Abstract
Cl−/H+ antiporters of the CLC superfamily transport anions across biological membranes in varied physiological contexts. These proteins are weakly selective among anions commonly studied, including Cl−, Br−, I−,NO3−, and SCN−, but appear to be very selective against F−. The recent discovery of a new CLC clade of F−/H+ antiporters, which are highly selective for F− over Cl−, led us to investigate the mechanism of Cl−-over-F− selectivity by a CLC Cl−/H+ antiporter, CLC-ec1. By subjecting purified CLC-ec1 to anion transport measurements, electrophysiological recording, equilibrium ligand-binding studies, and x-ray crystallography, we show that F− binds in the Cl− transport pathway with affinity similar to Cl−, but stalls the transport cycle. Examination of various mutant antiporters implies a “lock-down” mechanism of F− inhibition, in which F−, by virtue of its unique H-bonding chemistry, greatly retards a proton-linked conformational change essential for the transport cycle of CLC-ec1.
Membrane transport proteins of the CLC family are used by many organisms for a range of biological tasks that require movement of Cl− or other inorganic anions across cell membranes. These include electrical polarization of skeletal muscle1,2, acidification of mammalian endosomes and possibly lysosomes3–5, delivery of NO3− to plant vacuoles6, and electrical shunting of virtual proton pumps for extreme acid resistance in enteric bacteria7. Originally identified as Cl− channels8, CLC proteins are now understood to fall into two mechanistically distinct subclasses: anion channels and proton-coupled anion antiporters9,10. Work over many years establishes that both of these CLC subtypes, while rigorously rejecting cations and multivalent anions, are rather weakly selective among small monovalent anions, with Cl−, Br−, I−, SCN−, and NO3− permeating at rates typically within an order of magnitude of one another11–15. The F− ion conspicuously deviates from this trend, however. F− transport has been tested in only two Cl−-utilizing CLCs - a mammalian Cl− channel16 and a bacterial Cl−/H+ antiporter13 - and in both it is undetectable. The anomalous position of F− among the halides is further highlighted by a recently discovered clade of prokaryotic CLC F−/H+ antiporters17,18, which display substantial selectivity for F− over Cl−. This is chemically intriguing, since F−, being more strongly hydrated than Cl−, presents a greater difficulty to the protein in stripping off its water-shell, as it must do to discriminate between these similar anions19, which differ by less than 0.5 Å in ionic radius.
The dearth of information on F− in membrane biology motivates a re-examination of this anion’s behavior in conventional, Cl−-based CLC proteins, to scrutinize their handling of F− more closely than has been previously done. CLC-ec1, a bacterial Cl−/H+ antiporter, is studied here, since among CLCs it is uniquely amenable to membrane biochemistry, x-ray crystallography, and functional reconstitution by electrophysiological recording and ion flux methods in synthetic phospholipid membranes. We find that although CLC-ec1 fails to transport F−, it binds this anion similarly to the biological substrate Cl−. Moreover, a CLC mutation that ablates H+ coupling produces robust F− transport. Crystal structures of F− bound to various CLC-ec1 variants offer an explanation for this striking alteration of transport selectivity in terms of a “lock-down” mode of CLC-ec1 induced by F− occupancy of the anion transport pathway, in which a putative transport cycle intermediate is trapped by this strongly H-bonding halide.
Results
The recent discovery of the CLCF exporters – the first example of membrane proteins biologically purposed to transport F− ion17 – propels us to examine CLC-ec1, a long-studied “conventional” Cl−/H+ antiporter, for interactions with F−. Why is F− unique among small monovalent anions in its inability to move through this protein? The strong selectivity of CLC-ec1 against F− is apparent from anion efflux from liposomes reconstituted with the antiporter (Fig 1a). In these experiments, liposomes are pre-loaded with 300 mM KF or KCl and suspended in a solution containing 1 mM of the test anion and 300 mM K-isethionate (an impermeant monovalent anion). Efflux down the 300-fold halide gradient, initiated by the K+ ionophore valinomycin (Vln) to allow counterion movement, is followed by the appearance of the anion in the external solution, as monitored with ion-specific electrodes. Cl− efflux is rapid, with single-CLC turnover of ~400 Cl− ions/sec at pH 6, as previously documented20,21. A slow efflux of F− is also observed, but this leak is not discernibly faster than seen with protein-free liposomes. This background leak is due to the high lipid permeability of neutral HF, which as a permeant weak acid (pKa = 3.4) is non-negligibly present under these conditions. A generous estimate places an upper limit on the F− transport rate of 4 F− ions/sec, less than 1% of the Cl− rate (Supplementary Results, Supplementary Table 1).
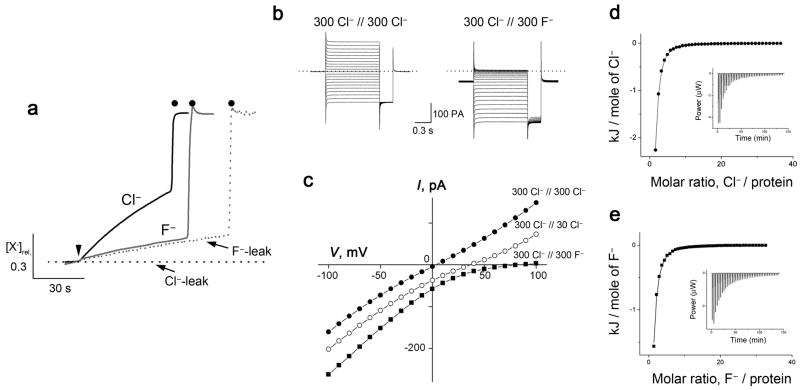
Figure 1. F− transport and binding in wildtype CLC-ec1.
a. Cl− and F− fluxes for wildtype CLC-ec1. Transport of each ion was triggered by Vln (arrowhead) and terminated by β-OG (filled circles). Cl− and F− leakage traces were determined with protein-free liposomes. Anion concentration [X−]rel is normalized to final level after detergent addition. b. Representative current traces in planar bilayers for symmetrical Cl− (left) and Cl−/F− bi-ionic (right) conditions. Dotted line indicates zero-current level. c. I–V relationships from separate bilayers in indicated ionic conditions. Reversal potentials for symmetrical, 10-fold gradient, and bi-ionic conditions from 4–22 bilayers each were: −0.5 ± 0.3, 34.9 ± 0.5, and 82 ± 5 mV, respectively. d,e. ITC data for equilibrium binding of Cl− and F− to wildtype CLC-ec1, with solid curves representing KD of 1 mM, 0.6 mM for F− and Cl−, respectively.
The transporter’s selectivity against F− is illustrated further by electrical recording of ionic currents in CLC-doped planar phospholipid bilayers (Fig 1b,c). Under bi-ionic conditions, with 300 mM Cl− on one side of the membrane and an equal concentration of F− on the other, the current-voltage relation is recorded. A reversal potential of ~80 mV - the voltage at which current is zero - shows that F− permeability is at most 3% of that of Cl− 14. This metric is only an upper limit on selectivity against F−, because we do not know if H+ movements are coupled to F, as they are to Cl−. Nevertheless, these recordings buttress the conclusion based on liposome flux experiments that F− is poorly transported, if at all, by CLC-ec1.
F− binding to the anion transport pathway
Though not transported, F− binds to the protein nearly as well as Cl−, as demonstrated by isothermal calorimetry (Fig 1d,e, Supplementary Results, Supplementary Table 2). Both anions show a dissociation constant of ~ 1 mM, in harmony with previous measurements for Cl− 21–23. The site where F− binds is apparent in a crystal structure at 2.9 Å resolution of CLC-ec1 in Cl−-free solution containing 100 mM F− (Fig 2a, Supplementary Table 2). The protein conformation is essentially identical to the Cl−-occupied antiporter (Cα rmsd 0.3 Å, Supplementary Fig 1). Prominent positive density appears in Fo-Fc maps in both subunits of the homodimer at the central Cl−-binding site, the mechanistic heart of the antiporter where anion and proton pathways converge24. While it is tempting to assign this density to a bound F− ion based on its location at a site known to be normally Cl−-occupied and “anion-hungry”25,26, F−cannot be distinguished from water on the basis of x-ray scattering alone. It is unlikely that this density represents a crystallographically ordered water molecule, however, since the protein groups surrounding it are inconsistent with dipolar interactions with water, and since a comparable-resolution crystal structure in the absence of halide ions shows no difference density in this region (Fig 2b). This comparison is thus suggestive of F− occupancy at the central site but cannot be considered a definitive demonstration of this.
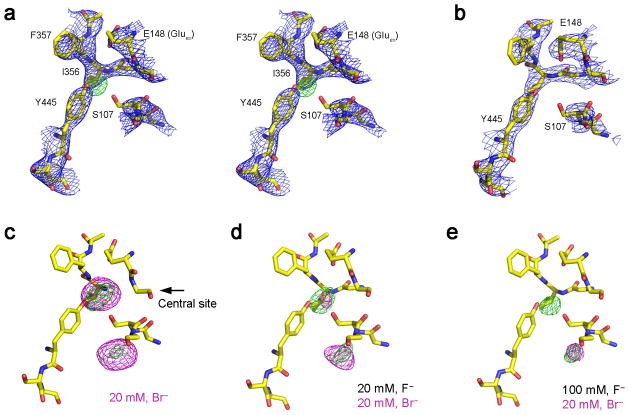
Fig. 2. Crystal structures of wildtype CLC-ec1 with F−.
a. Stereoview of crystal structure of CLC-ec1 near central anion binding site with 100 mM F−. 2Fo-Fc map contoured at 1.5 σ (blue mesh) and positive ion-omit map (Fo-Fc) at 3.5 σ (green mesh). Structures of CLC-ec1 with varying anion compositions: (b) without halide, (c) 20 mM Br−, (d) 20 mM Br−+20 mM F−, (e) 20 mM Br−+100 mM F−. Fo-Fc maps contoured at 3.5 σ (green mesh) and Br− anomalous difference map at 4.5 σ (magenta mesh).
If F− binds to the CLC-ec1 transport pathway, we should be able to infer its occupancy crystallographically by competition with Br−, a functionally faithful Cl− substitute14,21,22,27. Crystals were formed in 20 mM Br−, far above its dissociation constant22,23 in the presence or absence of F−, and Br− at the central transport site was detected by its anomalous difference density. The result is clear. In the absence of F−, prominent Br− density appears at the central site as expected22 (Fig 2c), but as F− is progressively raised to 20 mM and 100 mM, Br− density weakens and finally disappears (Fig. 2d,e). These results are seen in both subunits of the CLC homodimer in multiple crystals (Supplementary Table 4). Thus, F− and Br− compete for the central site, verifying our original suspicion that the electron density at this site in F−-bathed crystals in fact reflects F− occupancy. Weak Br− density also appears at a second location near the protein’s intracellular surface, where halides bind with very low affinity 22.
Mutations of the external gate promote F− transport
The results above imply that F− binds to the antiporter’s central anion site, but then transport stalls. Why should F−, uniquely among small monovalent anions, prevent the transport cycle from proceeding? To approach this question, we analyzed F−/Cl− selectivity in several well-characterized CLC-ec1 mutants. A key event in the CLC-ec1 antiport mechanism –indeed, the only conformational change known structurally - is the rotation of the conserved E148 “external glutamate” (Gluex) side chain. This residue acts as a gate separating the Cl−pathway from the extracellular solution. Its “open” rotamer, favored by protonation, allows rapid exchange between Cl− and sites within the protein. In its “closed” position, which is favored by deprotonation, the Gluex side chain rotates downward to block the pathway by coordinating the Cl− ion in the central site22,28,29. If this gate is removed by mutation, the resulting E148A protein still transports Cl− but is no longer H+-coupled9.
We observe with surprise that the E148A mutation leads to complete loss of selectivity against F−, which now permeates at an even higher rate than Cl− (Fig 3a, Supplementary Table 1). Equilibrium binding of F− again mirrors the known behavior of Cl−21,22, with each halide binding similarly, with higher affinity to the mutant than to WT protein (Fig 3b), although the increase in affinity for F− (5-fold) is less than that for Cl− (50-fold). Moreover, a crystal structure of E148A in 100 mM F− (Fig 3c) shows positive density in the F−-omit map at two sites: the central site and the “external” site that in WT is occupied by the carboxylate group of Gluex and is replaced by halide in the gate-deleted mutant. Crystallographic titration of Br− in the presence of F− again indicates F−/Br− competition at both of these sites (Fig 3d, Supplementary Table 4), and that therefore the densities represent F− at both sites, reprising the known binding of Cl− in this mutant22.
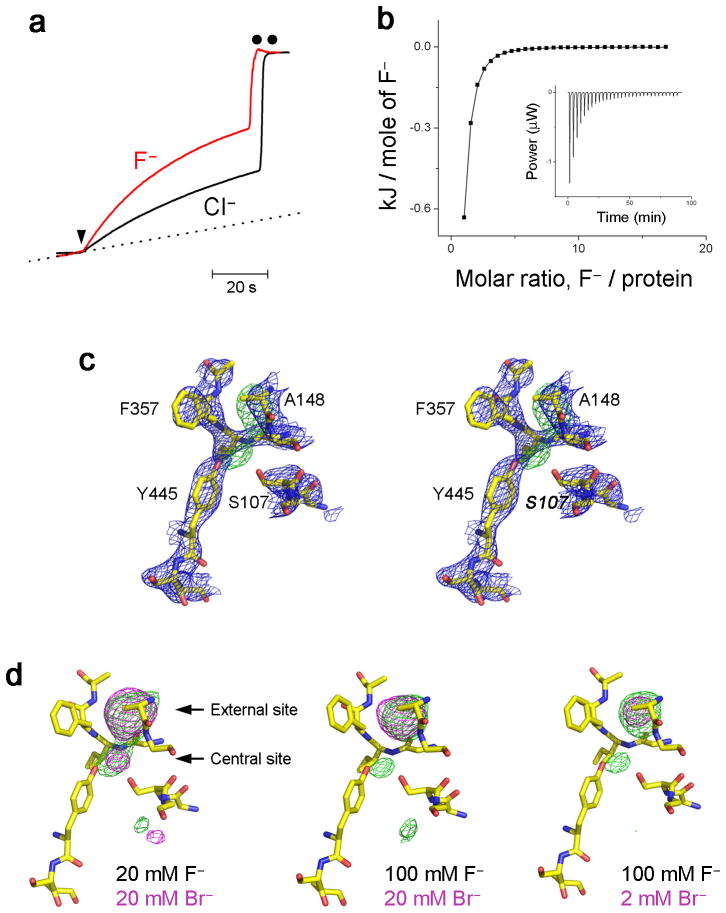
Fig. 3. F− handling by ungated mutant E148A.
a. Cl− and F− transport in E148A. Background F− leakage is indicated by dotted line. b. F− binding to E148A. ITC measurements were as in Fig 1, and isotherm fitted with KD = 0.18 mM. c. Stereoview of crystal structure of E148A in 100 mM F−. 2Fo-Fc map (blue mesh) and Fo-Fc map (green mesh) were contoured at 1.5 σ and 3.5 σ, respectively. d. Br−/F− competition in E148A: crystal structures were obtained in the presence of F−/Br− mixtures indicated.
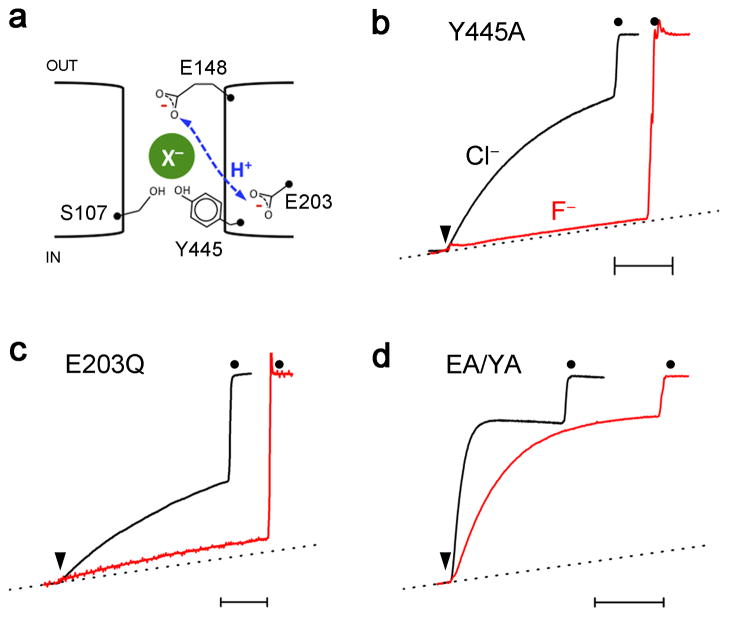
Robust F− transport in E148A is unexpected, since the external gate is commonly viewed as essential for conformational coupling between Cl− and H+, but not as a contributor to the antiporter’s ionic selectivity properties. Does this change in F−/Cl− selectivity reflect the lack of H+ coupling in E148A or a direct interaction of the bound anion with the Gluex side chain? This question may be approached by testing F− transport with mutations at other positions that also abolish H+ coupling to Cl− transport (Fig 4). We first examine Y445A, which disrupts the anion pathway’s inner gate and destabilizes Cl− at the central site, while leaving the external gate intact20,24. This mutant retains wildtype-like selectivity against F−. Second, we test E203Q, which specifically interferes with the proton pathway on its cytoplasmic side, distant from the external gate and from the Cl− pathway24,30,31. Again, transport-selectivity against F− is preserved in this mutant. Finally, F− is presented to E148A/Y445A, a double mutant with both inner and outer gates stripped off to create an always-open transmembrane pore32. This mutant, in which Cl− permeation is extremely fast (~20,000 s−1), also moves F− rapidly (6200 s−1), as reported in Supplementary Table 1. The first two mutants refute the idea that H+ coupling prevents − transport in WT protein, and the behavior of all the proteins taken together implicate the Gluex gate itself as the culprit that impedes F− transport.
Fig 4. H+ transport is not involved in F− inhibition.
a. Cartoon of elements governing H+-coupled anion antiport. X− represents anion binding at the central site and dashed line indicates H+-transit between E148 and E203. Normalized Cl− and F−efflux traces for Y445A (b), E203Q (c), and EA/YA (d, E148A/Y445A) mutants. Time bar is 20 sec.
A F−-dependent lock-down mechanism
How might this F−-specific inhibition of the WT transport cycle occur? One possibility suggests itself from the mechanism by which Gluex operates normally with Cl−21,29,33. At some point in the transport cycle, outlined in Fig 5a, the deprotonated Gluex carboxylate must move towards the central Cl− ion to accept a proton arriving from the intracellular side, although such close apposition between these two moieties has never been observed in previous CLC crystal structures. Once protonated, the side chain rotates up and away from the Cl− ion to open an aqueous pathway to the external solution. This picture emerged from CLC-ec1 variants with Gluex substituted by Gln, a surrogate for protonated Gluex22,28. All structures with Glu at this position show its side chain in the “closed” rotamer, with the transport pathway capped by the deprotonated carboxylate covering the bound Cl−. In contrast, in all previous crystal structures with the Gln substitution, the side chain adopts the “open” rotamer that connects the Cl−pathway to extracellular solvent. If, however, a bound F− ion, a strong H-bond acceptor, would stabilize the protonated Gluex carboxyl group in the closed position, the external gate would be locked down and transport would slow or stall.
Fig 5. Lock-down mechanism of F− inhibition.
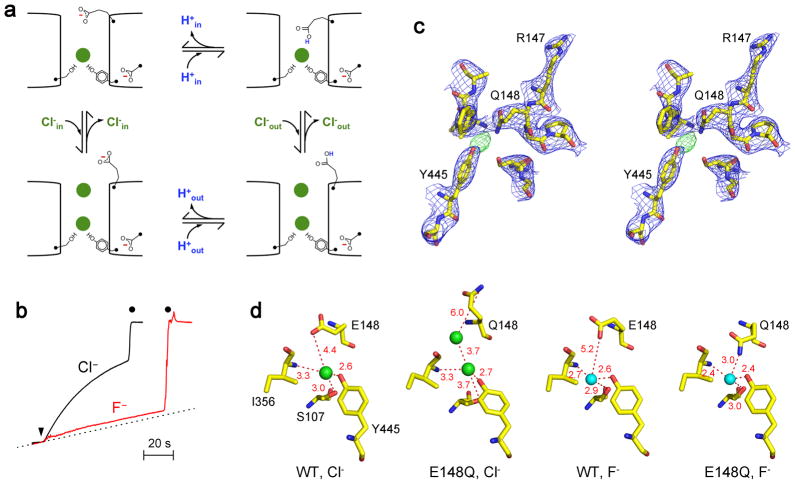
a. Outline of transport cycle proposed for CLC-ec1. This skeletal representation of previously proposed cycle33 emphasizing the configuration and protonation-state of the Gluex side chain. b. E148Q does not transport F−. Efflux data are shown for F− and Cl−, as in Fig 1a. c. Stereo view of E148Q crystal structure near the bound F− (green mesh). d. Comparison of anion coordination in WT and E148Q. Note the second Cl− ion in open conformation of E148Q (PDB #1OTU). Green and cyan spheres indicate Cl− and F−, respectively.
We tested this idea with the E148Q substitution, which transports Cl− unaccompanied by proton countermovement24. This mutant behaves like WT, binding but failing to transport F−(Fig 5b, Supplementary Table 2). This result, in striking contrast to the F− transporting, “gate-deleted” E148A mutant, is consistent with the lock-down mechanism posited above. A crucial test of this mechanism would be a E148Q crystal structure in F−; is the Gln side chain in the open or closed position? We solved structures from several E148Q crystals diffracting to 2.9–3.2 Å. The results are unambiguous (Fig 5c): While the overall structure is virtually identical to WT, the E148Q side chain adopts the closed conformation, never before observed for this mutant, but as anticipated by the lock-down mechanism. Moreover, the Gln amide group now directly coordinates the F− ion within H-bonding distance (3.0 Å), a result of the side chain moving 1.4–2.2 Å closer to the halide in the Cl− or F−-occupied WT structures (Fig 5d, Supplementary Fig 1). That it is the presence of F− rather than the absence of Cl− that brings the amide-bearing side chain into the closed configuration is reinforced by a halide-free crystal structure of E148Q22 wherein the side chain adopts its familiar open position, a result that we have reproduced with rigorously Cl− free conditions (Supplementary Table 3).
Discussion
This study is aimed at an oddity of a deeply studied member of the CLC superfamily of anion transport proteins: why F−, alone among small inorganic anions, is such a poor substrate for transport. While F− is irrelevant to the physiological role of CLC-ec1 in bacterial acid resistance7, the anion’s anomalous behavior enriches our still-incomplete view of the antiporter’s mechanism. We find that the selectivity of CLC-ec1 against F− reflects neither a low affinity at the crucial transport site nor a peculiarity associated with H+ coupling. Instead, the functional and crystallographic results implicate a strong, favorable interaction between F− and the Gluex side chain as a plausible explanation. According to this picture, upon binding to the transport pathway, F− freezes the protonated Gluex side chain in its “closed” configuration, thus locking down the transport cycle. We imagine that F−-specific inhibition arises from the tendency of this most electronegative ion to accept a H-bond from the protonated carboxyl group of Gluex. In the E148Q crystal structure, the permanently protonated isostere is apparently caught in flagrante in the act of H-bonding with the bound halide; this novel F−-stabilized configuration, we suggest, reflects a transport-cycle intermediate that is not captured in WT crystals, where the Gluex side chain lies farther away from the halide, perhaps because it is deprotonated in the crystals, which grow near pH 7. Since the normal Cl− transport cycle is complete in a few milliseconds, a F−-induced closed-rotamer dwell-time on the 0.1–1 second timescale, for instance, would naturally account for our observations on WT and mutant CLCs. Stabilization of the anion-Gluex interaction by a strong (~3 kcal/mol) H-bond to F− is plausible as the chemical basis of the antiporter’s selectivity against F−. The conservation of the external glutamate in CLCs suggests this lock-down mechanism might apply more generally, if other Cl−-transporting CLCs were also found to select against F−. To avoid possible confusion, we point out that the Gln side chain configuration reported here in E148Q is not analogous to the more deeply extended Gluex side chain configuration observed in a eukaryotic CLC structure29, wherein the carboxylate group occupies the central Cl− site itself, a configuration that has not been observed in any prokaryotic CLC structure.
Beyond providing a glimpse of a conjectured but previously unseen intermediate in the unusual CLC antiport cycle14,33, the crystal structures reported here are noteworthy for another reason. They supplement the rare examples34 of a macromolecular “free” F− site, wherein the anion is not in complex with a divalent metal cation, as it is in enolase, pyrophosphatase, and a F− riboswitch, to cite a few examples 35–37. Instead, in CLC-ec1, F− is coordinated much like Cl−, by the same dipolar moieties from the protein’s backbone and side chains without the participation of a metal. Moreover, F− in its binding site is mostly dehydrated, since the anion-occupied pathway offers little room for coordinating waters, and since the aqueous anion carries an unusually large hydration shell. Thus, the protein performs a thermodynamically difficult task by desolvating F− as the ion leaves water and engages with the transport pathway. In light of this, the seemingly innocuous observation of similar F− and Cl− binding affinity takes on a deeper meaning. In shedding water upon binding, F− unavoidably suffers a severe free energy penalty, 30 kcal/mol larger than Cl− for full dehydration38. Therefore, to match Cl− in equilibrium binding affinity, F− in the central site must enjoy additional favorable interactions that mitigate this desolvation handicap. The lock-down mechanism proposed here envisions this compensatory interaction arising in part from strong H-bonding between the Gluex carboxylic acid and the F− anion.
Online methods
Protein purification
Analytical grade chemicals and reagents were purchased from Sigma-Aldrich or as otherwise specified, and lipids were from Avanti Polar Lipids. For ion flux experiments, hexahistidine-tagged CLC-ec1 constructs were purified on cobalt-affinity and size-exclusion columns as described13. For bilayer recordings, which require extremely pure preparations free of outer membrane porin contamination39, protein was expressed in a porin-depleted E.coli strain (ΔlamB ompF::Tn5ΔompAΔompC)40. The 40 mM DM extract loaded onto the cobalt affinity column was sequentially washed with purification buffer (PB; 100 mM NaCl, 20 mM tris-HCl, pH 7.5, 5 mM DM) containing 0.1% (w/v) triton-X114, 20 mM imidazole, or 1 M NaCl, and CLC-ec1 was eluted with 400 mM imidazole. The eluate was diluted 10-fold into HS buffer (10 mM NaCl, 10 mM Mes-NaOH, pH 6.5, 5 mM DM) and loaded onto a 2-mL Poros HS cation-exchange column equilibrated with HS buffer. The column was washed with salt steps of 100 mM and 200 mM NaCl in HS buffer, and eluted with a step to 500 mM NaCl. Concentrated protein (~10 mg/mL) was then diluted 50-fold in HQ buffer (10 mM NaCl, 10 mM Mes-NaOH, pH 5.5, 5 mM DM), and loaded into a Poros HQ anion-exchange column. CLC-ec1 running through the column at this pH was collected and concentrated to ~10 mg/mL. Proteoliposomes were immediately prepared at high protein density. A micellar solution of Chaps (35 mM), protein (600 μg/mL), and a phospholipid mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylethanolamine (POPE, 15 mg/mL)/1-palmitoyl-2-oleoyl-phosphatidylglycerol (POPG, 5 mg/mL) was prepared in reconstitution buffer (450mM KCl, 25mM citrate-phosphate buffer, pH 7.5) and dialyzed for 2 days against reconstitution buffer. The resulting liposomes were frozen in aliquots and stored at −80°C until use.
Cl− and F− flux assay
Purified protein was reconstituted at a density of 0.5–5 μg per mg of E. coli polar lipid by dialyzing against 300 mM KCl or KF, 25 mM Mes-NaOH, pH 6.013, and Cl− or F− liposome-efflux assays were performed as described 20. Briefly, proteoliposomes were subjected to 3–5 freeze-thaw cycles and extruded through a 0.4 μm-polycarbonate membrane. External solution was replaced with flux buffer (1 mM KCl or KF, 300 mM K-isethionate, 25 mM Mes-NaOH, pH 6.0) immediately before each run by centrifuging proteoliposome samples through 1.5-mL G-50 columns equilibrated with flux buffer. Efflux of Cl− or F− was initiated by addition of Vln (1 μg/mL), and anion concentration in the suspension was continuously monitored by Ag/AgCl or LaF3/EuF3 electrodes; total trapped Cl− or F− was measured at the end of experiment by disrupting liposomes with 30 mM n-octyl-β-D-glucoside (β-OG). Electrode responses were calibrated in each experiment with known additions of KCl or KF (50–75 μM).
Planar bilayer recording
Electrical recordings of CLC-ec1 currents were made by fusing proteoliposomes into planar lipid bilayers (POPE/POPG, 3/1 in n-decane)30. The “cis” side of the bilayer is defined as the solution to which liposomes were added, and the opposite “trans” side is defined as zero voltage. Ag/AgCl electrodes immersed in 1 M KCl wells were connected to the recording solutions via 1.5% agar bridges containing 0.3 M KCl or NaF. Current-voltage (I–V) relationships were acquired in various ionic conditions: symmetrical Cl− (300 mM KCl, 20 mM Mes-NaOH, pH 5.5), 10-fold Cl− gradient (cis 300 mM KCl, 20 mM Mes-NaOH, pH 5.5; trans 30 mM KCl, 20 mM Mes-NaOH, pH 5.5), or Cl−/F− bionic conditions (cis 300 mM KCl, 20 mM Mes-NaOH, pH 5.5; trans 300 mM NaF, 20 mM Mes-NaOH, pH 5.5). After the conductance stabilized, 1-sec voltage stimuli from -100 mV to 100 mV in 10 mV increments were applied from a holding potential of zero voltage. Currents were low-pass filtered at 500 Hz and sampled at 10 kHz using a PC-505B amplifier (Warner) and pCLAMP software (Axon Instruments). Reversal potentials were interpolated from I–V curves and corrected for liquid junction potentials (<2mV).
Isothermal titration calorimetry
Thermodynamic binding parameters of Cl− or F− to CLC-ec1 were determined with a microcalorimeter (Nano ITC, TA Instruments, 300 μL sample volume). For obtaining the required initial Cl−-free conditions, protein was purified on a cobalt column in low-Cl− buffer (100 mM Na/K tartrate, 2 mM NaCl, 20 mM tris-SO4, 400 mM imidazole-SO4, pH 7.5, 5 mM DM), followed by size-exclusion chromatography in zero-Cl− buffer (100 mM Na/K tartrate or Na-isethionate, 10 mM Mes-NaOH, pH 6.0 (or 10 mM tris-SO4, pH 7.5), 5 mM DM). Protein (150–250 μM) was titrated with 1–μL injections of 5–25 mM Cl− or F− at 25°C. Data were fitted with single-site isotherms using NanoAnalyze 2.1.9 software.
X-ray crystallography
Since Cl− occupies the anion transport pathway with submillimolar binding affinity, Cl−contamination during crystallization can undermine attempts to localize F− in CLC-ec1. Accordingly, PEG stock solutions were treated with a mixed-bed ion-exchange resin (Amberlite MB-150) to reduce the Cl− content (~0.1 mM) of commercial PEG. In this way, we could confirm by direct measurement that our nominally Cl−-free crystallization solutions contained < 10 μM Cl−.
All crystallographic experiments used “ΔNC” constructs31 truncated at N and C termini, which consistently give crystals diffracting in the range 2.8–3.2 Å. For crystallizing CLC-ec1 with F, proteins were first purified as in ITC experiments. After removal of the His-tag, CLC-ec1/Fab complexes were formed28 and further purified on a size-exclusion column equilibrated with 0–100 mM NaF, 0–20 mM NaBr, 10 mM tris-SO4, pH 7.5, 5 mM DM. Protein was concentrated to ~10 mg/mL and mixed with an equal volume of crystallization solution in a hanging-drop vapor-diffusion chamber. Crystals, appearing in 20–28% (w/v) PEG 400, 20–100mM Na/K tartrate, pH 7.0 – 9.5 in 5–10 days at 22°C, were cryo-protected by slowly increasing PEG concentration in the mother liquor to ~35 %, followed by freezing in liquid N2. Similar conditions were used for halide-free structures, except that the final size-exclusion step was in halide-free solution, and crystallization solutions contained 20 mM Na/K tartrate or 100 mM K-isethionate.
Datasets were collected at beamline 8.2.1 or 8.2.2, Advanced Light Source, Lawrence Berkeley National Laboratory, at X-ray wavelength of 1 Å, or at 0. 919 Å for Br− anomalous diffraction. Data were integrated and scaled using HKL2000, and initial models were obtained by molecular replacement against 4ENE or 1OTS using Phaser in the CCP4 software suite. Models were rigid-body refined in REFMAC5 and further refined in Phenix. PDB accession codes for each crystal are reported in Supplementary Table 3.
Supplementary Material
Acknowledgments
We are grateful to Dr L. Tamm for providing the porin-deleted E coli strain and to the beamline scientists for their expertise and help at the Advanced Light Source, Lawrence Berkeley Labs.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Author contributions
HHL, RLS designed and executed experiments and wrote the paper; CM, designed experiments and wrote the paper.
References
- 1.Palade PT, Barchi RL. On the inhibition of muscle membrane chloride conductance by aromatic carboxylic acids. J Gen Physiol. 1977;69:879–896. doi: 10.1085/jgp.69.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinmeyer K, et al. Inactivation of muscle chloride channel by transposon insertion in mytotonic mice. Nature. 1991;354:304–308. doi: 10.1038/354304a0. [DOI] [PubMed] [Google Scholar]
- 3.Gunther W, Luchow A, Cluzeaud F, Vandewalle A, Jentsch TJ. ClC-5, the chloride channel mutated in Dent’s disease, colocalizes with the proton pump in endocytotically active kidney cells. Proceedings of the National Academy of Sciences USA. 1998;95:8075–8080. doi: 10.1073/pnas.95.14.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graves AR, Curran PK, Mindell JA. The Cl−/H+ antiporter CLC-7 is the primary chloride permeation pathway in lysozomes. Nature. 2008;453:788–792. doi: 10.1038/nature06907. [DOI] [PubMed] [Google Scholar]
- 5.Steinberg BE, et al. A cation counterflux supports lysosomal acidification. The Journal of cell biology. 2010;189:1171–1186. doi: 10.1083/jcb.200911083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeAngeli A, et al. AtCLCa, a proton/nitrate antiporter, mediates nitrate accumulation in plant vacuoles. Nature. 2006;442:939–942. doi: 10.1038/nature05013. [DOI] [PubMed] [Google Scholar]
- 7.Iyer R, Iverson TM, Accardi A, Miller C. A biological role for prokaryotic ClC chloride channels. Nature. 2002;419:715–718. doi: 10.1038/nature01000. [DOI] [PubMed] [Google Scholar]
- 8.Jentsch TJ, Steinmeyer K, Schwarz G. Primary structure of Torpedo marmorata chloride channel isolated by expression cloning in Xenopus oocytes. Nature. 1990;348:510–514. doi: 10.1038/348510a0. [DOI] [PubMed] [Google Scholar]
- 9.Accardi A, Miller C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl− channels. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- 10.Miller C. ClC chloride channels viewed through a transporter lens. Nature. 2006;440:484–489. doi: 10.1038/nature04713. [DOI] [PubMed] [Google Scholar]
- 11.Fahlke C, Durr C, George AL. Mechanism of ion permeation in skeletal muscle chloride channels. J Gen Physiol. 1997;110:551–564. doi: 10.1085/jgp.110.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rychkov GY, Pusch M, Roberts ML, Jentsch TJ, Bretag AH. Permeation and block of the skeletal muscle chloride channel, ClC-1, by foreign anions. J Gen Physiol. 1998;111:653–665. doi: 10.1085/jgp.111.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maduke M, Pheasant DJ, Miller C. High-level expression, functional reconstitution, and quaternary structure of a prokaryotic ClC-type chloride channel. J Gen Physiol. 1999;114:713–722. doi: 10.1085/jgp.114.5.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguitragool W, Miller C. Uncoupling of a CLC Cl−/H+ exchange transporter by polyatomic anions. J Mol Biol. 2006;362:682–690. doi: 10.1016/j.jmb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 15.De Stefano S, Pusch M, Zifarelli G. Extracellular determinants of anion discrimination of the Cl-/H+ antiporter protein CLC-5. J Biol Chem. 2011;286:44134–44144. doi: 10.1074/jbc.M111.272815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahlke C. Ion permeation and selectivity in ClC-type chloride channels. Am J Physiol Renal Physiol. 2001;280:F748–757. doi: 10.1152/ajprenal.2001.280.5.F748. [DOI] [PubMed] [Google Scholar]
- 17.Baker JL, et al. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stockbridge RB, et al. Fluoride resistance and transport by riboswitch-controlled CLC antiporters. Proc Natl Acad Sci U S A. 2012;109:15289–15294. doi: 10.1073/pnas.1210896109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hille B. Ion Channels of Excitable Membranes. 3. Sinauer Associates; 2001. [Google Scholar]
- 20.Walden M, et al. Uncoupling and turnover in a Cl−/H+ exchange transporter. J Gen Physiol. 2007;129:317–329. doi: 10.1085/jgp.200709756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picollo A, Malvezzi M, Houtman JC, Accardi A. Basis of substrate binding and conservation of selectivity in the CLC family of channels and transporters. Nat Struct Mol Biol. 2009;16:1294–1301. doi: 10.1038/nsmb.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobet S, Dutzler R. Ion binding properties of the CLC chloride selectivity filter. EMBO J. 2005;25:24–33. doi: 10.1038/sj.emboj.7600909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Picollo A, Xu Y, Johner N, Berneche S, Accardi A. Synergistic substrate binding determines the stoichiometry of transport of a prokaryotic H+:Cl− exchanger. Nature Struct Mol Biol. 2012;19:525–531. doi: 10.1038/nsmb.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Accardi A, et al. Separate ion pathways in a Cl−/H+ exchanger. J Gen Physiol. 2005;126:563–570. doi: 10.1085/jgp.200509417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faraldo-Gomez JD, Roux B. Electrostatics of ion stabilization in a ClC chloride channel homologue from Escherichia coli. J Mol Biol. 2004;339:981–1000. doi: 10.1016/j.jmb.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 26.Jayaram H, Robertson JL, Wu F, Williams C, Miller C. Structure of a slow CLC Cl/H+ antiporter from a cyanobacterium. Biochemistry. 2011;50:788–794. doi: 10.1021/bi1019258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Accardi A, Lobet S, Williams C, Miller C, Dutzler R. Synergism between halide binding and proton transport in a CLC-type exchanger. J Mol Biol. 2006;362:691–699. doi: 10.1016/j.jmb.2006.07.081. [DOI] [PubMed] [Google Scholar]
- 28.Dutzler R, Campbell EB, MacKinnon R. Gating the selectivity filter in ClC chloride channels. Science. 2003;300:108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- 29.Feng L, Campbell EB, Hsiung Y, MacKinnon R. Structure of a eukaryotic CLC transporter defines an intermediate state in the transport cycle. Science. 2010;330:635–641. doi: 10.1126/science.1195230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim HH, Miller C. Intracellular proton-transfer mutants in a CLC Cl−/H+ exchanger. J Gen Physiol. 2009;133:131–138. doi: 10.1085/jgp.200810112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim HH, Shane T, Miller C. Intracellular proton access in a Cl−/H+ antiporter. PLoS biology. 2012;10:e1001441. doi: 10.1371/journal.pbio.1001441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jayaram H, Accardi A, Wu F, Williams C, Miller C. Ion permeation through a Cl−-selective channel designed from a CLC Cl−/H+ exchanger. Proc Natl Acad Sci USA. 2008;105:11194–11199. doi: 10.1073/pnas.0804503105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller C, Nguitragool W. A provisional transport mechanism for a CLC-type Cl−/H+ exchanger. Phil Trans Roy Soc B. 2008;364:175–180. doi: 10.1098/rstb.2008.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cametti M, Rissanen K. Highlights on contemporary recognition and sensing of fluoride anion in solution and in the solid state. Chem Soc Rev. 2013;42:2016–2038. doi: 10.1039/c2cs35439j. [DOI] [PubMed] [Google Scholar]
- 35.Qin J, Chai G, Brewer JM, Lovelace LL, Lebioda L. Fluoride inhibition of enolase: crystal structure and thermodynamics. Biochemistry. 2006;45:793–800. doi: 10.1021/bi051558s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samygina VR, et al. Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies. J Mol Biol. 2007;366:1305–1317. doi: 10.1016/j.jmb.2006.11.082. [DOI] [PubMed] [Google Scholar]
- 37.Ren A, Rajashankar KR, Patel DJ. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature. 2012;486:85–89. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hummer G, Pratt LR, Garcia AE. Free energy of ionic hydration. J Phys Chem. 1996;100:1206–1215. [Google Scholar]
- 39.Accardi A, Kolmakova-Partensky L, Williams C, Miller C. Ionic currents mediated by a prokaryotic homologue of CLC Cl− channels. J Gen Physiol. 2004;123:109–119. doi: 10.1085/jgp.200308935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prilipov A, Phale PS, Van Gelder P, Rosenbusch JP, Koebnik R. Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS microbiology letters. 1998;163:65–72. doi: 10.1111/j.1574-6968.1998.tb13027.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.