Abstract
Objective
To define shared and unique features of SLE nephritis in mouse models of proliferative and glomerulosclerotic renal disease.
Methods
Perfused kidneys from NZB/W F1, NZW/BXSB and NZM2410 mice were harvested before and after nephritis onset. Affymetrix based gene expression profiles of kidney RNA were analyzed using Genomatix Pathway Systems and Ingenuity Pathway Analysis software. Gene expression patterns were confirmed using real-time PCR.
Results
955, 1168 and 755 genes were regulated in the kidneys of nephritic NZB/W F1, NZM2410 and NZW/BXSB mice respectively. 263 genes were regulated concordantly in all three strains reflecting immune cell infiltration, endothelial cell activation, complement activation, cytokine signaling, tissue remodeling and hypoxia. STAT3 was the top associated transcription factor, having a binding site in the gene promoter of 60/263 regulated genes. The two strains with proliferative nephritis shared a macrophage/DC infiltration and activation signature. NZB/W and NZM2410 mice shared a mitochondrial dysfunction signature. Dominant T cell and plasma cell signatures in NZB/W mice reflected lymphoid aggregates; this was the only strain with regulatory T cell infiltrates. NZW/BXSB mice manifested tubular regeneration and NZM2410 mice had the most metabolic stress and manifested loss of nephrin, indicating podocyte loss.
Conclusions
These findings identify shared inflammatory mechanisms of SLE nephritis that can be therapeutically targeted. Nevertheless, the heterogeneity of effector mechanisms suggests that individualized therapy might need to be based on biopsy findings. Some common mechanisms are shared with non-immune–mediated renal diseases, suggesting that strategies to prevent tissue hypoxia and remodeling may be useful in SLE nephritis.
Introduction
Lupus nephritis is a devastating complication of SLE for which current treatment is insufficiently effective and excessively toxic. Histologic analysis of renal biopsies does not always predict outcome or response to therapy. Furthermore, the study of pathogenic mechanisms of SLE nephritis is hampered by the small amount of biopsy material, the invasiveness of repeat biopsies and by therapeutic interventions begun prior to biopsy. It therefore remains essential to study animal models [1] in which the whole organ can be studied without the confounding effects of medications.
In these studies, we used transcriptional profiling to define similarities and differences in the renal inflammatory process between three well-characterized models of SLE nephritis. Female NZB/W F1 mice develop high titers of IgG2a anti-dsDNA autoantibodies and proliferative glomerulonephritis similar to class IV human lupus nephritis [1], [2]. NZM2410 mice, while genetically similar to NZB/W, express high levels of IL-4 and IgG1 and IgE autoantibodies; they develop rapidly progressive glomerulosclerosis with scant lymphocytic infiltrate [3]. Male NZW/BXSB mice carry the Yaa (Y linked autoimmune acceleration) locus containing a reduplication of the Tlr7 gene [4]. They develop anti-RNA and anti-cardiolipin antibodies and proliferative glomerulonephritis with severe tubulointerstitial inflammation [5]. Not surprisingly, differences in both pathogenic mechanisms and responses to immunologic interventions have been observed in the three models [6], [7]. These differences parallel the emerging appreciation of heterogeneity in human SLE nephritis [8], [9].
Our first goal was to identify shared expression profiles between the three models during active untreated nephritis; these define major driving forces in disease pathogenesis and may help in the development of broadly applicable treatment strategies. We next wished to identify profiles associated with proliferative nephritis, a subtype with a poor prognosis. Our final goal was to determine whether different strains with similar or different histologic lesions have unique features that could guide the choice of lupus model to study specific features of disease.
While a core set of regulated genes was shared among the three models, there were many differences, even between the two genetically similar strains and between the two strains with proliferative disease. Unique gene expression profiles accounted for approximately one third of the expression profile in each strain and revealed individual features of potential pathogenic significance. Our findings reflect the heterogeneity of renal responses to immune complex deposition and inflammation and suggest that targeting the appropriate effector mechanism in individual patients may improve the treatment of SLE nephritis.
Materials and Methods
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the Feinstein Institute (Protocol Number: 2007-054). All surgery was performed under ketamine/xylazine anesthesia, and all efforts were made to minimize suffering.
Mouse models
Kidneys were harvested after cardiac perfusion with 60 ml of sterile saline. A detailed description of the samples used for microarray analysis and derivation of the gene sets of interest has been previously published [10].
NZBW: NZB/NZW F1 female kidneys were harvested at the age of 6 (n = 7) or 16 (n = 8) weeks (no serum autoantibodies, immune complex deposition, or proteinuria) and 36–40 weeks (established proteinuria >300 mg/dl for >2 wk, n = 10) [11].
NZW/BXSB: Male (NZW×BXSB) F1 kidneys were obtained at the age of 8 weeks (no serum autoantibodies or proteinuria n = 4), 17 weeks (autoantibodies but no proteinuria, n = 6) and 18–21 weeks (established proteinuria >300 mg/dl for >2 wk and histologic glomerular score >2, n = 12; 6 randomly used for microarray).
NZM2410: NZM2410 kidneys were obtained at 6–8 weeks (no autoantibodies, renal immune complex deposition or proteinuria, n = 5) and 22–30 weeks (proteinuria >300 mg/dL for 7–10 days, n = 7; 5 randomly used for microarray). NZM2410 mice were harvested early after proteinuria onset as most die within 14 days [12].
Histologic assessment of kidneys
H & E stained sections were scored for glomerular and interstitial damage by a single blinded observer using a semi-quantitative scale from 0–4 as previously described [11].
Paraffin embedded sections were dewaxed and deparaffinized. Endogenous peroxidase activity was blocked with 3%H2O2 and antigen retrieval performed in 10 mM citrate buffer pH6. Slides were stained with at anti-Ki67 (Dako, Carpinteria, CA) or PCNA (Invitrogen Life Technologies, Grand Island, NY) followed by biotin conjugated anti-rat Ig and ABC developer (Vectastain, Burlingame, CA).
RNA purification and microarray hybridization
RNA extraction, cDNA synthesis, hybridization, microarray processing, data normalization and filtering were performed as previously described [10], [13]. Microarray gene expression data were normalized and batch-corrected together, and quality controls were used before further analyses. Significantly regulated genes were analyzed by creating biological literature-based networks using Genomatix Pathway System software (GePS) (www.genomatix.de). Canonical pathways were analyzed using Ingenuity Pathway Analysis software (IPA) (www.ingenuity.com). Principal component analysis was performed using ArrayTrack™ software (http://www.fda.gov/ArrayTrack). Gene expression datasets are on Gene Expression Omnibus (GEO) at http://www.ncbi.nlm.nih.gov/geo/ (accession numbers GSE32583, GSE44691 and #GSE49898).
Quantitative Real Time PCR (qPCR)
We selected to validate by qRT-PCR 158 genes including some that were highly significant, some that had a lower fold change and several genes involved in pathways of interest such as mitochondrial function, ER stress and control of circadian rhythm. A number of cytokine genes that are not expressed in normal kidneys and did not pass the cut-off on the arrays, were also tested by qRT-PCR.
Validation of selected genes was performed by qPCR as previously described (LightCycler480, Roche Diagnostics - [11], [13]; primers on request). Data were analyzed using a comparative cycle threshold (2-ΔΔCT) method, normalized to β-actin and expressed as fold induction relative to a pre-nephritic calibrator of the same strain [11], [13].
Statistical Analysis
The TIGR MultiExperiment Viewer (TMEV) application [14 #5174] was used for statistical analysis of microarray and qPCR data. For the array study, statistical unpaired analyses for each comparison were performed using the Significance Analysis of Microarrays (SAM) method and the following criteria: q-value ≤0.001 and fold change ≥1.4 for the upregulated genes and ≤0.7 for the downregulated genes. We used more stringent filter criteria than in our mouse-human comparison [10] so as to capture the maximum number of genes with differential expression. To avoid ambiguity between species, mouse genes were converted to the corresponding human orthologs using the NCBI homolog (Build 64) and Genomatix annotated ortholog databases (Table S1).
For the qPCR data, unpaired SAM and t-test were performed. Unsupervised hierarchical clustering with bootstrap procedures was performed using Euclidean metrics with average or complete linkage and visualized using TMEV. Data were scaled to the mean of the young mice in each strain, given a value of 1. Genes regulated between two groups with a fold change of >2 and q-value <0.05 were considered significant.
BrDU staining of renal cells
Groups of 3-6 NZB/W and NZW/BXSB mice were fed BrDU as previously described [13] for 12–15 days. Kidneys were perfused and harvested as above and flow cytometry was performed as previously described [13].
Results
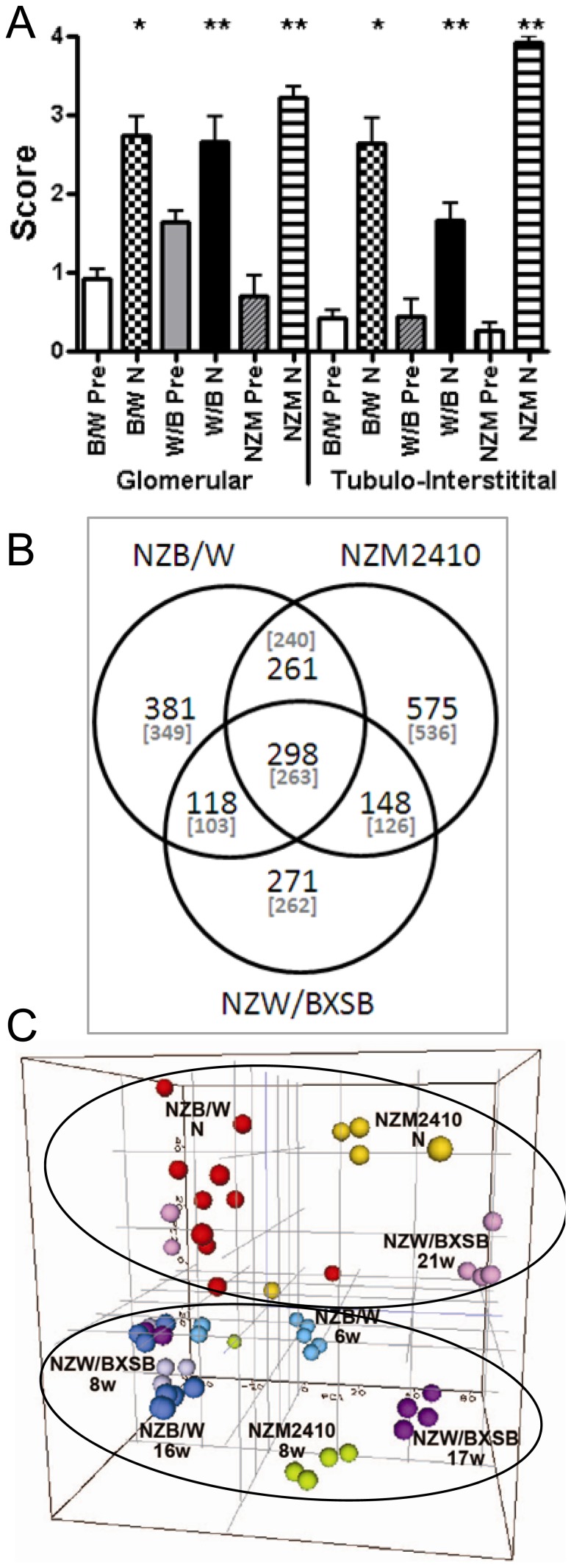
The clinical characteristics and histologic renal appearance of the three strains have previously been reported [11], [12], [15], [16]; renal damage scores are displayed in Figure 1A . Differentially regulated genes were identified by comparing the renal profiles of the nephritic mice to their prenephritic control groups. 955 (442 down, 513 up), 755 (208 down, 547 up) and 1168 (628 down, 540 up) genes were differentially regulated in NZB/W, NZW/BXSB and NZM2410 models respectively ( Figure 1B and Table S1). Principal component analysis of all genes expressed in the 3 strains showed a clear separation of the nephritic vs. non-nephritic mice as well as differences between strains ( Figure 1C ).
Figure 1. A. Glomerular and interstitial damage scores in pre-nephritic (Pre) and nephritic (N) mice (mean + SD) of NZB/W (B/W), NZW/BXSB (W/B) and NZM2410 NZM) strains (* p<0.001; ** p<0.01). The high tubulointerstitial score in the NZM2410 strain reflects severe tubular atrophy. B. Shared and unique gene expression profiles of each of the three strains. Parentheses indicate the number of genes with a human ortholog. C. 3D principal component analysis from the 14780 genes passing the cutoff value (see Materials and Methods) after normalization and batch correction of the arrays from the 3 mouse strains together.
Profiles shared by all three strains
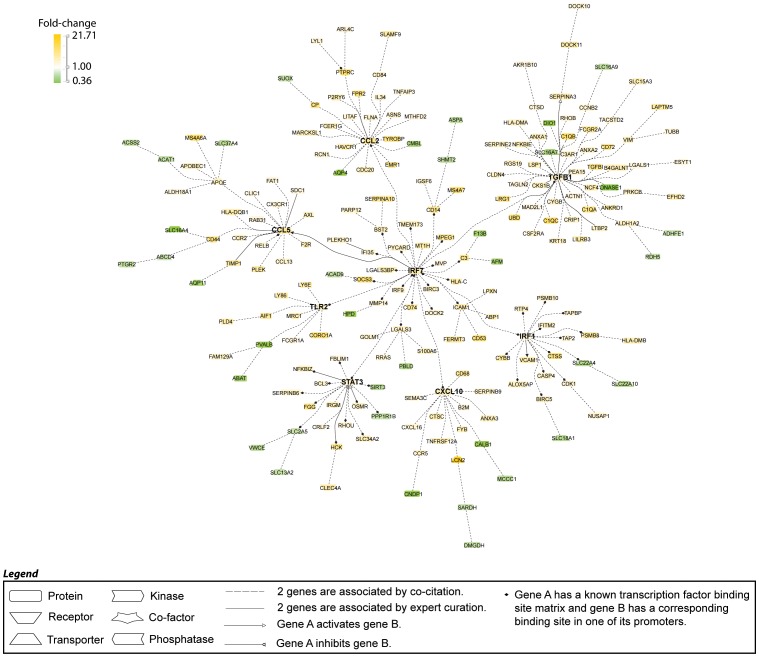
263 genes (67 down and 196 up) were regulated in the same direction in all 3 strains. Transcriptomic network analysis using Genomatix Pathway System was performed to identify the major nodes along with their gene clusters ( Figure 2 and data not shown). These nodes reflect transcripts associated with macrophage activation (CD68, activating Fc receptors, FPR2, C type lectins, cathepsins, HLA-DM), classical complement pathway proteins, endothelial activation (VCAM-1, ICAM-1), metabolic stress and proteasome activity (HPD, CNDP1, UBD), tissue remodeling and fibrosis (TIMP-1, MMP14, TGFB1) and tubular damage (HAVCR1, LCN2). Three cytokines, IL1f6 (IL-36), IL-34 and TGFβ, were shared by all three strains as well as a limited group of chemokines and chemokine receptors, including CCL2, CCL5, CCL9 and their receptors CCR2 and CCR5 as well as CXCL10 and CXCL16. Other genes of interest include the TWEAK receptor FN14, whose expression on intrinsic renal cells promotes glomerulonephritis [17], ANKRD1, which is associated with proteinuria in SLE nephritis [18], TLRs 2 and 13, and several proteinase inhibitors of the Serpin family involved in the coagulation pathway. SMPDL3B, the renal target of Rituximab [19] was also upregulated in all three strains. Transcription factor analysis identified STAT3 as the top transcription factor having binding sites in the promoter regions of 60 of the 263 genes (Table S2), followed by transcription factors involved in interferon signaling (IRF1 and IRF7) and RELB ( Table 1 , Figure 2 ). Accordingly, we identified 48, 34 and 32 of a panel of 195 interferon inducible genes [20], [21], [22] in NZB/W, NZM2410 and NZW/BXSB strains respectively (not shown). Consistent with these findings, the top ingenuity pathways included antigen processing and presentation, dendritic cell maturation, complement activation, and pathways reflecting innate and adaptive immune activation ( Table 1 ).
Figure 2. Literature-based analysis of genes shared among all three strains using Genomatix Pathway System (GePS) software.
263 human gene orthologs were regulated in the same direction in the nephritic vs. prenephritic kidneys in NZB/W, NZM2410 and NZW/BXSB. The picture shows the 204 genes that were co-cited in PubMed abstracts in the same sentence. Orange represents the genes that are upregulated and green represents the genes that are downregulated in nephritic compared to prenephritic mice.
Table 1. Top 10 canonical pathways significantly regulated sorted by Benjamini-Hochberg Multiple Testing corrected p-value (p-value<0.05), as assessed by IPA (Ingenuity Pathway Analysis).
| Canonical pathways(number of genes regulated in the pathway/number of genes in the pathway) | B-H Multiple testing corrected p-value | Regulated molecules in the pathway |
| From the 3 mouse model overlap (263 genes) * STAT3, IRF1, RELB , IRF7 , IRF9 † | ||
| Antigen Presentation Pathway (8/40) | 9.70E-06 | B2M, HLA-DMA, HLA-C, HLA-DMB, PSMB8, CD74, TAP2, TAPBP |
| Dendritic Cell Maturation (14/207) | 1.90E-04 | B2M, HLA-DMA, ICAM1, FCGR2A, TYROBP, NFKBIE, RELB, IL36A, HLA-DMB, HLA-DQB1, FCGR1A, TLR2, HLA-C, FCER1G |
| OX40 Signaling Pathway (7/94) | 4.09E-03 | B2M, HLA-DMA, HLA-C, NFKBIE, FCER1G, HLA-DMB, HLA-DQB1 |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses (9/106) | 4.09E-03 | TLR2, IRF7, C3, C1QC, C1QA, C1QB, CCL5, C3AR1, PRKCB |
| Graft-versus-Host Disease Signaling (6/50) | 4.09E-03 | HLA-DMA, HLA-C, FCER1G, IL36A , HLA-DMB, HLA-DQB1 |
| Complement System (5/35) | 5.22E-03 | C3, C1QC, C1QA, C1QB, C3AR1 |
| Altered T Cell and B Cell Signaling in Rheumatoid Arthritis (8/92) | 5.22E-03 | TLR2, HLA-DMA, TGFB1, RELB, FCER1G, IL36A, HLA-DMB, HLA-DQB1 |
| Allograft Rejection Signaling (6/95) | 5.73E-03 | B2M, HLA-DMA, HLA-C, FCER1G, HLA-DMB, HLA-DQB1 |
| Role of Hypercytokinemia / hyperchemokinemia in the Pathogenesis of Influenza (5/44) | 5.73E-03 | CXCL10, CCR5, CCL2, IL36A , CCL5 |
| Cytotoxic T Lymphocyte-mediated Apoptosis of Target Cells (6/85) | 7.40E-03 | B2M, HLA-DMA, HLA-C, FCER1G, HLA-DMB, HLA-DQB1 |
| From the NZB/W only (349 genes) * CEBPB, IRF4, FLI1, IRF8, IKZF1 † | ||
| CTLA4 Signaling in Cytotoxic T Lymphocytes (14/98) | 3.06E-06 | FYN, PTPN6, PIK3R5, PIK3C2G, CD3D, CD28, LCK, SYK, LAT, PPM1L, CD86, HLA-DOB, PTPN22, LCP2 |
| CD28 Signaling in T Helper Cells (13/132) | 3.47E-04 | FYN, PTPN6, PIK3R5, PIK3C2G, CD3D, CD28, LCK, SYK, LAT, CD86, HLA-DOB, VAV1, LCP2 |
| PKCθ Signaling in T Lymphocytes (11/143) | 5.25E-03 | FYN, CD28, LCK, LAT, PIK3R5, PIK3C2G, CD86, HLA-DOB, VAV1, CD3D, LCP2 |
| Natural Killer Cell Signaling (10/116) | 5.25E-03 | FYN, LCK, PTPN6, SYK, LAT, PIK3R5, PIK3C2G, SH3BP2, VAV1, LCP2 |
| T Cell Receptor Signaling (10/109) | 5.25E-03 | BTK, FYN, CD28, LCK, LAT, PIK3R5, PIK3C2G, VAV1, CD3D, LCP2 |
| Role of NFAT in Regulation of the Immune Response (13/198) | 6.23E-03 | FYN, CD79B, PIK3R5, PIK3C2G, CD3D, BTK, CD28, LCK, SYK, LAT, CD86, HLA-DOB, LCP2 |
| iCOS-iCOSL Signaling in T Helper Cells (10/123) | 6.41E-03 | CD28, LCK, IL2RG, LAT, PIK3R5, PIK3C2G, HLA-DOB, VAV1, CD3D, LCP2 |
| B Cell Development (5/33) | 9.83E-03 | IL7R, SPN, CD79B, CD86, HLA-DOB |
| Crosstalk between Dendritic Cells and Natural Killer Cells (8/95) | 9.83E-03 | CSF2RB, TLR4, CD28, IL2RG, LTB, CD86, CD83, HLA-F |
| Primary Immunodeficiency Signaling (6/62) | 9.83E-03 | IL7R, BTK, LCK, IL2RG, ADA, CD3D |
| From the NZM2410 only (536 genes) * JUN , NFKB1, MYC, EGR1, WT1 † | ||
| Putrescine Degradation III (5/30) | 2.64E-02 | ALDH1B1, ALDH1A1, SAT2, SMOX, ALDH9A1 |
| Aryl Hydrocarbon Receptor Signaling (14/161) | 2.64E-02 | ALDH1B1, NFKB2, NFKB1, CCND1, ALDH9A1, MYC, GSTT1, GSTM2, ALDH1A1, JUN, CDKN1A, DHFR, GSTO2, NFE2L2 |
| LPS/IL-1 Mediated Inhibition of RXR Function (18/239) | 2.64E-02 | ECSIT, ALDH1B1, NR1H4, IL1R1, ALDH9A1, SOD3, CHST15, GSTT1, GSTM2, ALDH1A1, JUN, IL1RN, ACSL5, ALAS1, HS6ST2, FABP1, SMOX, GSTO2 |
| TCA Cycle II (Eukaryotic) (5/41) | 7.08E-02 | IDH3G, ACO2, DLST, SDHC, ACO1 |
| Guanine and Guanosine Salvage I (2/9) | 8.55E-02 | PNP, HPRT1 |
| Cysteine Biosynthesis/Homocysteine Degradation (2/8) | 8.55E-02 | CBS, CTH |
| Role of IL-17F in Allergic Inflammatory Airway Diseases (6/47) | 1.03E-01 | IGF1, CCL7, CREB3, IL17RC, NFKB2, NFKB1 |
| Tryptophan Degradation X (Mammalian, via Tryptamine) (4/29) | 1.03E-01 | ALDH1B1, ALDH1A1, SMOX, ALDH9A1 |
| Valine Degradation I (4/35) | 1.45E-01 | HIBADH, BCKDHA, ACAD8, ACADSB |
| Dopamine Degradation (4/37) | 1.45E-01 | ALDH1B1, ALDH1A1, SMOX, ALDH9A1 |
| From the NZW/BXSB only (262) * LHX1 † | ||
| Cell Cycle Control of Chromosomal Replication (6/31) | 1.50E-03 | MCM3, MCM6, MCM2, CDT1, DBF4, MCM7 |
| Hepatic Fibrosis / Hepatic Stellate Cell Activation (10/146) | 2.24E-02 | MYL9, COL1A1, LY96, MYH14, ACTA2, IGFBP3, IL6 , PDGFB, PDGFRB, COL3A1 |
| Atherosclerosis Signaling (8/136) | 6.32E-02 | COL1A1, MSR1, PLA2G5, CD36, SERPINA1, IL6 , PDGFB, COL3A1 |
| Complement System (4/35) | 6.32E-02 | C5AR1, C8A, C2, C8G |
| DNA Double-Strand Break Repair by Homologous Recombination (3/17) | 9.33E-02 | LIG1, POLA1, BRCA1 |
| LXR/RXR Activation (7/136) | 1.57E-01 | LY96, MSR1, APOH, CD36, SERPINA1, IL6 , GC |
| Role of BRCA1 in DNA Damage Response (5/65) | 1.57E-01 | RFC4, RBL1, BRCA1, CHEK1, RFC3 |
| Intrinsic Prothrombin Activation Pathway (3/35) | 3.93E-01 | COL1A1, F13A1, COL3A1 |
| Role of CHK Proteins in Cell Cycle Checkpoint Control (4/57) | 4.36E-01 | RFC4, BRCA1, CHEK1, RFC3 |
| Estrogen Biosynthesis (3/49) | 4.96E-01 | CYP2D6, CYP2F1, HSD17B2 |
| From the NZB/W with NZM2410 overlap (240 genes) * | ||
| Mitochondrial Dysfunction (10/174) | 1.30E-02 | SDHA, NDUFS5, SDHB, NDUFS1, SOD2, UQCRC2, LRRK2, NDUFB10, COX15, AIFM1 |
| Valine Degradation I (4/35) | 3.03E-02 | ECHS1, AUH, ALDH6A1, BCKDHB |
| TCA Cycle II (Eukaryotic) (4/41) | 3.81E-02 | SDHA, SUCLA2, SDHB, IDH3B |
| Oleate Biosynthesis II (Animals) (3/18) | 4.06E-02 | FADS2, ALDH6A1, FADS1 |
| Ethanol Degradation II (4/43) | 4.50E-02 | ALDH4A1, ACSL3, PECR, ALDH7A1 |
| D-glucuronate Degradation I (2/13) | 4.50E-02 | CRYL1, DCXR |
| Fatty Acid β-oxidation I (4/45) | 4.89E-02 | ACSL3, ECHS1, AUH, HADH |
| Methylmalonyl Pathway (2/12) | 4.89E-02 | PCCA, MCEE |
| Arginine Degradation I (Arginase Pathway) (2/13) | 4.89E-02 | ALDH4A1, ARG2 |
| Molybdenum Cofactor Biosynthesis (2/15) | 4.89E-02 | GPHN, NFS1 |
| From the NZM2410 with NZW/BXSB overlap (126 genes) * | ||
| Leucine Degradation I (2/26) | 2.54E-01 | IVD, ACADM |
| LXR/RXR Activation (5/136) | 2.54E-01 | TNFRSF1A, LPL, CLU, ABCA1, CYP51A1 |
| Asparagine Degradation I (1/4) | 3.65E-01 | ASPG |
| Thiamin Salvage III (1/5) | 3.65E-01 | TPK1 |
| Sertoli Cell-Sertoli Cell Junction Signaling (5/195) | 4.68E-01 | TUBB6, CLDN1, TNFRSF1A, CLDN16, CLDN7 |
| Triacylglycerol Degradation (2/32) | 4.68E-01 | LPL, MGLL |
| Cell Cycle Control of Chromosomal Replication (2/31) | 4.68E-01 | CDC6, MCM4 |
| Methionine Salvage II (Mammalian) (1/9) | 4.68E-01 | BHMT2 |
| Fatty Acid β-oxidation I (2/45) | 4.68E-01 | IVD, ACADM |
| LPS/IL-1 Mediated Inhibition of RXR Function (5/239) | 4.68E-01 | ALDH1L2, ACOX2, TNFRSF1A, ACOX3, ABCA1 |
| From the NZW/BXSB with NZB/W overlap (103 genes) * SPI1 † | ||
| Hepatic Fibrosis / Hepatic Stellate Cell Activation (7/146) | 1.27E-02 | COL1A2, FN1, IL10RA, MYH9, EGF, IFNGR1, MMP2 |
| Caveolar-mediated Endocytosis Signaling (5/85) | 1.49E-02 | ITGB2, ITGAM, CD48, EGF, ITGAX |
| Atherosclerosis Signaling (5/136) | 6.73E-02 | IL33, COL1A2, ITGB2, F3, SELPLG |
| Inhibition of Matrix Metalloproteases (3/40) | 6.73E-02 | THBS2, MMP2, LRP1 |
| IL-8 Signaling (6/205) | 6.73E-02 | ITGB2, ITGAM, EGF, IKBKE, MMP2, ITGAX |
| Leukocyte Extravasation Signaling (6/201) | 6.73E-02 | ITGB2, NCF1, ITGAM, THY1, MMP2, SELPLG |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses (4/106) | 1.29E-01 | IFIH1, CLEC7A, TLR7, CLEC6A |
| Activation of IRF by Cytosolic Pattern Recognition Receptors (3/72) | 1.31E-01 | IFIH1, IKBKE, IFIT2 |
| Extrinsic Prothrombin Activation Pathway (2/20) | 1.31E-01 | F5, F3 |
| Colorectal Cancer Metastasis Signaling (6/258) | 1.31E-01 | TLR7, EGF, IFNGR1, MMP2, ADCY7, LRP1 |
Genes tested by RT-PCR are highlighted in bold and underlined.
(number of genes regulated in the same direction).
top transcription factors for each analysis as assessed by GePS.
Profiles shared by two strains, based on the defined filter criteria
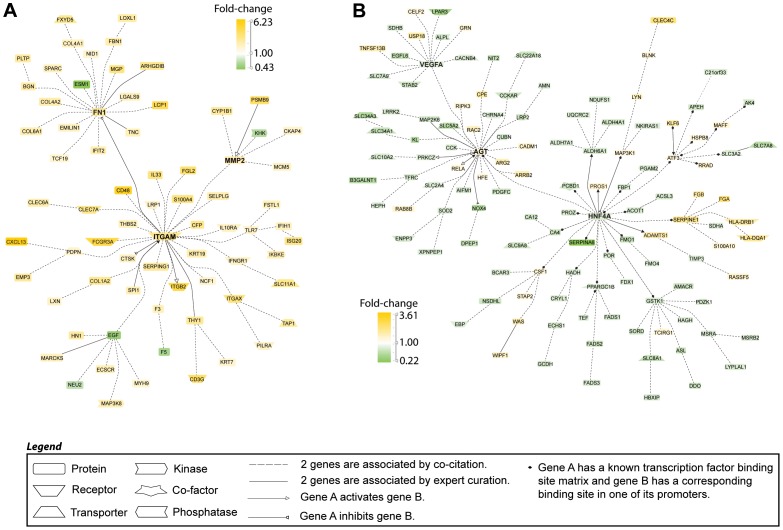
As described in the methods section, we applied stringent filter criteria to capture the maximum number of genes with differential expression, allowing us to identify the most important LN related genes in the development of the disease in the different mouse models. This filter does however exclude those genes with a trend towards differences in expression. Table 2 shows the regulation and associated significance in each mouse model of selected genes of interest. 103 genes were preferentially regulated (16 down and 87 up) in the NZB/W and NZW/BXSB strains with proliferative glomerulonephritis. These included the two chains of the CD11b molecule as well as other macrophage/DC expressed genes (IKBKE, FCGR3A, CXCL13) [11], [15]. These strains also had increased collagen type IV expression indicating expansion of basement membrane, as well as of collagen type 1, fibronectin, thrombospondin-2 and MMP2, indicating interstitial fibrosis and remodeling ( Figure 3A ). Transcription factor analysis revealed SPI-1 (PU.1) as the top transcription factor ( Table 1 ), consistent with its role in inflammatory macrophage function and antigen presentation [23].
Table 2. Expression and significance of selected genes.
| NZB/W | NZM2410 | NZW/BXSB | ||||
| Gene name | Fold-change | q-value | Fold-change | q-value | Fold-change | q-value |
| Genes shared by two strains, based on the defined filter criteria | ||||||
| IKBKE | 1.72 | 0.0000 | 1.638 | 0.0012 | 1.43 | 0.0000 |
| FCGR3A | 3.87 | 0.0000 | 1.654 | 0.0015 | 2.06 | 0.0005 |
| CXCL13 | 6.22 | 0.0000 | 1.781 | 0.0079 | 2.50 | 0.0000 |
| COL4A1 | 1.59 | 0.0000 | 1.131 | 0.1691 | 1.76 | 0.0000 |
| FN1 | 1.99 | 0.0000 | 1.614 | 0.0029 | 1.94 | 0.0000 |
| THBS2 | 1.62 | 0.0006 | 1.726 | 0.0112 | 1.59 | 0.0000 |
| MMP2 | 1.66 | 0.0000 | 1.423 | 0.0101 | 1.87 | 0.0000 |
| PROS1 | 1.64 | 0.0000 | 1.53 | 0.0006 | 1.34 | 0.0005 |
| FGA | 3.61 | 0.0000 | 4.53 | 0.0005 | 1.44 | 0.2188 |
| FGB | 2.57 | 0.0004 | 4.82 | 0.0005 | 1.83 | 0.0273 |
| SERPINE1 | 2.93 | 0.0000 | 5.83 | 0.0005 | 1.72 | 0.0043 |
| SERPINA6 | 0.22 | 0.0000 | 0.04 | 0.0000 | np | |
| KRT20 | 2.37 | 0.0030 | 17.06 | 0.0000 | 2.18 | 0.0000 |
| CLDN1 | 1.45 | 0.1531 | 2.89 | 0.0000 | 1.79 | 0.0000 |
| CLDN7 | 1.56 | 0.0039 | 2.71 | 0.0004 | 1.55 | 0.0000 |
| CLU | 1.62 | 0.0048 | 3.04 | 0.0000 | 1.96 | 0.0000 |
| CXCL1 | 1.84 | 0.0341 | 5.47 | 0.0000 | 2.35 | 0.0000 |
| CXCL2 | 1.67 | 0.0250 | 4.77 | 0.0000 | 2.93 | 0.0000 |
| Genes unique to one model, based on the defined filter criteria | ||||||
| CD3D | 2.28 | 0.0000 | 1.61 | 0.0057 | 1.31 | 0.0023 |
| PTPN22 | 2.60 | 0.0000 | 2.03 | 0.0139 | 0.86 | 0.2124 |
| FYN | 1.48 | 0.0000 | 1.09 | 0.3296 | 1.09 | 0.1870 |
| LCP2 | 1.42 | 0.0000 | 1.26 | 0.0062 | 1.24 | 0.0058 |
| HLA-DOB | 1.78 | 0.0000 | np | np | ||
| LCK | 1.73 | 0.0000 | 1.43 | 0.0042 | 1.30 | 0.0027 |
| SYK | 1.70 | 0.0000 | 1.15 | 0.1163 | 1.29 | 0.0004 |
| HELLS | 1.67 | 0.0011 | 1.80 | 0.0016 | 3.38 | 0.0000 |
| MCM2 | 1.33 | 0.0024 | 1.24 | 0.0212 | 1.67 | 0.0002 |
| MCM3 | 1.39 | 0.0004 | 1.22 | 0.0315 | 1.78 | 0.0000 |
| MCM6 | 1.30 | 0.1408 | 1.39 | 0.0084 | 2.01 | 0.0000 |
| MCM7 | 1.33 | 0.0000 | 1.37 | 0.0000 | 1.47 | 0.0003 |
| AURKB | Np | np | 1.81 | 0.0000 | ||
| COL5A1 | 1.43 | 0.0250 | 1.03 | 0.4772 | 2.04 | 0.0000 |
| COL5A2 | 1.02 | 0.9999 | 0.83 | 0.0397 | 1.68 | 0.0000 |
| COL6A2 | 1.41 | 0.0097 | 1.12 | 0.4009 | 1.85 | 0.0000 |
| COL6A3 | 1.15 | 0.4089 | 1.05 | 0.9999 | 2.03 | 0.0003 |
| NFKB1 | 1.27 | 0.0017 | 1.43 | 0.0000 | 1.34 | 0.0000 |
| MYC | 1.72 | 0.0015 | 2.50 | 0.0006 | 1.50 | 0.0013 |
| EGR1 | 0.85 | 0.9999 | 3.16 | 0.0006 | 0.63 | 0.2291 |
| JUN | 1.10 | 0.4309 | 2.17 | 0.0000 | 0.85 | 0.2382 |
| SOD3 | 0.80 | 0.0250 | 0.59 | 0.0005 | 0.94 | 0.1747 |
The genes passing the defined filter criteria are highlighted in bold. np: genes not passing the Affymetrix negative controls cut-off. In italic are the genes not significantly regulated (q-value>0.05).
Figure 3. Literature-based analysis of limited gene expression patterns using Genomatix Pathway System (GePS) software.
A. 103 shared genes were regulated in the same direction in the nephritic vs. prenephritic kidneys in NZW/BXSB and NZB/W mouse models. The picture shows the 72 that were co-cited in the same sentence of PubMed abstracts. B. 240 genes were regulated in the same direction in the nephritic vs. prenephritic kidneys in NZB/W and NZM2410 mouse models. The picture shows the 124 genes that were co-cited in PubMed abstracts in the same sentence. Orange represents the genes that are upregulated and green represents the genes that are downregulated in nephritic compared to prenephritic mice.
NZM2410 mice shared 240 genes with the NZB/W strain ( Figure 3B ) and 126 genes with NZW/BXSB. NZB/W and NZM2410 mice shared genes associated with the coagulation and fibrinolytic cascades (Protein S, fibrinogen and plasminogen activator inhibitor - SERPINE) with HNF4A being the top transcription factor ( Tables 1 and 2 ). Decreased expression of Serpina6 (corticosteroid binding protein) was also shared between these two strains ( Table 2 ). The top shared ingenuity pathway was mitochondrial dysfunction ( Table 1 ). Upregulated genes shared between NZM2410 and NZW/BXSB included epithelial cell proteins (Keratin20, Claudin1, Claudin7, CLU) and the chemokines CXCL1 and CXCL2 that both bind to CXCR2 ( Table 2 ).
Unique gene profiles, based on the defined filter criteria
Each strain also expressed unique genes during nephritis, comprising 37, 35 and 46% of the total genes in the expression profiles of NZB/W, NZW/BXSB and NZM2410 kidneys respectively. In NZB/W mice, the major signal was derived from T cells, with 4 of the top 5 canonical pathways involving T cell activation ( Tables 1 and 2 ). High levels of immunoglobulin transcripts in this strain reflected plasma cell infiltration. The top transcription factor was C/EBPβ a regulator of monocyte/macrophage proliferation and differentiation [24]. In NZW/BXSB mice there was evidence of chromosomal replication/chromatin remodeling (HELLS, MCM2-7, AURKB) suggesting cellular proliferation ( Table 2 ). In addition, increased expression of collagen V and VI genes indicated a greater degree of interstitial fibrosis than in the other two strains ( Table 2 ). The top transcription factor, LHX1, has essential roles in multiple steps of epithelial tubular morphogenesis during kidney organogenesis [25]. NZM2410 mice differentially regulated genes involved in multiple metabolic pathways, indicating cellular stress. Top transcription factors in this strain included JUN, a mediator of TGFβ induced fibrosis related to ER stress [26], NFKB, MYC and EGR1 ( Tables 1 and 2 ).
Validation by RT-PCR
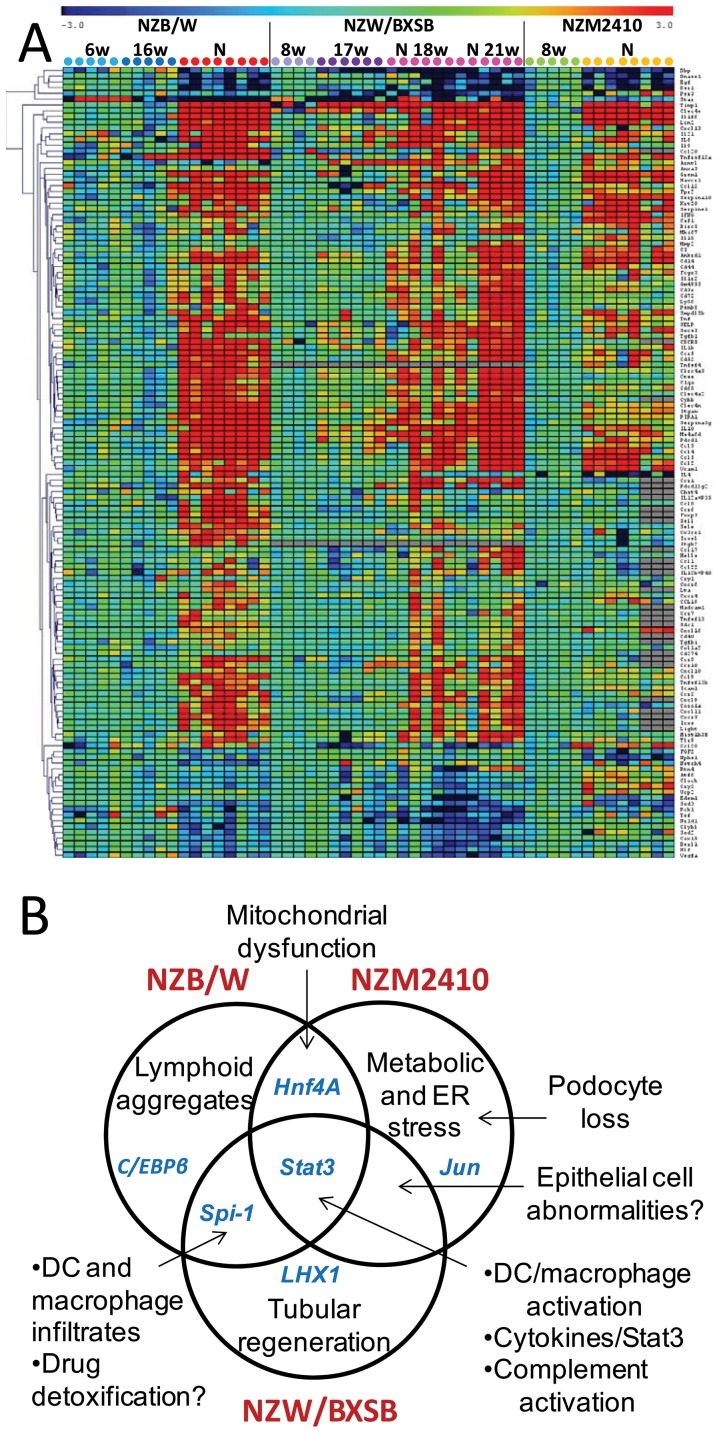
After excluding genes that were not expressed on the arrays, the concordance between the microarray data and qPCR data was 85.0%, 87.8% and 93.9% in the NZW/BXSB, NZM2410 and NZB/W strains respectively ( Figure 4A , Table S3). An increased sensitivity of the PCR assays accounted for almost all the observed discordance in NZB/W and NZW/BXSB mice whereas more than half of the discordance in NZM2410 mice was in genes that were <2 fold regulated on the microarrays. The higher discordance rates between microarray and real time PCR when low stringency cut-offs (<2 fold differences) are applied has previously been reported [27] but a higher stringency results in decreased specificity.
Figure 4. A. One way cluster analysis of genes with significantly altered expression in the PCR validation set (See Table S3). Gene expression was scaled to the mean of pre-nephritic controls for each strain. Significantly up or downregulated (>2 fold) genes by SAM with q value <0.05 are shown (corresponding to 137 genes). B. Summary of the unique and shared pathogenic pathways identified in the kidneys of the three mouse strains.
All three strains manifested significant regulation of 42 genes including LCN2, C3, IL1F6, C type lectins 4a3, 4e and 4N, ANKRD1, CCL2, CCL5, CXCL13, FCGR3, SERPINA10, ITGAM, TWEAK receptor, DNASE1, TIMP1 and VCAM-1 as demonstrated by microarray analysis. Because several cytokines either were not on the chip or did not pass the cut-off value, these were tested by PCR. IL6 and IL-10 signal through STAT3; these cytokines were tested and were upregulated in all three strains. Similarly, upregulation of IL-1β, TNFα and IFNγ was found in all three strains. Increased expression of IL17 was found in 50-85% of the mice in all three strains but reached significance only in the NZW/BXSB strain. The NZM2410 strain did not express IL-12p40, IL-21 or BAFF but was the only strain with increased expression of IL-18.
We next confirmed that the two strains with proliferative nephritis shared a profile consistent with inflammatory cell infiltration and endothelial activation. Genes in this set included chemokines, chemokine receptors, the actin cytoskeleton remodeling protein Coronin 1A, the vascular NADPH oxidase CYBB, LY86, CD40, BAFF, TLR9 and T cell molecules CD52, LIGHT, IL-21 and ICOS. Genes significantly downregulated in the kidneys of these two strains included the superoxide dismutases SOD2 and SOD3, the mitochondrial enzymes CLYBL and COX15, and estrogen related receptor β. Both strains manifested downregulation of the PAR domain basic leucine zipper transcription factors DBP, and HLF that control genes involved in lipid metabolism and detoxification [28]; downregulation of TEF, a gene with similar function was seen in NZW/BXSB. The NZM2410 mouse had less upregulation of CXC chemokines and their receptors than the other two strains and despite upregulation of CC chemokines, had only minimally increased expression of several of the corresponding chemokine receptors, consistent with the significantly lesser degree of inflammatory cell infiltration in this strain.
Unique genes regulated in NZB/W mice included the chemokine/chemokine receptor pair CCL20 and CCR6. Since we have previously identified Tregs in renal lymphoid aggregates of NZB/W mice [16] and Tregs express CCR6 we tested for FOXP3 expression. FOXP3 was upregulated only in NZB/W mice and correlated significantly with CCR6 expression (r value 0.87, p<0.001). In contrast, there was no correlation of CCR6 expression with IL-17A expression (not shown). Similarly, L selectin, a marker expressed on T cells in lymphoid aggregates [16] and CD138, a plasma cells marker, were found only in the NZB/W strain. Downregulation of nephrin was found only in NZM2410 mice reflecting podocyte loss associated with glomerulosclerosis. This strain also expressed ER stress markers ATF6 and RTN4. We confirmed the cell proliferation profile in the NZW/BXSB strain, with upregulation of the chromatin remodeling protein HELLS (helicase) and KI67. This strain also expressed the Th2 cell attracting chemokines CCL1, CCL17 and CCL22, perhaps reflecting the high degree of dendritic cell infiltration in this strain. A summary of the unique and shared pathogenic pathways identified in the three strains is shown in Figure 4B .
Identification of proliferating cells In NZW/BXSB mice
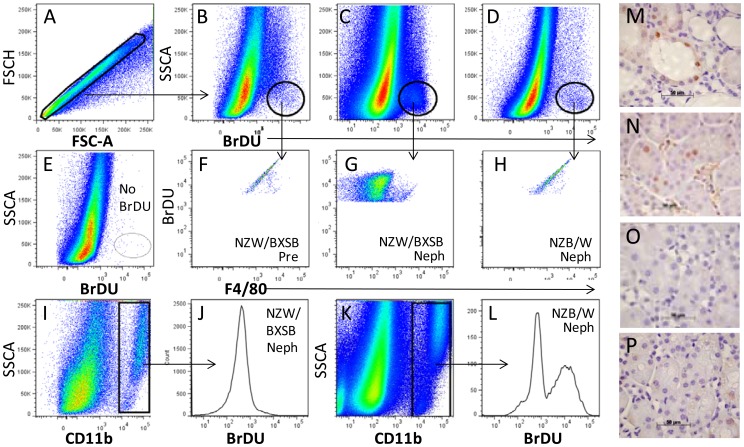
To determine which cells were proliferating in nephritic NZW/BXSB mice we analyzed renal cells by flow cytometry after BrDU feeding. We found a significant increase in CD11b-/BrDU+ cells in nephritic compared with either young NZW/BXSB (4.1+/− 2.0 vs. 0.3+/− 0% total renal cells; p<0.05) or nephritic NZB/W mice (0.7+/− 0.3%; p<0.05 – Figure 5A–E ) or NZM2410 mice (not shown). The BrDU+ cells were negative for lymphoid and macrophage/DC markers by flow cytometry ( Figure 5F–H and not shown). In contrast, BrDU uptake in NZB/W kidneys was restricted to CD11b+ cells ( Figure 5I–L ). To localize the proliferating cells in nephritic NZW/BXSB kidneys we stained them with anti-Ki67 and anti-PCNA and showed a pattern consistent with tubular proliferation/regeneration ( Figure 5M–P ).
Figure 5. A–L. Flow cytometry analysis of proliferating cells. After gating for singlets (A), whole kidney cells from prenephritic (Pre - B, F) and nephritic (Neph - C, G, I, J) NZW/BXSB and nephritic NZB/W (D, H, K, L) mice were analyzed for BrDU incorporation. A BrDU+/F4/80− population is seen only in nephritic NZW/BXSB mice (F–H). Proliferating CD11b+/F4/80+ macrophages are observed only in nephritic NZB/W mice (I–L). A non-BrDU treated control is shown in E. M–P. Immunohistochemistry of kidneys from nephritic NZW/BXSB (M, N), 8 week NZW/BXSB (O), and nephritic NZB/W (P) mice stained with antibodies to Ki67 (M) and PCNA (N–P). 40× magnification. Data are representative of 3 mice per stain.
Discussion
Lupus nephritis is a serious complication of SLE in which glomerular immune complex deposition triggers a cascade of inflammatory events that can lead to irreversible renal damage. Heterogeneity in the degree and type of peri-glomerular and tubulointerstitial inflammation, macrophage infiltration, and non-immune responses to inflammation and hypoxia may all contribute to the current difficulty in achieving or predicting treatment responses.
Because human renal tissue is rarely obtained either before treatment or on more than one occasion, mouse models of lupus are useful for addressing mechanisms of tissue damage in lupus nephritis. We have previously reported an analysis of the shared molecular profiles between kidneys from three murine models and human lupus nephritis biopsies. Using a low stringency for differential gene expression we found that 25–32% of the regulated genes in mice were also regulated in SLE renal biopsies and that both shared and unique features of the murine models overlapped with the human disease [10]. A limitation of the prior study is that the human samples were obtained after the initiation of immunosuppressive therapy, which likely modulated their inflammatory profile. In this study we compared the full molecular profiles and the derived transcriptional networks of the three mouse models of SLE nephritis with each other so as to identify mechanisms for disease pathogenesis, potential biomarkers for disease phenotype and potential therapeutic targets that can be tested in the appropriate model.
Despite differences in histologic appearance among the three models, 23–36% of the total gene set was shared among the three strains, reflecting known pathogenic processes downstream of immune complex deposition such as complement activation, macrophage and endothelial cell activation, and processes involved in tissue damage and remodeling. Among the 263 genes regulated in all three models, STAT3 was the top transcription factor, having a binding site in the promoter regions of 60 of those 263. STAT3 acts as a master regulator of cell metabolism and is activated by many cytokines and growth factors including type I interferons, IL10, and the IL-6 family of cytokines; by real-time PCR we confirmed increased renal expression of both IL-6 and IL-10 in all three lupus strains as well as a panel of IFN-inducible genes. Activation of STAT3 is associated with many forms of renal injury and the JAK2/STAT3 pathway has been implicated in the progression of renal fibrosis in several models of renal disease [29]. A recent study showed that a JAK2 inhibitor reduces renal STAT3 phosphorylation and prolongs survival in 7 month old NZB/W mice [30]. Importantly, 30 and 25 of the 60 Stat3 regulated genes identified in the mice were regulated in glomeruli and tubulointerstitium of human SLE renal biopsies respectively (Table S2), suggesting that Jak inhibitors may be a potential therapeutic approach for human SLE nephritis.
The most highly upregulated cytokine was IL1F6 (IL-36A), a member of the IL-1 family that is expressed by epithelial cells in the distal convoluted tubules and cortical collecting ducts of mice with SLE [31]. IL1F6 induces expression of IL-6 in epithelial cells and mediates a decrease in adhesion and loss of cuboidal morphology in a renal epithelial cell line [31]. IL1F6 is pathogenic when expressed in the skin and is highly expressed in human psoriasis [32]. In all three strains the level of expression of IL1F6 as assessed by qPCR correlated significantly with LCN-2 and HAVCR1, two markers of proximal tubular damage (r values for NZM2410: 0.5562, p = 0.0021 and 0.8489, p<0.0001; r values for NZB/W: 0.3729, p = 0.0077 and 0.8186, p<0.0001; r values for NZW/BXSB: 0.9063, p<0.0001 and 0.9277, p<0.0001), showing that progressive nephritis is associated with damage to the entire tubular system. Importantly, significant upregulation of these three markers did not occur in the human biopsies [10], indicating a lesser degree of tubular damage in treated individuals; this is consistent with our previous findings that glomerular damage precedes tubulointerstitial damage in NZB/W mice, that upregulation of LCN-2 and HAVCR1 occurs only in the late stages of SLE nephritis in NZB/W and NZW/BXSB mice, and that there is marked downregulation of LCN-2, IL1F6 and HAVCR1 in the kidneys of mice in remission ([11], and manuscript in preparation).
Another shared upregulated cytokine was IL-34, an alternate ligand for the CSF1 receptor. This cytokine is expressed by neurons and keratinocytes and directs the differentiation of macrophages in brain and skin [33], [34]. In the skin, IL-34 is not required for infiltration or differentiation of inflammatory monocytes but it is necessary for the maintenance of Langerhans cells after the resolution of inflammation [33]. A small amount of IL-34 is constitutively expressed by kidney proximal tubules [34]. Whether there is a role for IL-34 either in the maintenance of local renal macrophages or in the propagation of renal inflammation is not yet known.
We also showed that the downregulation of DNAse1 previously described in NZB/W mice and in human Class IV lupus nephritis [35] occurs in all three murine models. While it is still not known how renal DNAse1 expression is regulated, its lack has been postulated to enhance the size of chromatin deposits in the kidney and therefore the amount of autoantibody deposition and the pro-inflammatory response to nucleic acids [36]. Other protective genes that were downregulated included the superoxide dismutase 2 that protects from tissue hypoxia [37], [38].
It was surprising that the two models with proliferative disease only shared a small number of additional genes. Since most of the renal tissue is derived from the interstitium, it is possible that some of the glomerular signature was diluted in the whole kidney sample. Analysis of isolated glomeruli should further address this possibility. Nevertheless, the shared expression of macrophage related genes in proliferative disease suggests that macrophage activation and interstitial accumulation contributes to the poor prognosis of this histologic subtype. Indeed, poor prognosis has been found to correlate best with macrophage infiltration and with total number of interstitial CD45+ cells in several studies of human SLE [39], [40]. We did not observe upregulation of genes that are highly expressed in human netting neutrophils [41], consistent with the low number of infiltrating neutrophils in these mouse strains.
Genes associated with coagulation/fibrinolysis were more highly expressed in NZB/W and NZM2410 mice than in NZW/BXSB. One of these, PAI, is associated with renal fibrosis; targeting of PAI protects from renal fibrosis in a variety of models [42]. Preliminary studies have indicated that inhibition of PAI is protective in the NZB/W model, an appropriate model to test this therapeutic intervention (Naiman et. al. Abstract 2681, American College of Rheumatology Annual Meeting, Nov 2012). Another downregulated gene in both NZB/W and NZM2410 mice was Serpina6, a corticosteroid binding globulin that is expressed in proximal tubules only in female kidneys [43] where it is a regulator of renal osmotic pressure.
Although the kidneys receive a large amount of blood flow, tubular blood flow is easily compromised because it depends on upstream blood flow through the glomeruli. Oxidative stress alters mitochondria and electrolyte transport efficiency and this in turn stimulates interstitial fibrosis and may induce or worsen hypertension [44].The NZB/W and NZM2410 strains shared a mitochondrial dysfunction signature; importantly, this signature was shared by human renal SLE biopsies [10]. Targeting oxidative stress may be another approach to protect the kidney in SLE patients and can be tested in the NZB/W and NZM2410 models.
We identified a set of epithelial genes shared between nephritic NZM2410 and NZW/BXSB mice. These included Claudin1, a protein associated with tight junctions of renal epithelial cells [45], Slp-2a that regulates tubular apical polarization [46] and clusterin, an inhibitor of tight junction disintegration by MMPs [47]. These strains also expressed high levels of the CXCR2 ligands CXCL1 and CXCL2. CXCL1 is produced by renal epithelial cells in response to HGF stimulation and helps to mediate tubule repair [48]. This signature, in sum suggests an alteration in differentiation state of the renal epithelium of these two strains.
Unique features of each model were also highly informative. The accumulation of lymphoid aggregates, that occurs in some human SLE patients [8], [49], was found predominantly in the NZB/W model and was manifested by expression of adhesion molecules, activated T cell markers and accumulation of regulatory T cells and plasma cells. A B cell signature similarly occurs in only a subset of human SLE renal biopsies ([49] and our unpublished data) although neither T nor B cell numbers were found to correlate with prognosis [40]. While all three models manifested features of metabolic stress, this was most marked in the NZM2410 model in which there was little accumulation of lymphoid or myeloid cells. Tubular regeneration was noted in the NZW/BXSB model. Finally, we observed a decrease in the circadian transcription factors, DBP, HLF and TEF in NZW/BXSB and, to a lesser extent in NZB/W mice. These transcription factors regulate proteins that contribute to detoxification of a variety of substrates including cyclophosphamide [28]. Downregulation of circadian transcription factors can be mediated by cytokines such as TNF [50] and TGFβ [51] and dysregulation of the metabolic clock may exacerbate inflammation [52]. Importantly, dysregulation of these factors may have clinical significance with respect to toxicity of drugs used to treat active nephritis. Our data suggests that more work is required to determine the effects of nephritis on renal circadian functions.
Activation of intrinsic protective mechanisms also occurred in the nephritic kidneys. Examples include upregulation of SOCS3, an inactivator of cytokines, SERPINA3G (Spi-2A) that protects from caspase mediated cell death [53], NOX2 that may protect against ischemia and oxidative stress [54] and clusterin that protects against renal fibrosis [55]. A challenge in SLE nephritis therapy is maintaining naturally protective pathways while antagonizing pro-inflammatory pathways, so as to promote healing rather than scarring.
Our approach shows that in addition to identifying broadly applicable pathogenic features of SLE nephritis, we can also predict the appropriate mouse model to study specific pathologic features of and therapeutic approaches for lupus nephritis. Our study identifies both immune and non-immune pathways of renal damage that could be independently targeted in a personalized fashion.
Supporting Information
List of regulated unique and shared genes (human orthologs) in the nephritic vs. prenephritic mice of the NZB/W, NZM2410 and NZW/BXSB strains.
(DOCX)
Stat3 regulated genes in NZB/W, NZM2410, NZW/BXSB and in human LN renal biopsies.
(DOCX)
Comparison of microarray data and qPCR data.
(DOC)
Funding Statement
This work was supported by NIH R01 DK085241-01. (http://www.nih.gov/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Theofilopoulos AN, Dixon FJ (1985) Murine models of systemic lupus erythematosus. Adv Immunol 37: 269–390. [DOI] [PubMed] [Google Scholar]
- 2. Schiffer L, Kumpers P, Davalos-Misslitz AM, Haubitz M, Haller H, et al. (2009) B-cell-attracting chemokine CXCL13 as a marker of disease activity and renal involvement in systemic lupus erythematosus (SLE). Nephrol Dial Transplant 24: 3708–3712. [DOI] [PubMed] [Google Scholar]
- 3. Singh RR, Saxena V, Zang S, Li L, Finkelman FD, et al. (2003) Differential contribution of IL-4 and STAT6 vs STAT4 to the development of lupus nephritis. J Immunol 170: 4818–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, et al. (2006) Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science 312: 1669–1672. [DOI] [PubMed] [Google Scholar]
- 5. Hang LM, Izui S, Dixon FJ (1981) (NZW x BXSB)F1 hybrid. A model of acute lupus and coronary vascular disease with myocardial infarction. J Exp Med 154: 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davidson A, Aranow C (2010) Lupus nephritis: lessons from murine models. Nat Rev Rheumatol 6: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Z, Bethunaickan R, Huang W, Ramanujam M, Madaio MP, et al. (2011) IFN-{alpha} Confers Resistance of Systemic Lupus Erythematosus Nephritis to Therapy in NZB/W F1 Mice. J Immunol 187: 1506–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peterson KS, Huang JF, Zhu J, D'Agati V, Liu X, et al. (2004) Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser-captured glomeruli. J Clin Invest 113: 1722–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giannico G, Fogo AB (2013) Lupus nephritis: is the kidney biopsy currently necessary in the management of lupus nephritis? Clin J Am Soc Nephrol 8: 138–145. [DOI] [PubMed] [Google Scholar]
- 10. Berthier CC, Bethunaickan R, Gonzalez-Rivera T, Nair V, Ramanujam M, et al. (2012) Cross-species transcriptional network analysis defines shared inflammatory responses in murine and human lupus nephritis. J Immunol 189: 988–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schiffer L, Bethunaickan R, Ramanujam M, Huang W, Schiffer M, et al. (2008) Activated renal macrophages are markers of disease onset and disease remission in lupus nephritis. J Immunol 180: 1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramanujam M, Bethunaickan R, Huang W, Tao H, Madaio MP, et al. (2010) Selective blockade of BAFF for the prevention and treatment of systemic lupus erythematosus nephritis in NZM2410 mice. Arthritis Rheum 62: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bethunaickan R, Berthier CC, Ramanujam M, Sahu R, Zhang W, et al. (2011) A unique hybrid renal mononuclear phagocyte activation phenotype in murine systemic lupus erythematosus nephritis. J Immunol 186: 4994–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, et al. (2006) TM4 microarray software suite. Methods Enzymol 411: 134–193. [DOI] [PubMed] [Google Scholar]
- 15. Kahn P, Ramanujam M, Bethunaickan R, Huang W, Tao H, et al. (2008) Prevention of murine antiphospholipid syndrome by BAFF blockade. Arthritis Rheum 58: 2824–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schiffer L, Sinha J, Wang X, Huang W, von Gersdorff G, et al. (2003) Short term administration of costimulatory blockade and cyclophosphamide induces remission of systemic lupus erythematosus nephritis in NZB/W F1 mice by a mechanism downstream of renal immune complex deposition. J Immunol 171: 489–497. [DOI] [PubMed] [Google Scholar]
- 17. Hotta K, Sho M, Yamato I, Shimada K, Harada H, et al. (2011) Direct targeting of fibroblast growth factor-inducible 14 protein protects against renal ischemia reperfusion injury. Kidney Int 79: 179–188. [DOI] [PubMed] [Google Scholar]
- 18. Matsuura K, Uesugi N, Hijiya N, Uchida T, Moriyama M (2007) Upregulated expression of cardiac ankyrin-repeated protein in renal podocytes is associated with proteinuria severity in lupus nephritis. Hum Pathol 38: 410–419. [DOI] [PubMed] [Google Scholar]
- 19. Fornoni A, Sageshima J, Wei C, Merscher-Gomez S, Aguillon-Prada R, et al. (2011) Rituximab targets podocytes in recurrent focal segmental glomerulosclerosis. Sci Transl Med 3: 85ra46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Der SD, Zhou A, Williams BR, Silverman RH (1998) Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A 95: 15623–15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, et al. (2003) Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A 100: 2610–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu J, Karypis G, Hippen KL, Vegoe AL, Ruiz P, et al. (2006) Genomic view of systemic autoimmunity in MRLlpr mice. Genes Immun 7: 156–168. [DOI] [PubMed] [Google Scholar]
- 23. Karpurapu M, Wang X, Deng J, Park H, Xiao L, et al. (2011) Functional PU.1 in macrophages has a pivotal role in NF-kappaB activation and neutrophilic lung inflammation during endotoxemia. Blood 118: 5255–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huber R, Pietsch D, Panterodt T, Brand K (2012) Regulation of C/EBPbeta and resulting functions in cells of the monocytic lineage. Cell Signal 24: 1287–1296. [DOI] [PubMed] [Google Scholar]
- 25. Pedersen A, Skjong C, Shawlot W (2005) Lim 1 is required for nephric duct extension and ureteric bud morphogenesis. Dev Biol 288: 571–581. [DOI] [PubMed] [Google Scholar]
- 26. Chiang CK, Hsu SP, Wu CT, Huang JW, Cheng HT, et al. (2011) Endoplasmic reticulum stress implicated in the development of renal fibrosis. Mol Med 17: 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Y, Barbacioru C, Hyland F, Xiao W, Hunkapiller KL, et al. (2006) Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gachon F, Olela FF, Schaad O, Descombes P, Schibler U (2006) The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab 4: 25–36. [DOI] [PubMed] [Google Scholar]
- 29. Matsui F, Meldrum KK (2012) The role of the Janus kinase family/signal transducer and activator of transcription signaling pathway in fibrotic renal disease. J Surg Res 178: 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu LD, Stump KL, Wallace NH, Dobrzanski P, Serdikoff C, et al. (2011) Depletion of autoreactive plasma cells and treatment of lupus nephritis in mice using CEP-33779, a novel, orally active, selective inhibitor of JAK2. J Immunol 187: 3840–3853. [DOI] [PubMed] [Google Scholar]
- 31. Ichii O, Otsuka S, Sasaki N, Yabuki A, Ohta H, et al. (2010) Local overexpression of interleukin-1 family, member 6 relates to the development of tubulointerstitial lesions. Lab Invest 90: 459–475. [DOI] [PubMed] [Google Scholar]
- 32. Blumberg H, Dinh H, Trueblood ES, Pretorius J, Kugler D, et al. (2007) Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J Exp Med 204: 2603–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, et al. (2012) Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity 37: 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, et al. (2012) IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol 13: 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zykova SN, Tveita AA, Rekvig OP (2010) Renal Dnase1 enzyme activity and protein expression is selectively shut down in murine and human membranoproliferative lupus nephritis. PLoS One 5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fismen S, Mortensen ES, Rekvig OP (2011) Nuclease deficiencies promote end-stage lupus nephritis but not nephritogenic autoimmunity in (NZB x NZW) F1 mice. Immunol Cell Biol 89: 90–99. [DOI] [PubMed] [Google Scholar]
- 37. Kelkka T, Laurila JP, Sareila O, Olofsson P, Laukkanen MO, et al. (2012) Superoxide dismutase 3 limits collagen-induced arthritis in the absence of phagocyte oxidative burst. Mediators Inflamm 2012: 730469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Richters L, Lange N, Renner R, Treiber N, Ghanem A, et al. (2011) Exercise-induced adaptations of cardiac redox homeostasis and remodeling in heterozygous SOD2-knockout mice. J Appl Physiol 111: 1431–1440. [DOI] [PubMed] [Google Scholar]
- 39. Hill GS, Delahousse M, Nochy D, Remy P, Mignon F, et al. (2001) Predictive power of the second renal biopsy in lupus nephritis: significance of macrophages. Kidney Int 59: 304–316. [DOI] [PubMed] [Google Scholar]
- 40. Hsieh C, Chang A, Brandt D, Guttikonda R, Utset TO, et al. (2011) Predicting outcomes of lupus nephritis with tubulointerstitial inflammation and scarring. Arthritis Care Res (Hoboken) 63: 865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Villanueva E, Yalavarthi S, Berthier CC, Hodgin JB, Khandpur R, et al. (2011) Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol 187: 538–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rerolle JP, Hertig A, Nguyen G, Sraer JD, Rondeau EP (2000) Plasminogen activator inhibitor type 1 is a potential target in renal fibrogenesis. Kidney Int 58: 1841–1850. [DOI] [PubMed] [Google Scholar]
- 43. Rinn JL, Rozowsky JS, Laurenzi IJ, Petersen PH, Zou K, et al. (2004) Major molecular differences between mammalian sexes are involved in drug metabolism and renal function. Dev Cell 6: 791–800. [DOI] [PubMed] [Google Scholar]
- 44. Palm F, Nordquist L (2011) Renal oxidative stress, oxygenation, and hypertension. Am J Physiol Regul Integr Comp Physiol 301: R1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martin-Martin N, Ryan G, McMorrow T, Ryan MP (2010) Sirolimus and cyclosporine A alter barrier function in renal proximal tubular cells through stimulation of ERK1/2 signaling and claudin-1 expression. Am J Physiol Renal Physiol 298: F672–682. [DOI] [PubMed] [Google Scholar]
- 46. Yasuda T, Saegusa C, Kamakura S, Sumimoto H, Fukuda M (2012) Rab27 effector Slp2-a transports the apical signaling molecule podocalyxin to the apical surface of MDCK II cells and regulates claudin-2 expression. Mol Biol Cell 23: 3229–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jeong S, Ledee DR, Gordon GM, Itakura T, Patel N, et al. (2012) Interaction of clusterin and matrix metalloproteinase-9 and its implication for epithelial homeostasis and inflammation. Am J Pathol 180: 2028–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ueland JM, Gwira J, Liu ZX, Cantley LG (2004) The chemokine KC regulates HGF-stimulated epithelial cell morphogenesis. Am J Physiol Renal Physiol 286: F581–589. [DOI] [PubMed] [Google Scholar]
- 49. Chang A, Henderson SG, Brandt D, Liu N, Guttikonda R, et al. (2011) In situ B cell-mediated immune responses and tubulointerstitial inflammation in human lupus nephritis. J Immunol 186: 1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Petrzilka S, Taraborrelli C, Cavadini G, Fontana A, Birchler T (2009) Clock gene modulation by TNF-alpha depends on calcium and p38 MAP kinase signaling. J Biol Rhythms 24: 283–294. [DOI] [PubMed] [Google Scholar]
- 51. Kon N, Hirota T, Kawamoto T, Kato Y, Tsubota T, et al. (2008) Activation of TGF-beta/activin signalling resets the circadian clock through rapid induction of Dec1 transcripts. Nat Cell Biol 10: 1463–1469. [DOI] [PubMed] [Google Scholar]
- 52. Hashiramoto A, Yamane T, Tsumiyama K, Yoshida K, Komai K, et al. (2010) Mammalian clock gene Cryptochrome regulates arthritis via proinflammatory cytokine TNF-alpha. J Immunol 184: 1560–1565. [DOI] [PubMed] [Google Scholar]
- 53. Liu N, Wang Y, Ashton-Rickardt PG (2004) Serine protease inhibitor 2A inhibits caspase-independent cell death. FEBS Lett 569: 49–53. [DOI] [PubMed] [Google Scholar]
- 54. Turgeon J, Haddad P, Dussault S, Groleau J, Maingrette F, et al. (2012) Protection against vascular aging in Nox2-deficient mice: Impact on endothelial progenitor cells and reparative neovascularization. Atherosclerosis 223: 122–129. [DOI] [PubMed] [Google Scholar]
- 55. Jung GS, Kim MK, Jung YA, Kim HS, Park IS, et al. (2012) Clusterin attenuates the development of renal fibrosis. J Am Soc Nephrol 23: 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of regulated unique and shared genes (human orthologs) in the nephritic vs. prenephritic mice of the NZB/W, NZM2410 and NZW/BXSB strains.
(DOCX)
Stat3 regulated genes in NZB/W, NZM2410, NZW/BXSB and in human LN renal biopsies.
(DOCX)
Comparison of microarray data and qPCR data.
(DOC)