree legend see Figure 1. Ca, colorectal adenoma; Cx, cervix; EPCAM; epithelial cell adhesion molecule; Eso, esophagus; Thy, thyroid; Ut, urothelial tract.
Laboratory methods
Immunohistochemistry for the MMR proteins was performed as previously described (1). In Family R, EPCAM deletion analysis was performed on DNA extracted from peripheral blood using a commercially available MLPA (multiplex ligation-dependent probe amplification kit; P072 version 6, MRC Holland, Amsterdam, The Netherlands) as per the manufacturer’s instructions. This kit contains oligonucleotide probes targeting EPCAM exons 3, 8, 9, and two probes in the intervening region between EPCAM and MSH2: one 3 kb downstream of EPCAM and one 2.5 kb upstream from the MSH2 gene. In Family A, the mutation was tested for using a deletion-specific polymerase chain reaction as previously described (1).
RESULTS
USA Family R
Overall, DNA testing for the EPCAM mutation in Family R showed that 22 individuals were positive, 16 were obligate mutation carriers, and 42 were negative (Figure 1). Fifteen family members received their results at the March 2010 FIS, and an additional 49 received their results through scheduled telephone counseling sessions. Carriers of the mutation shared this information with their children and siblings who had not been tested. Subsequently, additional samples were submitted for testing and the results are pending.
Table 1 depicts the cancer spectrum in Family R in those who have lived to at least age 20 and have a risk of mutation carrier-ship of ≥25%. This table includes those who were tested positive for EPCAM mutation, those who were tested negative, and those who were not tested. When those not tested are excluded, all but one of the individuals with CRC either tested positive or is an obligate gene carrier.
Table 1.
Cancer spectrum of USA family R members age 20 + and syndrome risk ≥ 25%
| Cancer sites | EPCAM family genetic test results | |||
|---|---|---|---|---|
| Pos + OGCa | Negative | Not tested | Total | |
| Colon | 22 | 1 | 27 | 50 |
| Skin | 2b | 1 | 2 | 5 |
| Small intestine | 1c | 1d | 1c | 3 |
| Stomach | 1c | 0 | 1 | 2 |
| Unknown primary | 1 | 0 | 2 | 3 |
| Breast | 0 | 2 | 5c | 7 |
| Upper uroepithelial | 0 | 0 | 3c | 3 |
| Lymphoma | 0 | 0 | 3 | 3 |
| Lung | 0 | 0 | 2 | 2 |
| Brain | 0 | 0 | 2 | 2 |
| Prostate | 0 | 1 | 1 | 2 |
| Pancreas | 0 | 0 | 1 | 1 |
| Testicle | 0 | 0 | 1 | 1 |
| Urinary bladder | 0 | 0 | 1 | 1 |
| Female organ? | 0 | 0 | 1c | 1 |
| Endometrium | 0 | 1 | 0 | 1 |
| Leukemia | 0 | 1 | 0 | 1 |
| No cancer | 13 | 29 | 201 | 243 |
| Total individuals | 36 | 37 | 250 | 323 |
EPCAM, epithelial cell adhesion molecule; OGC, obligate gene carrier.
Includes those tested positive plus Obligate Gene Carriers (child tested positive).
Both individuals also diagnosed with colon cancer.
One individual also diagnosed with colon cancer.
This individual also diagnosed with melanoma.
Of the 57 individuals tested, 20 are positive and 37 are negative.
Noteworthy is the fact that 50 Family R members had CRC with a mean age at diagnosis of 49 years (range 20–76 years). It is of interest that, unlike other LS families, only a single patient had endometrial cancer and this individual was found to be negative for the EPCAM mutation. There were no cases of ovarian cancer. Three family members had small bowel cancer; of these, one tested positive for the EPCAM mutation, one tested negative, and the third has not been tested. The tumor tissue from the small bowel cancer from the confirmed EPCAM mutation carrier was available and showed loss of MSH2 and MSH6 expression. Three family members had upper uroepithelial tract cancer; none of them has been tested.
Dutch Family A
For the Dutch Family A, clinical data were collected for all family members who underwent genetic testing (Table 2). All 10 patients with CRC were carriers of the mutation. The mean age at diagnosis of their first CRC was 47 years (range 18–70 years). Within this group, the only extracolonic cancers detected were one MSH2-deficient duodenal cancer, one MSH2-proficient urothelial carcinoma, two breast, one lung, and one pancreas carcinoma. These latter four tumors were not available for further analysis. No endometrial cancers were observed.
Table 2.
Cancer spectrum of dutch family A members age 20 + and syndrome risk ≥25%
| Cancer sites | EPCAM family genetic test results | |||
|---|---|---|---|---|
| Pos + OGC* | Negative | Not tested | Total | |
| Colon | 10 | 0 | 5 | 15 |
| Skin | 0 | 0 | 0 | 0 |
| Esophagus | 0 | 0 | 1 | 1 |
| Duodenal | 1a | 0 | 0 | 1 |
| Stomach | 0 | 0 | 0 | 0 |
| Unknown primary | 0 | 0 | 0 | 0 |
| Breast | 2a | 0 | 0 | 2 |
| Upper urothelial tract | 1b | 0 | 0 | 1 |
| Lymphoma | 0 | 0 | 0 | 0 |
| Lung | 1 | 0 | 0 | 1 |
| Brain | 0 | 0 | 0 | 0 |
| Prostate | 0 | 0 | 0 | 0 |
| Pancreas | 1a | 0 | 0 | 1 |
| Testicle | 0 | 0 | 0 | 0 |
| Urinary bladder | 0 | 0 | 0 | 0 |
| Endometrium | 0 | 0 | 0 | 0 |
| Leukemia | 0 | 0 | 0 | 0 |
| Thyroid | 0 | 1c | 0 | 1 |
| Cervix | 0 | 1 | 0 | 1 |
| No cancer | 31 | 31 | 25 | 87 |
| Total individuals | 41 | 32 | 31 | 104 |
EPCAM, epithelial cell adhesion molecule; OGC, obligate gene carrier.
Individuals also diagnosed with colon cancer.
Also diagnosed with colorectal adenomas.
Also diagnosed with cervical cancer.
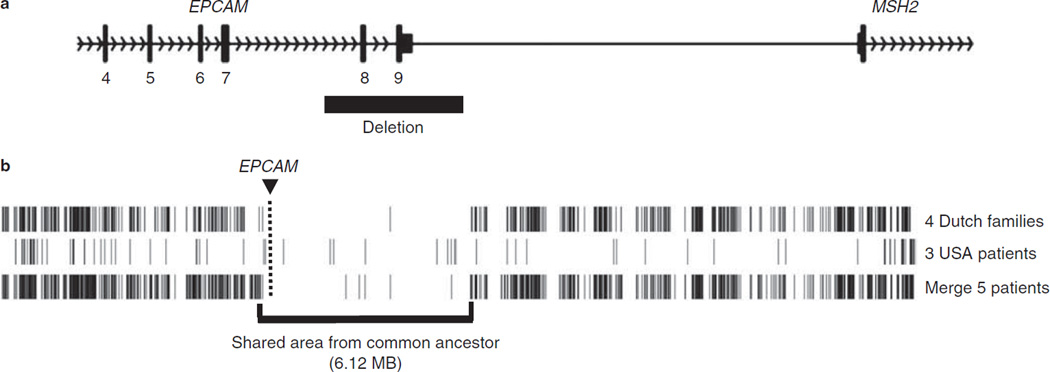
Common ancestor
Both families share the 3′ end EPCAM deletion c.859-1462_* 1999del. Single nucleotide polymorphism array-based genotyping showed that a large genomic region surrounding the deletion (haploblock) was shared by members of both Family R and Family A (Figure 3). These findings indicate that the EPCAM mutations in Family R and Family A originate from a common ancestor. The same mutation has been detected in 15 other Dutch probands (5,7).
Figure 3.
EPCAM founder mutation. (a) Schematic representation of the epithelial cell adhesion molecule (EPCAM) founder deletion seen in Families A and R. The deletion encompasses exons 8 and 9 of the EPCAM gene but does not extend into the MSH2 promoter. (b) Representation of Affymetrix SNP6.0 array data of the genomic region around EPCAM in four Dutch Families (including Family A) and three members of USA Family R. Haplotypes of chromosome 2 were derived from homozygous calls of the 75,933 single nucleotide polymorphisms (SNPs) on this chromosome, and positions of discordant homozygous calls (DHCs) were marked. The location of EPCAM is indicated by the box and line. For Family A and Family R, there is a shared region, which includes EPCAM (boxed). Comparing the Dutch family members and one member of Family R (the lane labeled “Merge 5 patients”) there is a stretch of 2,186 SNPs (6.12 Mb) with only five DHCs, strongly indicating a common ancestor. For the Dutch patients, there were two large stretches of 1,341 and 906 SNPs, without DHCs, separated by a single DHC. For Family R, there is a larger stretch of shared SNPs compared with Dutch Family A members. This stretch is 17.22 Mb, 9,850 SNPs long with 81 DHCs. The larger shared haploblock seen in Family R would be consistent with the founding of Family R in the United States by immigration of a single Dutch EPCAM mutation carrier after origination of the deletion in the Netherlands.
DISCUSSION
The extended kindreds described here are among the largest reported in medical genetics. The breadth and size of the kindreds, and the number of related cancers, is testament to the potential impact of a single mutation event. Over the years, genetic epidemiology research has resulted in a significant expansion of knowledge on what was initially termed the “cancer family syndrome”, (9) subsequently called hereditary nonpolyposis CRC, and now referred to as LS (11,12,21). Specifically, diagnostic parameters such as the occurrence of cancer of diverse anatomic sites, its pattern of cancer combinations, early age of cancer onset, pathological features, MMR penetrance, accelerated carcinogenesis, and survival may vary enormously within and between families. The Amsterdam Criteria (22,23) were developed two decades ago in an attempt to better describe and define LS families. However, only about 60% of families fulfilling the Amsterdam Criteria manifest MMR germline mutations consonant with LS (24). A subset of the remaining 40% include familial CRC type X (25), while the more recently discovered EPCAM mutation explains part of the remainder (1–6).
Site-specific CRC and EPCAM
The tumors of EPCAM deletion carriers show loss of expression of MSH2 and MSH6, a pattern also seen with MSH2 germline mutation carriers. Nowadays, the majority of clinical laboratories that are testing for MMR gene mutations test for EPCAM deletions as relevant probes for the EPCAM gene are included in the MLPA kit used for the analysis of MSH2 deletions. The medical and genetic literature shows that MSH2 mutation carriers develop extracolonic cancers, especially endometrial cancer, more frequently than carriers of MLH1 mutations (3,11,12). Deletions in EPCAM result in hypermethylation and tissue-restricted silencing of MSH2 (1), which is dependent on the expression pattern of the EPCAM gene. This finding probably explains the rare incidence of extracolonic cancers in the subset of EPCAM mutation carriers in both extended families described here and multiple other families described earlier (7). There is little experimental evidence on the relative expression of EPCAM in various tissues. We postulate that at least during early stages of endometrium carcinogenesis, EPCAM is not sufficiently expressed and therefore the MSH2 promoter is not efficiently silenced, leading to a lower endometrial cancer risk than observed in MSH2 mutation carriers. In our recent collaborative study of specific EPCAM deletion genotype/phenotype correlations, no endometrial cancer was observed in 62 female EPCAM deletion carriers with mutations distant from the MSH2 promoter (7). Endometrial cancer was observed in only 3 out of 30 carriers with larger EPCAM deletions that extend close to the MSH2 promoter. In line with this, the family history of the total 12 additional EPCAM deletion families shows no endometrial cancers (6,26). Unspecified gynecologic cancers were noted in only one of these families, which has a deletion extending close to the MSH2 promoter (6). A large collaborative study is needed to further refine the endometrial cancer risk with respect to the size and location of the EPCAM deletion. Nevertheless, the absence of endometrial cancer in the two large families of ancestral origin described here and the additional 33 female carriers with the same deletion described earlier (7) is highly significant and strongly supports the assumption that families with small EPCAM deletions have no increased risk for endometrial cancer. This finding lends support to practice-changing management for LS patients with such EPCAM deletions. Specifically, screening could be focused exclusively on CRC. Therefore, women could be spared surveillance and consideration for prophylactic hysterectomy (27).
Three cases of small bowel adenocarcinoma, two occurring in the duodenum and one in the jejunum, were seen in Families R and A. Other small bowel cancers, located in the duodenum and ileum, have been identified in EPCAM mutation carriers (1,4,7). The timing and frequency of small bowel cancer surveillance in EPCAM mutation carriers require further study before an informed recommendation can be made.
Genetic counseling implications
LS families with EPCAM mutations now have a rational explanation for their significantly increased cancer susceptibility. Therefore, instead of receiving an “uninformative negative” MSH2 mutation test result, they are given results based on a pathogenic EPCAM mutation. Family members found not to harbor the EPCAM mutation do not have an increased lifetime risk for cancer and thus can follow general population screening guidelines. Conversely, those found to have the EPCAM pathogenic mutation will be advised to follow the high-risk LS CRC surveillance recommendations. Having a definitive diagnosis has increased acceptance of LS cancer screening recommendations (11,12).
Limitations for implementing diagnosis and cancer control in LS
LS is frequently under-recognized due to small families, physicians not taking a detailed family history for cancer, or unawareness by patients of their own family history (28). Although routine microsatellite instability testing at the initiative of the pathologist in CRC diagnosed before age 50 can significantly enhance recognition of LS patients (29), this procedure has been implemented in only relatively few centers. In the next few years, many evolving genetic and genomic technologies devoted to cancer risk assessment will reach our daily clinical practice, heralding and finally implementing the era of personalized medicine (30). It will be important for clinicians to recognize the oft en profound cancer control potential when attention is paid to the family history including early age at cancer diagnosis and, when necessary, referral is made to genetic centers that offer high levels of expertise for DNA testing when appropriate. This permits the physician to apply appropriate surveillance programs to patients who require this, and to avoid inappropriate surveillance in at-risk family members who did not inherit the pathological allele. Family R and Family A, with the identification of their EPCAM mutation, is one such example.
The presence of a small 3′ EPCAM deletion has resolved the high incidence of microsatellite instability-high tumors in two large LS families that were unexplained for decades. The lack of endometrial cancer in these families marks further understanding of the differences in the LS phenotype that will lead to more effective cancer control. The discovery of EPCAM deletions and their impact on hereditary cancer is likely the tip of the proverbial iceberg with respect to mutations that affect known disease causing genes indirectly. Large, well-characterized families as described here can be instrumental in unraveling such novel genetic mechanisms, which will increase our understanding of gene regulation and may provide a multitude of families with important information for surveillance (11,12).
Study Highlights.
WHAT IS CURRENT KNOWLEDGE
-
✓
Lynch syndrome is a complex disease with a wide phenotype caused by mutations in several genes. Some of the variations seen in the Lynch syndrome phenotype can be attributed to which gene is mutated. However, the gene-specific risks for Lynch syndrome are still under investigation.
-
✓
Lynch syndrome’s cancer phenotype may differ due to colorectal cancer (CRC) predominance with deficit of extracolonic cancer resulting from deletion at the 3′ end of the epithelial cell adhesion molecular (EPCAM) gene, resulting in transcription read-through with silencing of MSH2 through hypermethylation of its promoter.
WHAT IS NEW HERE
-
✓
Epithelial cell adhesion molecule (EPCAM) mutation was identified in two highly extended Dutch families, both of whom harbored the same deletion at the 3′ end of EPCAM, showing that the deletion originated from a common ancestor.
-
✓
Findings of almost exclusive colorectal cancer (CRC) with virtually no extracolonic cancer show a likely paradigm change in cancer screening of certain families with the EPCAM mutation.
-
✓
Lynch syndrome patients carrying this EPCAM deletion are not at increased risk for the extracolonic cancers seen in Lynch syndrome. Therefore, cancer control in this group should focus on CRC.
Acknowledgments
Financial support: The work on Family R was supported by revenue from Nebraska cigarette taxes awarded to Creighton University by the Nebraska Department of Health and Human Services. The contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of the State of Nebraska or the Nebraska Department of Health and Human Services. Dr Henry Lynch’s work is partially funded through the Charles F. and Mary C. Heider Chair in Cancer Research, which he holds at Creighton University. The work on Dutch Family A was financially supported by the Sacha Swarttouw-Hijmans foundation and the Dutch Cancer Society Grant 2009-4335.
Footnotes
CONFLICT OF INTEREST
Guarantor of the article: Henry T. Lynch, MD.
Specific author contributions: Study concept and design, acquisition, analysis, and interpretation of data, manuscript drafting, obtained funding, study supervision, and family contact and education: Henry T. Lynch; study concept and design, acquisition, analysis, and interpretation of data, manuscript drafting, technical and material support, and family contact and education: Douglas Riegert-Johnson; acquisition, analysis, and interpretation of data, critical manuscript revision, and family contact and education: Carrie Snyder, Jane F. Lynch, and Janine Davies; analysis and interpretation of data and technical and material support: Jill Hagenkord; analysis and interpretation of data and critical manuscript revision: C. Richard Boland; analysis and interpretation of data: Jennifer Rhees; acquisition, analysis, and interpretation of data, critical manuscript revision, and technical and material support: Stephen N. Thibodeau; acquisition, analysis, and interpretation of data and technical and material support: Lisa A. Boardman; acquisition, analysis, and interpretation of data, critical manuscript revision, and technical and material support: Roland P. Kuiper; acquisition, analysis, and interpretation of data, critical manuscript revision, and technical and material support: Nicoline Hoogerbrugge; study concept and design, acquisition, analysis, and interpretation of data, manuscript drafting, obtained funding, and study supervision: Marjolijn J.L. Ligtenberg.
Potential competing interests: None.
REFERENCES
- 1.Ligtenberg MJL, Kuiper RP, Chan TL, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3 ´ exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 2.Kobelka CE. Silencing is not-so golden: a new model for inheritance of Lynch syndrome. Clin Genet. 2009;75:522–523. [Google Scholar]
- 3.Kovacs ME, Papp J, Szentirmay Z, et al. Deletions removing the last exon of TACSTD1 constitute a distinct class of mutations predisposing to Lynch syndrome. Hum Mutat. 2009;30:197–203. doi: 10.1002/humu.20942. [DOI] [PubMed] [Google Scholar]
- 4.Niessen RC, Hofstra RMW, Westers H, et al. Germline hypermethylation of MLH1 and EPCAM deletions are a frequent cause of Lynch syndrome. Genes Chromosomes Cancer. 2009;48:737–744. doi: 10.1002/gcc.20678. [DOI] [PubMed] [Google Scholar]
- 5.Kuiper RP, Vissers LELM, Venkatachalam R, et al. Recurrence and variability of germline EPCAM deletions in Lynch syndrome. Hum Mutat. 2011;32:407–414. doi: 10.1002/humu.21446. [DOI] [PubMed] [Google Scholar]
- 6.Rumilla K, Schowalter KV, Lindor NM, et al. Frequency of deletions of EPCAM (TACSTD1) in MSH2-associated lynch syndrome cases. J Mol Diagn. 2011;13:93–99. doi: 10.1016/j.jmoldx.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kempers MJE, Kuiper RP, Ockeloen CW, et al. Risk of colorectal and endometrial cancers in EPCAM deletion-positive Lynch syndrome: a cohort study. Lancet Oncol. 2011;12:49–55. doi: 10.1016/S1470-2045(10)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch PM, Lynch HT, Harris R, et al. Heritable colon cancer and solitary adenomatous polyps. In: Nieburgs HE, editor. Prevention and Detection of Cancer; 3d International Symposium on Detection and Prevention of Cancer; 1976; New York. New York: Marcel Dekker, Inc.; 1978. pp. 1573–1589. [Google Scholar]
- 9.Lynch HT, Harris RE, Bardawil WA, et al. Management of hereditary site-specific colon cancer. Arch Surg. 1977;112:170–174. doi: 10.1001/archsurg.1977.01370020064007. [DOI] [PubMed] [Google Scholar]
- 10.Lynch HT. Family information service and hereditary cancer. Cancer. 2001;91:625–628. doi: 10.1002/1097-0142(20010215)91:4<625::aid-cncr1044>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Lynch HT, de la Chapelle A. Genomic medicine: hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 12.Lynch HT, Lynch PM, Lanspa SJ, et al. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch HT, Kimberling W, Albano WA, et al. Hereditary nonpolyposis colorectal cancer (Lynch syndromes I and II). I. Clinical description of resource. Cancer. 1985;56:934–938. doi: 10.1002/1097-0142(19850815)56:4<934::aid-cncr2820560439>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 14.Lynch HT, Schuelke GS, Kimberling WJ, et al. Hereditary nonpolyposis colorectal cancer (Lynch syndromes I and II). II. Biomarker studies. Cancer. 1985;56:939–951. doi: 10.1002/1097-0142(19850815)56:4<939::aid-cncr2820560440>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 15.Peltomäki P, Aaltonen L, Sistonen P, et al. Genetic mapping of a locus pre-disposing to human colorectal cancer. Science. 1993;260:810–812. doi: 10.1126/science.8484120. [DOI] [PubMed] [Google Scholar]
- 16.Lindblom A, Tannergard P, Werelius B, et al. Genetic mapping of a second locus predisposing to hereditary nonpolyposis colorectal cancer. Nat Genet. 1993;5:279–582. doi: 10.1038/ng1193-279. [DOI] [PubMed] [Google Scholar]
- 17.Fishel R, Lescoe MK, Rao MRS. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 18.Leach FS, Nicolaides NC, Papadopoulos N, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 19.Bronner CE, Baker SM, Morrison PT, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary nonpolyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 20.Papadopoulos N, Nicolaides NC, Wei Y-F, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 21.Boland CR, Troncale FJ. Familial colonic cancer without antecedent poly-posis. Ann Intern Med. 1984;100:700–701. doi: 10.7326/0003-4819-100-5-700. [DOI] [PubMed] [Google Scholar]
- 22.Vasen HFA, Mecklin J-P, Meera Khan P, et al. The International Collaborative Group on hereditary nonpolyposis colorectal cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 23.Vasen HFA, Watson P, Mecklin J-P, et al. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative Group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 24.Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26:5783–5788. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindor NM, Rabe K, Petersen GM, et al. Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: familial colorectal cancer type X. JAMA. 2005;293:1979–1985. doi: 10.1001/jama.293.16.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guarinos C, Castillejo A, Barberá VM, et al. EPCAM germ line deletions as causes of Lynch syndrome in Spanish patients. J Mol Diagn. 2010;12:765–770. doi: 10.2353/jmoldx.2010.100039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmeler KM, Lynch HT, Chen L-M, et al. Prophylactic surgery to reduce the risk of gynecologic cancers in the Lynch syndrome. N Engl J Med. 2006;354:261–269. doi: 10.1056/NEJMoa052627. [DOI] [PubMed] [Google Scholar]
- 28.Singh H, Schiesser R, Anand G, et al. Underdiagnosis of Lynch syndrome involves more than family history criteria. Clin Gastroenterol Hepatol. 2010;8:523–529. doi: 10.1016/j.cgh.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overbeek LI, Hermens RP, van Krieken JH, et al. Electronic reminders for pathologists promote recognition of patients at risk for Lynch syndrome: cluster-randomised controlled trial. Virchows Arch. 2010;456:653–659. doi: 10.1007/s00428-010-0907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDonald DJ, Blazer KR, Weitzel JN. Extending comprehensive cancer center expertise in clinical cancer genetics and genomics to diverse communities: the power of partnership. JNCCN. 2010;8:615–624. doi: 10.6004/jnccn.2010.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]