Summary
The obesity epidemic has intensified efforts to understand the mechanisms controlling adipose tissue development. Adipose tissue is generally classified as white adipose tissue (WAT), the major energy storing tissue, or brown adipose tissue (BAT), which mediates non-shivering thermogenesis. It is hypothesized that brite adipocytes (brown in white) may represent a third adipocyte class. The recent realization that brown fat exist in adult humans suggests increasing brown fat energy expenditure could be a therapeutic strategy to combat obesity. To understand adipose tissue development, several groups are tracing the origins of mature adipocytes back to their adult precursor and embryonic ancestors. From these studies emerged a model that brown adipocytes originate from a precursor shared with skeletal muscle that expresses Myf5-Cre, while all white adipocytes originate from a Myf5-negative precursors. While this provided a rational explanation to why BAT is more metabolically favorable than WAT, recent work indicates the situation is more complex because subsets of white adipocytes also arise from Myf5-Cre expressing precursors. Lineage tracing studies further suggest that the vasculature may provide a niche supporting both brown and white adipocyte progenitors; however, the identity of the adipocyte progenitor cell is under debate. Differences in origin between adipocytes could explain metabolic heterogeneity between depots and/or influence body fat patterning particularly in lipodystrophy disorders. Here, we discuss recent insights into adipose tissue origins highlighting lineage-tracing studies in mice, how variations in metabolism or signaling between lineages could affect body fat distribution, and the questions that remain unresolved.
Keywords: lineage tracing, white adipose tissue (WAT), brown adipose tissue (BAT), brite or beige adipocytes, Myf5, adipocyte progenitor/precursor, lipodystrophy
Adipose tissue is complex and heterogeneous
Obesity is a risk factor for many diseases including type 2 diabetes, cardiovascular disease, and cancer. Obesity occurs when energy intake exceeds energy expenditure causing adipose tissue to overgrow. However, body fat distribution patterns are highly variable between individuals and not all fat is equal with some depots having more favorable metabolic properties than others [1]. Some lipodystrophies also present as fat distribution disorders characterized by regional lipoatrophy with other depots being spared or even expanding [2, 3]. With a worldwide obesity epidemic well underway, the study of adipose tissue growth, distribution, and regulation is a major research focus. Yet the developmental origins of fat, the determinants of body fat distribution, and the signaling mechanisms that control fat growth remain poorly defined.
In mammals, fat is typically classified by morphological appearance as being either white adipose tissue (WAT) or brown adipose tissue (BAT). WAT (the primary site of energy storage) is mostly composed of adipocytes containing a large unilocular lipid droplet. WATs are found throughout the body; however, the distribution of mass between each depot varies in the population as a function of genetics, age, and for some depots, sensitivity to hormones and glucocorticoids. WAT location is often classified as being visceral (in the trunk cavity) or subcutaneous (below the skin). There is not a uniformly applied system for describing the anatomical location of each WAT depot and therefore depot names often vary between studies. According to [4], WAT depots in rodents include the anterior subcutaneous WATs (asWATs) including interscapular and axillary WAT (located in the scapular region), inguinal WAT (ingWAT; attached dorsally along the pelvis to the thigh of the hindlimb), perigonadal WAT (pgWAT; surrounding the uterus and ovaries in females and the epididymis and testes in males), retroperitoneal WAT (rWAT; located within the abdominal cavity along the dorsal wall of the abdomen behind the kidney but not attached to the kidney or mixed with perirenal brown fat), and mesenteric WAT (mWAT; lining the surface of the intestines) (Figure 1). Although the primary function of WAT is energy storage, it also functions as an endocrine organ secreting hormones and cytokines such as leptin and adiponectin that regulate feeding and metabolism [5]. Epidemiological studies have found that the accumulation of visceral fat (determined by high waist-to-hip ratio) associates with metabolic disease (i.e. insulin resistance, type 2 diabetes, dyslipidemia, hypertension, atherosclerosis, hepatic steatosis, and cancer) while the accumulation of subcutaneous WAT associates with improved insulin sensitivity and low risk for developing type 2 diabetes [6-14].
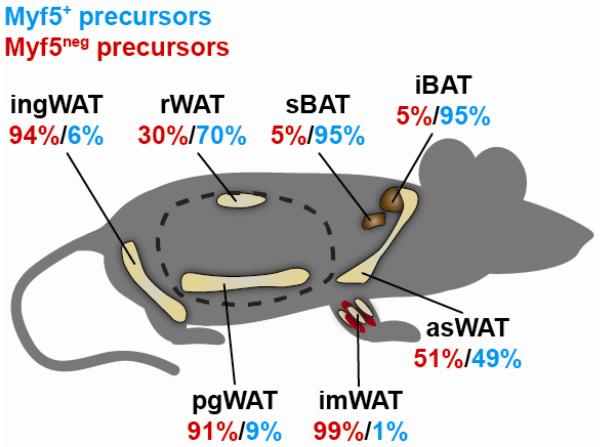
Figure 1. The Myf5 lineage contribution to the precursor pool in each fat depot varies with its anatomical location.
The contribution of the Myf5 lineage to the adipocyte precursor cell compartment (defined as CD31−CD45−Terr119−CD29+CD34+Sca1+ cells) was recently determined by lineage tracing with Myf5-Cre;R26R-YFP mice. More than 95% of the precursors in brown fat are labeled with Myf5-Cre. In the anterior subcutaneous WATs (including interscapular and axial WATs) nearly 50% of the precursors trace to Myf5+ precursors, and in the rWAT (a visceral WAT), the Myf5 precursors give rise to approximately 70% of the adipocyte precursor cell pool. In contrast, the Myf5 lineage contributes very little to the adipocyte precursor pool in ingWAT and pgWAT, which are 90-95% Myf5neg. The contribution of the Myf5 lineage to the intramuscular adipogenic precursor pool (defined with slightly different cell surface markers) is very low. Dotted line indicates the abdominal cavity. References provided in the text.
Brown adipocytes contain multiple smaller (multilocular) lipid droplets, are rich in mitochondria, and reside in depots that are highly innervated and vascularized. In rodents, BAT is located in discrete depots in interscapular (iBAT), sub-scapular (sBAT), and cervical (cBAT) regions of the upper anterior side of the trunk and neck (Figure 1) [4]. BAT also grows around parts of the aorta and kidneys [15]. These depots are often called “classical” BAT to distinguish them from brown-adipocyte-like cells, called brite adipocytes, which reside within some WATs (discussed below). In contrast to energy storing white adipocytes, brown adipocytes are specialized to expend energy to generate heat in a process called adaptive thermogenesis [16]. BAT is stimulated by the sympathetic nervous system following exposure to cold temperature and regulates the acute non-shivering thermogenesis response as well as the adaptive cold acclimatization response following chronic cold exposure. Thermogenesis is meditated by uncoupling protein 1 (UCP1), which embeds in the inner mitochondrial membrane, and produces heat by dissipating the proton electrochemical gradient over the inter-mitochondrial membrane space without generating ATP [17]. BAT stores energy for thermogenesis as perilipin coated lipid droplets and glycogen granules [18, 19] and upon stimulation rapidly increases glucose and FA uptake to replenish its supplies. The high glucose uptake capacity of BAT makes it readily detectable by 18F-fluorodeoxyglucose positron emission tomography (FDG-PET) [17, 20, 21]. While long thought to be critical only in rodents and newborn humans, the recent realization that BAT functions in adult humans (made possible by its sensitivity to FDG-PET) [22-26] raises the possibility that therapeutically controlling BAT growth and/or energy expenditure could be a strategy to combat obesity [27-34].
Interest is also growing in a third potential class of adipocyte called a brite adipocyte (also known as a “beige”, “inducible brown”, or “recruitable brown” adipocyte) [19, 21, 35-41]. This mysterious type of adipocyte exists among classical white adipocytes and is morphologically indistinguishable from its neighboring white adipocytes in the basal or unstimulated state. However, upon stimulation by chronic cold exposure (or other mechanisms that mimic beta-adrenergic stimulation) they become multilocular and begin expressing UCP1 [19, 21, 40, 42]. The presence of brite adipocytes in the WAT of mice is not homogeneous. For example, many adipocytes in ingWAT or rWAT become multilocular and induce UCP1 following stimulation; however, only a few UCP1+ brite adipocytes arise in pgWAT in the same mice [19, 40]. Brite adipocyte content varies between mouse strains and correlates with overall strain sensitivity to high fat diet [43-47]. Whether brite adipocytes form by trans-differentiation of existing white adipocytes or arise from a unique preadipocyte lineage is under debate (discussed below) [19, 40, 41, 48-51]. Perhaps the biggest unresolved issue pertaining to brite adipocytes is whether these cells actually contribute significantly to thermogenesis. For example, while UCP1 mRNA is induced in brite adipocytes several hundred-fold relative to white adipocytes (which barely expresses UCP1) [38, 52, 53], the total UCP1 expression is still an order of magnitude lower than that detected in brown adipose tissue [52]. In fact, the maximum thermogenic capacity of brite fat has been estimated at only ~10% of classical brown fat in mice. A debate is underway as to whether human brown fat is more similar to brite fat or to classical brown fat in mice. Early reports argue that what is currently being called human brown fat is more similar to murine brite fat than to classical murine brown fat [48, 49]. However, recent studies that more extensively profile different layers of adipose tissue in newborns and adults reveals that humans have brown fat deposits—particularly in the neck—that have a classic brown fat signature [54, 55]. Interestingly, there appears to be a gradient of fat cell types in the neck, with deep neck fat being classical BAT, intermediate cells possibly being more brite-like, and the most peripheral adipocytes being classical white adipocytes [55]. These findings are important considering the growing emphasis on developing therapeutic strategies to induce the “browning” of WAT in humans [35, 56-59] because they suggest that the studies of classical brown fat in mice could also have important therapeutic implications in humans. The physiological significance and regulatory mechanisms of brown versus brite fat in humans clearly needs to be determined.
Each individual fat depot is complex, composed not only of mature adipocytes but also of adipocyte precursor cells, fibroblasts, nerves, vascular cells, macrophages, and other cell types (collectively called the stromal vascular fraction or SVF). Adding to their complexity is the fact that adipose tissues are functionally heterogeneous. Although the sharpest functional divergence is clearly between energy storing WAT and energy-expending BAT, functional divergence is also evident between WATs best exemplified by the risk associated with excess visceral fat versus subcutaneous fat. However, such simple distinctions are oversimplifications because differences in lipogenic activity, cell dynamics, proliferative and differentiation capacity, and gene expression even between categorically similar WAT depots have been reported [60-67]. Heterogeneity likely exists even within a single WAT as suggested by studies showing that neighboring adipocytes can respond differently to genetic perturbations or drugs [68-72]. The existence of such heterogeneity within and between fat tissues suggests broad conclusions should not be based on a limited survey of select fat depots, and that each depot should be considered separately.
Does adipocyte tissue heterogeneity reflect different developmental origins of adipocytes, variances in the developmental cues a particular adipocyte may experience during differentiation, or extracellular influences on the mature adipocytes unique to the local cell or tissue environment? Answering these questions could lead to new anti-obesity therapies, improve the understanding of lipodystrophies, and increase the prospect of using adipocyte progenitor cells for cell-based therapeutics [17, 73, 74].
The origin of adipocytes
Central to understanding the complexities and heterogeneity of adipose tissue is to understand where the path to becoming an adipocyte begins. Moreover, strategies targeting nascent adipocytes could be useful in fighting obesity; however the identity of the adipocyte progenitor cells is just beginning to be unraveled. As obesity progresses, adipose tissues grow by hypertrophy (increasing the size of their adipocytes) and by hyperplasia (increasing the number of adipocytes) [75-79]. A yearly adipocyte turnover rate for lean and obese humans of 10 percent has been reported [79], but because mature adipocytes do not divide, a resident adipocyte precursor cell must exist.
Undifferentiated adipocyte progenitor cells were long assumed to reside in the stromal vascular fraction, but only recently have cell surface markers been used to distinguish adipocyte precursor cells from non-adipogenic stromal cells by Fluorescence Activated Cell Sorting (FACS) [37, 80-82]. One study identifies a CD45−CD31−Ter119−(Lin−) CD29+Sca1+CD34+CD24+ population that contains adipocyte progenitors capable of reconstituting a functional white adipose tissue when transplanted into lipodystrophic mice [80]. Similar cell surface markers have been used to isolate adipocyte precursors from brown fat and skeletal muscle [37, 83, 84]. Whether prospectively isolated adipocyte precursors originating from different fat depots are functionally similar is an open question. Moreover, whether a true adipocyte stem cell exists that can serially reconstitute an entire fat tissue including the same complement of stem and committed progenitor cells is unclear. Notably, depot-specific differences have been reported in adipocyte precursor cell surface marker expression levels, gene expression profiles, proliferation and differentiation capacity, and response to high fat diet [81, 85-87]. Strain-dependent differences in adipocyte precursor cell number have also been observed [85]. It has been argued that using cell surface markers to isolate adipose precursors is problematic because these markers lack functional relevance and cannot be used to localize adipocytes in vivo [88]. While valid, alternative approaches rely on identifying adipocyte precursor cell functional marker genes and using the promoter to drive expression of a reporter to genetically mark lineages (to date, a truly specific adipocyte precursor cell functional marker remains elusive) [21, 89, 90]. Both approaches are in fact quite complimentary; however, in clinical practice the most practical method to isolate adipocyte precursor cells from human lipoaspirates is by cell sorting based on surface marker expression [73]. Protocols to prospectively isolate human adipocyte precursor cells are currently being optimized.
Lineage tracing on the other hand is the only technique that can conclusively indicate the developmental origin of an adipocyte or reveal which tissue-resident progenitors actually become mature adipocytes, and therefore these studies can only be performed in mice. Many groups are performing lineage-tracing analyses using transgenic and Cre-Lox technology [Table 1]. Hypothesizing that Peroxisome Proliferator-Activated Receptor γ (PPARγ) expression is an early marker of adipocyte commitment, one study marked PPARγ-expressing stromal vascular fraction cells with GFP (using a PPARγ-tetracycline transactivator (tTA) knock-in mouse together with the tetracycline response element (TRE)-H2B-GFP reporter system) [82]. This labeled an adipocyte precursor population that expresses cell surface markers that are similar to the prospectively isolated adipocyte precursors described above suggesting that early precursors express PPARγ [80]. Interestingly, these GFP marked precursors appear to commit to the adipocyte lineage in embryonic development or shortly after birth, and localize in vivo to the adipose tissue mural cell compartment of the vasculature [82]. Using platelet-derived growth factor receptor β (PDGFRβ) expression (which marks mural cells in many tissues) to isolate and fractionate stromal vascular fraction cells confirmed that the PDGFRβ+ mural cells in adipose tissue, but not in other tissues like kidneys, skeletal muscle, and cardiac muscle, are enriched for adipogenic precursors. In fact, when PDGFRβ–Cre is combined with the Rosa 26 reporter (R26R) -LacZ reporter (which permanently labels PDGFRβ–Cre expressing cells and their descendants) rWAT and ingWAT are homogeneously marked [82]. The PDGF receptors (PDGFRα and β) are platelet-derived growth factor sensing receptor tyrosine kinases that activate several down-stream signaling pathways including the MAPK, PI3K and PLCγ pathways [91]. In contrast, PPARγ-GFP lineage tracing only labels a subset of mural cells, indicating that while all the adipocytes appear to arise from mural cells, only a subset of the mural cells are adipogenic [82]. What determines the identity of the adipogenic population is not known. These observations are consistent with previous ultra-structural findings suggesting the adipose vasculature may function as the adipocyte progenitor cell niche [92-94].
Table 1. Summary of recent lineage tracing studies of pre- and mature adipocytes.
| Genetic approach | Promoter location1 |
Rationale | Reported result2 | References3 | |
|---|---|---|---|---|---|
|
White
adipocytes |
sox10-Cre;
R26R-eYFP |
Transgene | Sox10 expresses in pre- and migratory neural crest cells at all rostro-caudal levels |
Mature adipocytes in the salivary gland labeled positive; pgWAT and ingWAT labeled negative |
[103, 139] |
|
PPARγ-tTA;TRE-Cre;
R26R-LacZ |
Knock-in | PPARγ is a nuclear receptor expressed in all tissues and is critical for adipogenesis |
IngWAT and rWAT positive; some positive SVF cells | [82] | |
| aP2-GFP | Transgene | aP2 (FABP4) expresses in mature adipocytes and is a target of PPARγ |
ingWAT positive | [82] | |
| aP2-Cre;R26R-LacZ | Transgene | Described above | ingWAT and pgWAT positive | [82] | |
|
PDGFRβ-Cre;
R26R-LacZ |
Transgene | PDFGRβ is expressed in vascular mural cells | ingWAT and rWAT positive | [82, 140] | |
|
myf5-Cre;
R26R-eYFP |
Knock-in | Myf5 is a muscle differentiation transcription factor that is first expressed in the dermomyotome at E8.0 and expresses in adult satellite cells |
ingWAT and pgWAT negative | [116, 117] | |
|
wnt1-Cre;
R26R-eYFP |
Transgene | Restrictive marker of migrating neural crest cells | Cephalic WAT positive; pgWAT and ingWAT negative. 70% of adipocyte precursors in cephalic WAT positive; all ingWAT adipocyte precursors negative |
[104, 141] | |
|
LysM-Cre;
R26R-LacZ |
Knock-in | The lysozyme M is exclusively expressed in macrophages and granulocytes |
5-20% of pgWAT adipocytes positive | [110, 111] | |
|
VE-cadherin-Cre; R26R-LacZ |
Knock-in | VE-cadherin is expressed in endothelial cells, is involved in cell adhesion, and is essential for vascular system development |
ingWAT and pgWAT positive | [96, 98] | |
|
VE-cadherin-Cre;
R26R-eGFP |
Knock-in | Described above | Subset of mature adipocytes in the pgWAT positive | [96, 98] | |
|
VE-cadherin-CreERT2;
R26R-LacZ |
Knock-in | VE-cadherin-CreERT2 is a tamoxifen-inducible version of the VE-cadherin-Cre |
Numerous adipocytes in the ingWAT, pgWAT and BAT positive |
[97, 98] | |
| ZFP423-GFP | Knock-in | Zfp423 is a transcriptional regulator broadly expressed but essential for preadipocyte commitment |
ingWAT positive | [95] | |
|
pax3-Cre;
R26R-mTmG |
Knock-in | Pax3 is a muscle differentiation transcription factor first expressed in dorsal neural crest and somites; it cooperates with Myf5 to drive muscle development |
50% of the adipogenic SVF cells from asWATs positive |
[123, 124] | |
| myf5-Cre;R26R-LacZ | Knock-in | Described above | Mature adipocytes from asWATs and rWAT positive; Few mature adipocytes from ingWAT and pgWAT positive; 50-60% of adipogenic SVF cells from asWAT and rWAT positive; <2% from the ingWAT and pgWAT positive |
[83, 117] | |
| myf5-Cre;R26R-eYFP | Knock-in | Described above | 50-70% of the adipocyte precursors from asWATs and rWAT positive; <10% of the adipocyte precursors from pgWAT and ingWAT positive |
[83, 117] | |
|
vav1-Cre;
R26R-mTmG |
Knock-in | Vav1 is a proto-oncogene that expresses in the hematopoietic and lymphoid systems |
All adipocyte precursors and mature adipocytes from ingWAT, pgWAT, rWAT and mWAT negative |
[100, 112] | |
| tie2-Cre;R26R-mTmG | Transgene | Tie2 is an angiopoietin receptor specific to endothelial cells and some hematopoietic cells |
All adipocyte precursors and mature adipocytes from ingWAT, pgWAT, rWAT and mWAT negative |
[100, 142] | |
|
VE-cadherin-Cre;
R26R-mTmG |
Knock-in | Described above | All adipocyte precursors and mature adipocytes from ingWAT, pgWAT, rWAT and mWAT negative |
[96, 100] | |
|
PDFGRα-Cre;
R26R-mTmG |
Transgene | PDGFRα is expressed in most mesenchymal cells can activate the MAPK, PI3K and PLCγ signaling pathways |
All adipocyte precursors and mature adipocytes from ingWAT, pgWAT, rWAT and mWAT positive |
[100, 143] | |
|
Brown
adipocytes |
En1-Cre;R26R-LacZ | Knock-in | Engrailed 1 is a homeobox transcription factor that expresses in cells of the central dermomyotome |
BAT positive | [114, 144] |
| myf5-Cre;R26R-eYFP | Knock-in | Described above | BAT positive | [116, 117] | |
|
pax7-CreERT2; R26R-LacZ |
Knock-in | Pax7 is expressed in medial and central cells of the dermomyotome and in adult satellite cells |
BAT positive | [121, 122] | |
| pax3-cre;R26R-mTmG | Knock-in | Described above | All adipogenic cells from the SVF of BAT positive | [123, 124] | |
| myf5-Cre;R26R-LacZ | Knock-in | Described above | BAT positive; >99% of adipogenic BAT SVF cells positive |
[83, 117] | |
| myf5-Cre;R26R-eYFP | Knock-in | Described above | >95% of BAT adipocyte precursors positive | [83, 117] | |
|
VE-cadherin- Cre;R26R-LacZ |
Knock-in | Described above | BAT positive | [96, 98] | |
|
VE-cadherin-CreERT2;
R26R-LacZ |
Knock-in | Described above | Many brown adipocytes positive | [97, 98] | |
|
Brite
adipocytes |
myf5-Cre;R26R-eYFP | Knock-in | Described above | Brite adipocytes in ingWAT negative | [116, 117] |
|
PDGFRα-CreERT2;
R26R-tdTomato |
Knock-in |
PDGFRα-CreERT2 is a tamoxifen-inducible version of the PDGFRα-CreER |
90% of the CL316,243 induced brite adipocytes in pgWAT positive |
[50, 145] | |
| myf5-Cre;R26R-LacZ | Knock-in | Described above | CL316,243 induced brite adipocytes in asWATs and rWAT positive; brite adipocytes induced in ingWAT negative |
[83, 117] | |
|
VE-cadherin-CreERT2;
R26R-LacZ |
Knock-in | Described above | Rosiglitazone induced bright adipocytes in ingWAT positive |
[97, 98] | |
|
UCP1-CreERT2;
R26R-tdRFP |
Knock-in | UCP1 is the most accepted functional marker of brown adipocytes |
Brite and white adipocytes interconverts in ingWAT depending on the temperature |
[51] | |
|
Adipocyte
precursors in skeletal muscle |
myf5-Cre;R26R-eYFP | Knock-in | Described above | negative | [84, 117] |
| myf5-Cre;R26R-eYFP | Knock-in | Described above | negative | [37, 117] | |
| myf5-Cre;R26R-mTmG | Knock-in | Described above | negative | [117, 124] | |
|
Pax3-Cre; R26R- mTmG |
Knock-in | Described above | negative | [123, 124] |
R26R: Rosa26 reporter.
tTA: tetracycline transactivator.
TRE: tetracycline response element.
SVF: stromal vascular fraction.
The theory that adipocytes arise from a vascular bed has recently been expanded to include the hypothesis that adipocytes actually originate from a subset of endothelial cells. For example, transgenic mice driving GFP expression from the promoter of the zing finger protein 423 (zfp423), a transcription factor involved early in preadipocyte commitment, not only labels mural cells, but also some endothelial cells in ingWAT and iBAT as determined by co-expression with the endothelial cell marker CD31 and PDGFRβ [88, 95]. Interestingly, the zfp423-GFP marked cells are different from those marked with PPARγ-GFP, and exhibit depot-specific differences in the expression of surface markers [80, 82, 95]. Thus, zfp423 could be a functional marker of white and brown preadipocytes early in commitment. However, the specificity of zfp423 to preadipocytes is unclear because it is also expressed in skeletal muscle and brain tissues at comparable levels [95]. A related study also arrives at an endothelial-cell-of-origin model using the endothelial cell specific VE-cadherin-Cre knock-in allele with the R26R-LacZ reporter, which reportedly marks adipocytes in pgWAT, ingWAT and iBAT [96-98]. However, another study found that conditionally deleting PPARγ with the endothelial specific receptor tyrosine kinase-Cre (Tie2-Cre) did not affect PPARγ expression in fat or adipose tissue mass [99]. Moreover, when the same constitutive VE-cadherin-Cre driver or Tie2-Cre is used in combination with the R26R-mTmG reporter (in which Cre turns off expression of membrane-targeted Tomato fluorescent protein and induces expression of membrane-targeted GFP) no adipocytes expressed GFP indicating that VE-cadherin or Tie2 was not expressed in their lineage [100]. These contradictory findings might suggest that only subsets of adipocytes are of endothelial cell origin. This needs further investigation.
Accurate lineage tracing can be challenging in adipocytes and this warrants a brief discussion of certain considerations. For starters, in any lineage tracing study drivers expressed from an endogenous locus/promoter are preferred because they more likely reflect the physiological expression pattern. One factor to consider specifically when tracing mature adipocytes is their paucity of cytoplasm. This poses a particular problem when analyzing tissue sections for the expression of cytoplasmic reporters, such as LacZ or YFP/GFP. The mTmG reporter overcomes this caveat because the fluorescent reporters are membrane targeted and therefore concentrate the reporter at the cell membrane surrounding the predominantly lipid-filled interior of the adipocyte. It is also wise to employ careful co-staining with known markers to distinguish between signals emanating from the miniscule membrane/cytoplasmic region of the adipocyte from signals originating in the abutting stromal vascular fraction cells, especially in highly vascularized depots.
While progress is being made in identifying the white adipocyte progenitors residing in mature adipose tissues, much less is understood about the embryonic origins of these progenitors. Previously all adipocytes were thought to arise from a common mesodermal precursor [17], but in recent years lineage tracing studies appear to have challenged this assumption and the emerging view—perhaps not surprisingly—is that white adipocyte ancestry is complex. An early clue suggesting white adipocytes might have different ancestries comes from the pathology of certain partial lipodystrophies. For example patients with familial partial lipodystrophy type 2 (FPLD2; OMIM #151660 – also called Dunnigan syndrome) suffer from the gradual atrophy of adipose tissue in the limbs and parts of the trunk while adipose tissue in the face, neck, and intra-abdominal areas are spared and often increase in mass. Similarly, in congenital infiltrating lipomatosis of the face (CIL-F) excessive adipose tissue only infiltrates the facial soft tissue forming a lipomatous tumor that causes severe disfigurement [101, 102]. Such regional characteristics suggest a distinct adipogenic route in the cephalic region. Consistent with this hypothesis, lineage-tracing studies in mice indicate that Sox10-Cre, which expresses in pre-migratory and migratory cells of neural crest, mark adipocytes in the head but not in the ingWAT and pgWAT [103]. A related lineage tracing study using the Wnt1-Cre, which is induced during neural crest formation, finds that craniofacial progenitors and adipocytes but not subcutaneous or visceral adipocytes trace to a neural crest cell of origin [104]. This study also finds that the neural crest adipogenic lineage is the predominant contributor to a pool of adipogenic progenitors residing in both the cephalic fat and muscle. Curiously, the number of neural crest -derived progenitors declines with age and are replaced with an unidentified lineage in what has been called a lineage-succession model [104]. Interestingly, these two distinct adipogenic lineages have similar metabolic properties indicating that their divergent origin does not impart any unique functional feature that distinguishes them. Bones and skeletal muscles in the head are known to originate from the neural crest [105]. Thus, one possibility is that some adipocytes could share a common origin with their neighboring organs and this might reflect an important developmental relationship.
It has been reported that some adipogenic cells originate in the bone marrow, and that after bone marrow transplants in rodents, hematopoietic cells become adipocytes in iBAT, asWAT, omental WAT, and mWAT [106-108]. Moreover, it has also been reported that a substantial number of adipocytes in the pgWAT are labeled with lysozyme 2-Cre in LysM-Cre;R26R-LacZ mice in which Cre expression marks the myeloid lineage [109-111]. However, this too is up for debate because Vav1 oncogene-Cre (Vav1-Cre), which marks hematopoietic lineages [112] fails to label any adipocytes or adipocyte progenitors [100].
The Myf5 lineage contribution to fat
While the vision of white adipocyte origins remains blurred, what seemed to be increasing in clarity was that brown adipocytes arise from a distinct mesenchymal precursor cell shared with skeletal muscle, but not with any white adipocyte. Evidence suggesting that BAT and WAT could have distinct developmental origins first came from a lineage tracing analysis of central dermomyotome cells expressing the transcription factor Engrailed1 (a pattern formation related Homeobox-family member) with an En1-CreER knock-in allele [113]. Using the R26R-LacZ reporter this study revealed that iBAT but not ingWAT or pgWAT shares a common origin with skeletal muscle and dermis [114][Radhika Atit, personal communication]. Using microarrays to compare the gene expression profiles of preadipocytes during in vitro differentiation, a subsequent study determined that brown preadipocytes isolated from a mix of iBAT, sBAT and cBAT but not white preadipocytes from pgWAT have a myogenic-like transcriptional signature (including expression of myogenic factor 5 (Myf5) and myogenic differentiation antigen (MyoD) [115]. This model seemed all-but-confirmed by a report that traced the Myf5 lineage in fat [116]. In this study, the Myf5-Cre knock-in allele [117] was used to drive YFP expression from the R26R-YFP reporter. Using an anti-GFP antibody to detect YFP by immunofluorescence, it was shown that regionally localized iBAT and skeletal muscle, but not interscapular WAT expresses YFP [116]. YFP was also not detected in the brite adipocytes induced to form within ingWAT. This Myf5-Cre labeling pattern was confirmed by examining YFP mRNA expression. Other related work found that when Myf5-Cre drives expression of the R26R-diphteria toxin A-chain construct, the Myf5-lineage is ablated and this greatly reduces the classical BAT depots [118, 119] [Thomas Braun, personal communication]. Finally, lineage tracing performed with Pax7 (paired box7)-Cre or Pax3 (paired box3)-Cre knock-in alleles (Pax7 and Pax3 are transcription factors that cooperate with Myf5 to drive myogenesis) also label BAT [120-124]. The notion that BAT and skeletal muscle share a common origin distinct from WAT is logical because of the metabolic similarities between BAT and muscle (such as high mitochondrial content and increased energy expenditure) and thus from these studies emerged a model arguing that brown adipocytes can be delineated from white (and brite) adipocytes by their ancestral expression of Myf5 [17, 89].
The simplicity and logic behind the Myf5 model led to its widespread acceptance. Therefore, the recent finding that deleting phosphatase and tensin homolog (PTEN) with Myf5-Cre affects brown and white adipose tissue was quite surprising [83]. PTEN is a tumor suppressor and negative regulator of the insulin-PI3K-AKT-mTOR growth pathway [125]. In a genetic study aimed at understanding how this pathway regulates the growth of mesodermal derived tissues, the same Myf5-Cre knock-in allele used previously to trace the origin of brown adipocytes was used to conditionally delete PTEN. The myf5-cre;PTENfl/fl conditional knockout (PTENmyf5cKO) mice have severely overgrown interscapular, subscapular, and cervical BATs that are PTEN-deficient, consistent with brown adipocytes originating from a Myf5-positive (Myf5+) lineage. However, in contradiction to the model that all white adipocytes originate from a Myf5-negative (Myf5neg) precursor, PTENmyf5cKO mice also have hypertrophic anterior subcutaneous WATs and retroperitoneal WAT that are also PTEN deficient [83]. Even more surprising is that pgWAT and mesenteric WAT as well as posterior subcutaneous WATs including ingWAT are completely absent [83](Figure 2). This total body fat redistribution phenotype led to a comprehensive reevaluation of the Myf5+ lineage contribution to fat.
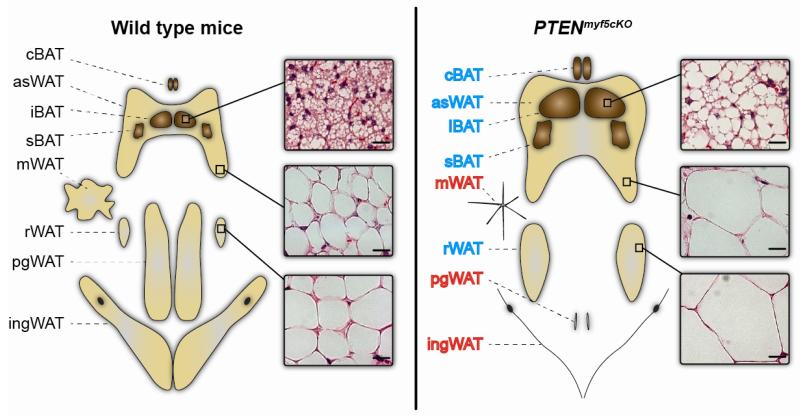
Figure 2. Deleting PTEN with Myf5-Cre models partial lipodystrophy.
Deleting PTEN with Myf5-Cre causes lipohypertrophy of the fat depots largely derived from Myf5+ adipocyte precursors (labeled in blue) and lipoatrophy of the fats that are largely Myf5neg (labeled in red). The overgrown fats are bigger because they contain more cells due to expansion of the Myf5+ adipocyte precursor cell population (not shown) and bigger cells as a result of increased lipid content (see histology insets). The absent fats are reduced to fibrotic tissue. This body fat distribution phenotype resembles the pathology of a rare human lipodystrophy syndrome called Madelung’s disease.
Originally, the Myf5 lineage had only been examined in a few adipose tissue depots. To revisit the Myf5+ lineage contribution to adipose tissue, the Myf5-Cre knock-in allele [117] was again combined with R26R-YFP reporter and separately with the R26R-LacZ reporter, and lineage tracing was performed on a broad selection of depots [83]. Here, Myf5-Cre was found to drive YFP mRNA expression not only in classical brown fat depots (iBAT, sBAT and cBAT) as expected, but also in anterior subcutaneous white fats (interscapular and axillary) and in rWAT. In contrast, very little YFP mRNA expression was detected in white adipocytes located in the ingWAT or pgWAT. These results were confirmed using Myf5-Cre;R26R-LacZ mice indicating that the Myf5-lineage contribution to fat is broader than previously thought. As mentioned above, En1-CreER had previously been shown to label dorsal dermis in addition to skeletal muscle and BAT; in fact, Myf5-cre also labels dorsal dermis [126] (J Sanchez-Gurmaches & DA Guertin) and reportedly (using a different Myf5-Cre knock-in allele) a portion of the ribs and cervical vertebrae [127]. Thus, it is perhaps not surprising that Myf5-Cre also labels white adipocytes in the same general anatomical location.
Interestingly, when stromal vascular fraction cells are prepared from various WAT depots isolated from Myf5-Cre;R26R-LacZ mice and differentiated in vitro, about half of the adipogenic cells from the asWATs or rWAT trace with Myf5-Cre compared to less-than 1% in ingWAT and pgWAT. This mixed lineage contribution is also detectable in the adipocyte precursor pool. Using Myf5-Cre;R26R-YFP and a FACS-based cell sorting strategy based on [80], it was found that most of the CD31−CD45−Ter119−CD29+Sca1+CD34+ cells isolated from BAT, nearly half of the adipocyte precursors isolated from the asWATs, and three-quarters of the adipocyte precursors isolated from rWAT, trace with Myf5-Cre. In contrast, less than 10% of the adipocyte precursors isolated from ingWAT and pgWAT are marked (Figure1). Gene expression analysis indicates that the Myf5-Cre tracing white adipocyte precursors do not express brown preadipocyte markers [83]. Consistent with these data, a recent study finds that the Pax3-Cre knock-in allele also labels adipogenic precursors isolated from the stromal vascular fraction of asWAT [124]. Interestingly, the Pax3-Cre driver also labels adipocyte precursor cells with a pattern similar to the Myf5-Cre driver (J Sanchez-Gurmaches and DA Guertin, unpublished). Why previous work failed to uncover the Myf5-lineage contribution to white fat could reflect the limited survey of fat depots, technical limitations, or strain differences.
A revised Myf5 lineage model and its implications
These new insights suggest a revised model of the Myf5-Cre lineage contributions to adipose tissue. We propose two possibilities: In Model (1) the Myf5+ (Pax3+) adipocyte precursors could originate in the central dermomyotome and mix with Myf5neg (Pax3neg) lineage(s) in the adipocyte precursor cell compartment of each depot (Figure 3). In BAT, most of the adipocyte precursors are Myf5+, in asWATs and rWAT the mix is more evenly distributed, while in other WATs such as ingWAT and pgWAT, a Myf5neg lineage(s) predominates (Figure 1). Based on this model, one might predict that the mature adipocyte pool in each depot would reflect this heterogeneity. Interestingly however, Myf5-cre lineage tracing of mature adipocytes with the R26R-LacZ reporter reveals a large fraction of the mature adipocytes in BAT, asWAT, and rWAT are LacZ positive, while only a few (<1%) mature adipocytes in ingWAT and pgWAT are positive [83]. Thus, despite the heterogeneity in the adipocyte precursor cell compartment, it appears that the Myf5+ lineage may selectively differentiate in the BAT, asWATs, and rWAT, while Myf5neg lineages selectively give rise to most adipocytes in the ingWAT and pgWAT. Importantly, exactly what cell types are contained within the adipocyte precursor pools as identified by cell surface markers, and how selective expansion of certain lineages might be determined is not known. Notably, the true adipocyte progenitor is likely contained within a subset of the adipocyte progenitor cell pool and it cannot yet be ruled out that Myf5neg precursors give rise to Myf5+ (Pax3+) precursors within that pool. Accordingly in Model (2), Myf5-Cre (and Pax3-Cre) expression could be induced during determination or adipocyte differentiation in some depots (Figure 3). Thus, it will be important to determine exactly when Myf5-Cre and Pax3-Cre express in the adipocyte lineages (e.g. with an inducible Cre), how many mature adipocytes in each depot arise from these lineages, and whether Myf5/Pax3 have functional roles in these adipocytes [128]. This requires further investigation.
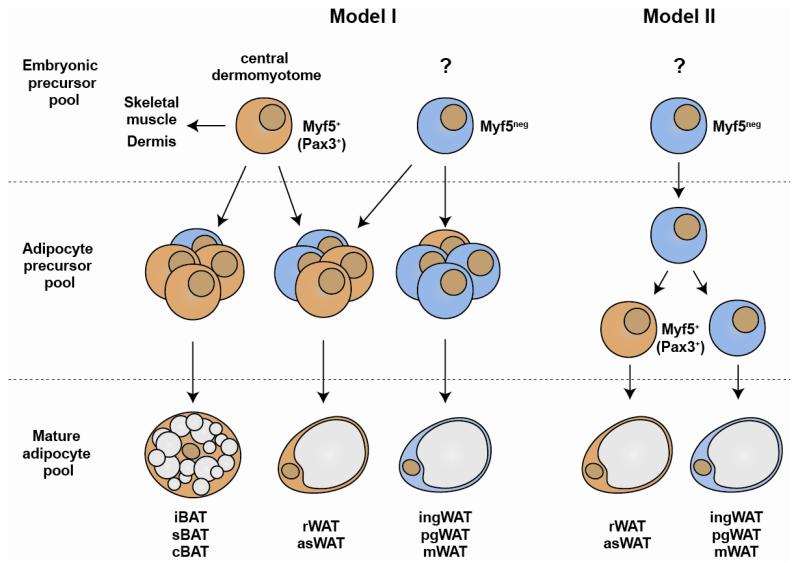
Figure 3. Where is Myf5-Cre expressed during adipocyte development?
At least two models could explain the origins of adipocytes that trace to Myf5-Cre+ precursors. (I) In the “common origin model”, Myf5+ (and possibly Pax3+) adipocyte precursors originate in the central dermomyotome during embryonic development and give rise to adipocyte progenitor cells residing in the stromal vascular fraction of each depot. Both Myf5+ and Myf5neg lineages contribute to the adipocyte precursor cell pool (lin−CD29+CD34+Sca1+) to varying degrees in each depot; however in BATs, asWATs, and rWAT the Myf5+ lineage precursors appear to be preferentially selected to become adipocytes while in the ingWAT, pgWAT, and mWAT the Myf5neg lineage precursors selectively differentiate. (II) Alternatively, in the “selective induction model” Myf5neg precursors residing in the stromal vascular fraction of asWAT and rWAT (and perhaps even in BAT) induce Myf5-Cre (and possibly Pax3) expression upon differentiation giving rise to Myf5+ precursors, while in the other WATs Myf5-Cre is not induced. Whether Myf5 protein has a functional role in these adipocyte lineages is unknown.
Previously, it made sense that the mitochondria rich and energy expending brown fat might share a common precursor with skeletal muscle, but what is the significance of arising from a Myf5+ (Pax3+) versus a Myf5neg (Pax3neg) precursor for a white adipocyte? One possibility is that progenitors and/or mature adipocytes arising from Myf5+ precursors could have a metabolic advantage (or disadvantage). Metabolic variations between distinct lineages could affect depot-specific fat growth and thus body fat patterning. Another possibility is that endocrine function could vary between lineages. Alternatively, there may be no functional differences between the adipocyte precursor cells arising from Myf5 positive or negative precursors. Precedence for such functional convergence exists. For example, it was recently shown that dermal precursors in the trunk arise from Myf5+ precursors while dermal precursors in the face arise from Myf5neg (Wnt1+) precursors, yet despite having different developmental origins, the mature dermal cells appear functionally similar based on marker profile, proliferation capacity, and ability to self-renew and differentiate [126]. Similarly, adipogenic precursors that arise from the neural crest appear functionally similar to mesoderm-derived adipogenic precursors [103, 104]. Thus, being of a different developmental origin does not automatically imply a functional difference.
While it remains to be seen whether natural variations in metabolism between white adipocytes of different origins contributes to body fat patterning, is it possible that a mutation could arise that targets a specific adipocyte lineage. Such a mutation could alter the growth, metabolism, or insulin sensitivity of that lineage, and it seems reasonable to speculate that such a scenario could be the basis of certain lipodystrophy disorders. Lipodystrophy can be inherited or more commonly acquired, and can affect all body fat (i.e. generalized lipoatrophy) or only specific regions (i.e. partial lipoatrophy). In many cases of partial lipodystrophy the spared regions increase in mass for example in Dunnigan-type familial partial lipodystrophy (described above) and Barraquer-Simons partial lipodystrophy (in which lipoatrophy begins in the head and neck and spreads to the chest, abdomen and upper extremities, while the adipose tissues in the lower abdomen, gluteal region, and lower extremities expand) [129]. Even in congenital generalized lipodystrophy or Berardinelli-Seip syndrome, which is characterized by the near-complete absence of adipose tissue, some fat depots are spared including those in the head, palms, and sole of the foot [2]. These regional variations in fat pathology have contributed to the hypothesis that adipocytes have distinct developmental origins.
Consistent with the idea that lineage-restricted mutations could cause lipodystrophy is the discovery that PTEN loss in the Myf5 lineage causes severe body fat redistribution [83]. In PTENmyf5cKO mice, body fat is redistributed such that Myf5+ adipocytes expand (causing lipohypertrophy of BAT, asWATs, and rWAT) at the expense of Myf5neg adipocytes (i.e. inguinal, perigonadal, and mesenteric WAT), which are reduced to fibrotic tissue (Figure 2). The hypertrophic fats have more cells due to expansion of the adipogenic precursors, and bigger cells caused by increased lipid content. PTENmyf5cKO mice also exhibit skeletal muscle atrophy, which appears to be caused by a systemic effect because PTEN expression is maintained at normal levels in most skeletal muscles by a post-transcriptional mechanism. Losing PTEN hyperactivates the major intracellular effectors of insulin including PI3K, AKT, and mTOR, which likely alters the metabolic balance in the Myf5 adipocyte lineage to favor energy uptake and storage. Interestingly, the PTENmyf5cKO mouse phenotypically resembles a rare partial lipodystrophy syndrome called multiple symmetric lipomatosis or Madelung’s disease [3]. Patients with Madelung’s disease suffer from mild to severe adipose tissue expansion in the head and neck with extension down the back (type I) and/or the shoulders, upper arms and abdomen (type II). The PTENmyf5cKO phenotype also resembles the buffalo hump syndrome often seen in HIV patients on aggressive anti-retroviral therapy, which is currently the most common lipodystrophy disorder in the United States [130]. Thus, the PTENmyf5cKO mouse is a novel model of partial lipodystrophy and its phenotype supports the idea that mutations targeting metabolism or the metabolic regulatory circuits in a particular adipocyte lineage could be the pathological basis of certain lipodystrophies.
Where do brite adipocytes come from?
In response to cold or β-adrenergic agonists, brite adipocytes appear within white fat and can comprise a significant fraction of the total adipocyte population depending upon the depot. While ingWAT and to a lesser extent rWAT are predisposed to browning based on their rapid response to acute cold challenge, other depots like pgWAT are more resistant and only show some browning after chronic cold challenge [4, 21, 41, 52]. Two models have been proposed to explain the origins of brite adipocytes. One posits that brite adipocytes arise by transdifferentiation of existing mature white adipocytes; the second argues that brite adipocytes arise de novo from a distinct precursor population [41, 48, 50, 131]. These models may not be mutually exclusive and depot-specific differences may exist (discussed below).
The brite adipocytes that arise in the largely Myf5neg lineage fats (e.g. pgWAT and ingWAT) are reportedly Myf5neg while the brite adipocytes arising in Myf5+ lineage fats (e.g. asWATs and rWAT) are Myf5+ [83, 116]. This is an important point because it is argued that all brite adipocytes are Myf5neg, which is true in ingWAT but not in rWAT or asWATs. Thus, brite adipocytes from different depots likely have different origins. Factors like the proximity to noradrenergic parenchymal nerve fibers, to the vasculature, or exposure to other micro-environmental conditions could determine which cells become brite [19, 40, 41]. Recently, several preadipocyte cell lines were sub-cloned from immortalized stromal cultures derived from ingWAT, and only some of these lines responded like a brite preadipocyte (called beige in this report) would be predicted too following stimulation (i.e. induce UCP1) [48]. These could be brite preadipocytes and although they were found to express distinct surface markers, lineage tracing was not performed, thus it is unclear if they represent a unique brite-adipocyte lineage. Brite/beige adipocytes from other white fat tissues were not examined [48].
Interestingly, another study finds that when mice are treated with a low dose of the β3-agonist CL316,243 that promotes proliferation but avoids inflammation, the brite adipocytes appearing in the pgWAT arise from a resident progenitor cell while the brite adipocytes induced in the ingWAT appear to arise by transdifferentiation [41, 50]. This suggests that the mechanism by which brite adipocytes arise varies with the depot and the duration of stimulation. Perhaps immediate responding brite adipocytes exist in ingWAT but not in pgWAT, which would explain why brite adipocytes only appear after chronic cold exposure in pgWAT, but are often readily detectable in ingWAT even at ambient temperature. The emerging nascent brite cell population in pgWAT traces to a platelet-derived growth factor receptor α (PDGFRα) positive precursor residing in the stromal vascular fraction (labeled with an inducible PDGFRα-CreERT2 allele). This precursor population lacks expression of the cell surface markers CD24, PDGFRβ, the endothelial marker IB4, and is negative for smooth muscle actin and PPARγ indicating it may be distinct from other precursors described above [50, 80, 82]. Originally, it was reported that the fate of these PDGFRα positive precursors is not restricted to brite fat because high-fat diet feeding instructs them to become white adipocytes [50]. A more recent lineage tracing study using a constitutive PDGFRα-Cre driver together with the R26R-mTmG reporter argues that all mature adipocytes in all WAT depots are in fact derivatives of PDGFRα+ precursors [100]. Mature adipocytes do not express PDGFRα [50] indicating these adipocytes are branded with PDGFRα-Cre as precursors. Whether PDGFRα has a functional role in the adipocyte precursor is unclear. However, PDGFR stimulation with ligand inhibits adipogenesis in 3T3-L1 cells [132] and expressing an activated mutant allele of PDGFRβ in pericytes or mesenchymal progenitor cells blocks white adipocyte differentiation [133], suggesting down-regulating PDGFR signaling might be essential for adipocyte differentiation. More information regarding PDGFR signaling can be found in [91].
A strong argument in favor of brite-to-white interconversion in the ingWAT was recently reported [51]. In this study, researchers used lineage tracing to mark the cold-induced brite adipocytes in the ingWAT (with a UCP1-CreER driver) then followed their fate upon returning the mice to warmer temperature. In response to warming (or “whitening”) the brite adipocyte characteristics disappeared but importantly the adipocytes that were labeled during cold-induced “brightening” retained their lineage mark and now expressed “white” characteristics. This is in contrast to classical brown adipocytes, which retain their brown characteristics even when unstimulated, and argues that brite adipocytes are not dedifferentiated or eliminated when no longer needed. Moreover, these same white adipocytes can be reconverted to brite adipocytes upon subsequent cold challenge. Thus, in the ingWAT interconversion between the white and brite state appears to be responsible for much of the cold-induced changes in adipocyte morphology. Futures studies are needed to determine whether depot, age, or strain specific differences exist in the interconversion rate and whether any lineages have inherent potential to become brite when stimulated.
Ectopic adipocytes in skeletal muscle
Ectopic fat accumulation in muscle is a pathological feature of several conditions including myopathies like Duchenne muscular dystrophy (OMIM #310200), obesity, and type 2-diabetes [134, 135]. Studies using FACS technology to prospectively isolate adipogenic precursor cells from rodent limb muscles are also beginning to expose the identity of muscle resident adipogenic precursors [37, 84, 136]. While the cell surface marker profiles differ slightly between the different protocols (e.g. CD45−CD31−CD34+Sca1−α7-integrin− in [84] and CD31−CD45−SM/C-2.6− in [136]), almost all of the adipogenic precursors identified in hind limb skeletal muscles express PDGFRα [84, 136]. These PDGFRα+ precursors are of mesenchymal origin and reside in the interstitial space between muscle fibers; however, they are not marked with Myf5-Cre or Pax3-Cre by lineage tracing and are distinct from satellite cells based on their localization [84, 124, 136]. Muscle-resident adipogenic cells can also be isolated using a CD45− Mac1−Sca1+ protocol [37]. In vitro, these cells can be induced to express high levels of UCP1 when stimulated with bone morphogenetic protein 7 (BMP7) suggesting they may function like inducible brown adipocytes. In fact, a histological examination of the thigh muscle revealed that brown adipocytes, defined as UCP1-positive multilocular cells, are interspersed in the muscle of mice [137]. It is interesting that muscle-associated adipogenic precursors originate from Myf5neg/Pax3neg lineages because this suggests that the adipogenic cells in muscle may have migrated from a different developmental location. It will important to analyze the Myf5+/Pax3+ lineage contribution to the muscle adipogenic precursor populations across several muscles to determine if the Myf5+ precursor contribution varies with anatomical location.
What can we learn from our ancestors?
The ability to safely store large amounts of energy in adipocytes was a critical adaptation in mammalian evolution. But as the energetic cost of obtaining calories has plummeted compared to evolutionary times, the remarkable energy storing capacity of adipose tissue is now being revealed, as are its vices. White fat depots are dispersed throughout the body, and while a typical healthy individual probably has around 80,000 calories stored in fat (equivalent to over one month of meals), the distribution of that fat between different depots is highly variable in the population. These variations are exaggerated among obese individuals. In general white adipose tissue biology is not well understood, but perhaps one of the most clinically relevant mysteries is why expansion of some depots (e.g. visceral white fat) correlates with high risk of metabolic disease, while expansion of other depots (e.g. subcutaneous white fat) is benign or protective. Although one reason is likely that visceral depots drain directly into the liver causing damage, there may be other underlying functional differences related to origin. Like white fat, the amount of brown fat also varies between individuals and its presence negatively correlates with body mass index [22, 28, 32, 33]. The existence of brown fat cells in adult humans may open doors to novel anti-obesity therapies that could function by increasing brown fat energy expenditure; however, the study of human brown fat is in its infancy.
Could lineage-tracing studies explain some of the variability between fat depots? Previously it was thought that all adipocytes descended from a common mesodermal precursor, but recent studies have argued that this assumption is incorrect. A current trending model is that brown, white, and brite adipocytes have distinct origins; however, evidence is mounting that this view is too simplistic (Figure 4). Although all adipocytes have a common precursor if traced far enough back, moving forward it will be important to determine when unique adipocyte lineage marks are established and whether early adipocyte ancestors convey unique properties to their descendant adipocytes that make them functionally different from adipocytes that arise from other lineages. Adipocyte lineages could vary in their developmental gene expression pattern, which could affect adipocyte metabolism, the regulatory circuits that control metabolism or endocrine function, or the ability of resident progenitor cells to respond to growth cues. How might this affect body fat patterning? One possibility is that variations in metabolism or insulin sensitivity between lineages could affect depot growth rate. Partial lipodystrophies in which body fat is redistributed may be extreme examples of this. Notably, the regional absence of fat tissue associated with many lipodystrophies may often be caused by intrinsic loss of adipocyte function. However, the PTENmyf5cKO model suggests the opposite scenario is also possible, that is a subset of regionally overactive fat tissues can prohibit the formation of fat depots in distal regions (Figure 2). To what extent adipocytes vary in their origin, and whether origin influences function, remains to be seen. One thing that is clear is that different fat depots are heterogeneous, and thus it seems prudent at this point to view each depot as a unique organ when experimenting.
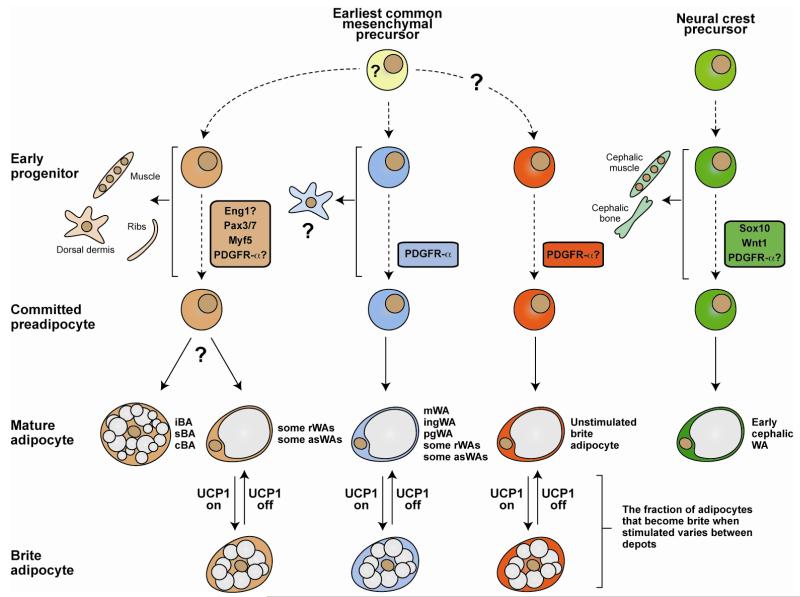
Figure 4. A model based on recent lineage tracing studies that explains some of the complexity of adipocyte ancestry.
A trending idea is that morphologically brown (multilocular), white (unilocular), and brite (interconvertible) adipocytes develop along unique lineages. However, recent studies in mice using Cre-Lox technology to irreversibly label cellular lineages argues that it is likely not this simple. An alternative view is that morphologically white adipocytes arise from multiple distinct lineages. One of these lineages is also marked by ancestral Myf5 expression and gives rise to morphologically brown adipocytes, a subset of morphologically white adipocytes (e.g. in the asWAT and rWAT), skeletal muscle cells, dermis, and ribs. Some of the Myf5-positive lineage derived white adipocytes become brite adipocytes when stimulated (e.g. Myf5-positive lineage white adipocytes in rWAT and asWAT). Other markers such as Engrailed-1 might also express early in this lineage. Whether the Myf5 lineage derived brown and white adipocytes arise from the same precursor is still unknown. Most white adipocytes in ingWAT and pgWAT develop from a Myf5-negative lineage (or possibly multiple lineages). Some of these Myf5-negative lineage white adipocytes can also become brite when stimulated (e.g. the Myf5-negative lineage white adipocytes that undergo interconversion in the ingWAT). Some brite adipocytes might also originate from a lineage distinct from the other white adipocytes although a Cre driver unique to a brite adipocyte lineage has not been found. Although white/brite adipocyte lineages are just beginning to be defined, PDGFRα appears to express in the ingWAT, pgWAT, rWAT, and mesenteric WAT lineages. Importantly, when each lineage mark actually expresses, when the lineages diverge during development, and the earliest common mesenchymal progenitor cell remain unclear and this is indicated by dotted lines and question marks. Another distinct white adipocyte lineage arises from Sox10 and Wnt1-expressing neural crest precursors in the cephalic region of young mice (notably, an unidentified lineage begins replacing neural crest derived cephalic adipocytes with age). These adipocytes label positive for PDGFR-α+ by antibody staining and therefore likely would trace with PDGFR-Cre, although this has not been shown. In sum, the emerging lineage tracing data presents a complex picture of adipocyte origins. iBA, sBA, cBA = interscapular, subscapular, and cervical brown adipocyte; asWA, rWA, pgWA, ingWA = anterior subcutaneous, retroperitoneal, perigonadal, inguinal white adipocyte. For more detailed information see [50, 51, 83, 100, 103, 114, 116, 127].
Is there an advantage to having adipocytes that arise from different developmental origins? One possibility is that having multiple origins might ensure that fat tissue still grows if one lineage becomes compromised (for example by a spontaneous mutation). Partial lipodystrophy syndromes again are examples of this in which regional lipoatrophy is often compensated for by lipohypertrophy of other depots. A second possibility is that some adipose tissue depots may grow in coordination with adjacent lean tissues, perhaps from a common precursor, to provide a locally available fuel reserve or other support service. This suggests a fate switching mechanisms could exist in a common precursor. Such a model has been proposed for brown adipose tissue and skeletal muscle development [116]. Adipose tissues of different origins could have other specialized functions, such as hormone production, responding to diet, or fueling exercise. In mice, adipose tissues appear at different times during embryonic and post-natal growth. Perhaps early growing fat depots arise as part of a developmental program, and these are metabolically more favorable depots. In contrast, the growth of less favorable fat tissues could arise from different precursors later perhaps in close correlation with caloric load. In mice for example, WATs that arise early such as subcutaneous WATs tend to be more metabolically favorable, while pgWAT arises later, is less vascularized, less innervated, and more static in response to cold challenge [41, 138].
What is the potential impact of understanding adipocyte origins for human health? As coping with the consequences of extreme adipose tissue growth is now a major burden on the health care system, therapeutic methods to restore normal energy balance are in great demand because exercise alone is no longer enough. Determining where adipocytes originate is an essential step towards elucidating the instructional program that functionally controls adipocyte development and function, and elucidating these pathways may reveal new mechanisms to block fat growth or increase the energy expenditure. On the flip side, adipose tissue is useful for reconstructive surgery and so knowing how to grow fat also has clinical value. Adipose tissue is a source of adult stem/progenitor cells that are obtainable by minimally invasive procedures, and there is clearly no shortage of them. Understanding the origins of adipose stem cells could be valuable in optimizing cell-based therapies. While the mouse is invaluable for lineage-tracing studies because of its genetic tractability, there are many differences between rodents and humans with respect to fat distribution. Protocols to isolate human adipocyte progenitor cells are under development and it will interesting and important to determine which functional markers are conserved.
HIGHLIGHTS.
Lineage tracing studies are revealing the complexity of adipocyte origins
The vasculature may be a niche for adipocyte progenitor cells
Myf5+ precursors give rise to brown adipocytes and a subset of white adipocytes
Different origins might explain depot heterogeneity and certain lipodystrophies
The significance of adipocyte origins, particularly in humans, remains unclear
Acknowledgements
We thank Michael Czech and Chien-Min Hung for critically reading the manuscript. D.A.G. is currently supported by The American Diabetes Association (ADA 1-13-BS-066), the NIH (R21CA161121), and the Pew Charitable Trusts. J.S.G. is supported by the Beatriu de Pinós program from the Generalitat de Catalunya.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lee MJ, Wu Y, Fried SK. Adipose tissue heterogeneity: implication of depot differences in adipose tissue for obesity complications. Molecular aspects of medicine. 2013;34:1–11. doi: 10.1016/j.mam.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Garg A. Acquired and inherited lipodystrophies. N Engl J Med. 2004;350:1220–1234. doi: 10.1056/NEJMra025261. [DOI] [PubMed] [Google Scholar]
- [3].Herbst KL. Rare adipose disorders (RADs) masquerading as obesity. Acta pharmacologica Sinica. 2012;33:155–172. doi: 10.1038/aps.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Walden TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. American Journal of Physiology-Endocrinology and Metabolism. 2012;302:E19–E31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- [5].Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nature Reviews Molecular Cell Biology. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Carey VJ, Walters EE, Colditz GA, Solomon CG, Willett WC, Rosner BA, Speizer FE, Manson JE. Body fat distribution and risk of non-insulin-dependent diabetes mellitus in women - The Nurses’ Health Study. American Journal of Epidemiology. 1997;145:614–619. doi: 10.1093/oxfordjournals.aje.a009158. [DOI] [PubMed] [Google Scholar]
- [7].Wang YF, Rimm EB, Stampfer MJ, Willett WC, Hu FB. Comparison of abdominal adiposity and overall obesity in predicting risk of type 2 diabetes among men. American Journal of Clinical Nutrition. 2005;81:555–563. doi: 10.1093/ajcn/81.3.555. [DOI] [PubMed] [Google Scholar]
- [8].Zhang C, Rexrode KM, van Dam RM, Li TY, Hu FB. Abdominal obesity and the risk of all-cause, cardiovascular, and cancer mortality - Sixteen years of follow-up in US women. Circulation. 2008;117:1658–1667. doi: 10.1161/CIRCULATIONAHA.107.739714. [DOI] [PubMed] [Google Scholar]
- [9].Baik I, Ascherio A, Rimm EB, Giovannucci E, Spiegelman D, Stampfer MJ, Willett WC. Adiposity and mortality in men. American Journal of Epidemiology. 2000;152:264–271. doi: 10.1093/aje/152.3.264. [DOI] [PubMed] [Google Scholar]
- [10].Pischon T, Boeing H, Hoffmann K, Bergmann M, Schulze MB, Overvad K, van der Schouw YT, Spencer E, Moons KGM, Tjonneland A, Halkjaer J, Jensen MK, Stegger J, Clavel-Chapelon F, Boutron-Ruault MC, Chajes V, Linseisen J, Kaaks R, Trichopoulou A, Trichopoulos D, Bamia C, Sieri S, Palli D, Tumino R, Vineis P, Panico S, Peeters PHM, May AM, Bueno-de-Mesquita HB, van Duijnhoven FJB, Hallmans G, Weinehall L, Manjer J, Hedblad B, Lund E, Agudo A, Arriola L, Barricarte A, Navarro C, Martinez C, Quiros JR, Key T, Bingham S, Khaw KT, Boffetta P, Jenab M, Ferrari P, Riboli E. General and Abdominal Adiposity and Risk of Death in Europe. New England Journal of Medicine. 2008;359:2105–2120. doi: 10.1056/NEJMoa0801891. [DOI] [PubMed] [Google Scholar]
- [11].Misra A, Garg A, Abate N, Peshock RM, StrayGundersen J, Grundy SM. Relationship of anterior and posterior subcutaneous abdominal fat to insulin sensitivity in nondiabetic men. Obesity Research. 1997;5:93–99. doi: 10.1002/j.1550-8528.1997.tb00648.x. [DOI] [PubMed] [Google Scholar]
- [12].Snijder MB, Dekker JM, Visser M, Bouter LM, Stehouwer CDA, Kostense PJ, Yudkin JS, Heine RJ, Nijpels G, Seidell JC. Associations of hip and thigh circumferences independent of waist circumference with the incidence of type 2 diabetes: the Hoorn Study. American Journal of Clinical Nutrition. 2003;77:1192–1197. doi: 10.1093/ajcn/77.5.1192. [DOI] [PubMed] [Google Scholar]
- [13].Qiao Q, Nyamdorj R. Is the association of type II diabetes with waist circumference or waist-to-hip ratio stronger than that with body mass index? European Journal of Clinical Nutrition. 2010;64:30–34. doi: 10.1038/ejcn.2009.93. [DOI] [PubMed] [Google Scholar]
- [14].Meisinger C, Doring A, Thorand B, Heier M, Lowel H. Body fat distribution and risk of type 2 diabetes in the general population: are there differences between men and women? The MONICA/KORA Augsburg Cohort Study. American Journal of Clinical Nutrition. 2006;84:483–489. doi: 10.1093/ajcn/84.3.483. [DOI] [PubMed] [Google Scholar]
- [15].Fitzgibbons TP, Kogan S, Aouadi M, Hendricks GM, Straubhaar J, Czech MP. Similarity of mouse perivascular and brown adipose tissues and their resistance to diet-induced inflammation, American journal of physiology. Heart and circulatory physiology. 2011;301:H1425–1437. doi: 10.1152/ajpheart.00376.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological reviews. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- [17].Tseng Y-H, Cypess AM, Kahn CR. Cellular bioenergetics as a target for obesity therapy. Nature Reviews Drug Discovery. 2010;9:465–481. doi: 10.1038/nrd3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Virtanen KA, van Marken Lichtenbelt WD, Nuutila P. Brown adipose tissue functions in humans. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbalip.2012.12.011. [DOI] [PubMed] [Google Scholar]
- [19].Cinti S. The adipose organ at a glance. Disease Models & Mechanisms. 2012;5:588–594. doi: 10.1242/dmm.009662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, Eychmueller A, Gordts PLSM, Rinninger F, Bruegelmann K, Freund B, Nielsen P, Merkel M, Heeren J. Brown adipose tissue activity controls triglyceride clearance. Nature Medicine. 2011;17:200–U293. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- [21].Frontini A, Cinti S. Distribution and Development of Brown Adipocytes in the Murine and Human Adipose Organ. Cell Metabolism. 2010;11:253–256. doi: 10.1016/j.cmet.2010.03.004. [DOI] [PubMed] [Google Scholar]
- [22].Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto N-J, Enerback S, Nuutila P. Brief Report: Functional Brown Adipose Tissue in Healthy Adults. New England Journal of Medicine. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- [23].van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JMAFL, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJJ. Cold-activated brown adipose tissue in healthy men. The New England journal of medicine. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- [24].Zingaretti MC, Crosta F, Vitali A, Guerrieri M, Frontini A, Cannon B, Nedergaard J, Cinti S. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. Faseb Journal. 2009;23:3113–3120. doi: 10.1096/fj.09-133546. [DOI] [PubMed] [Google Scholar]
- [25].Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, Iwanaga T, Miyagawa M, Kameya T, Nakada K, Kawai Y, Tsujisaki M. High Incidence of Metabolically Active Brown Adipose Tissue in Healthy Adult Humans Effects of Cold Exposure and Adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. American Journal of Physiology-Endocrinology and Metabolism. 2007;293:E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- [27].Yoneshiro T, Aita S, Mastushita M, Ogawa T, Okamatsu-Ogura Y, Kawai Y, Saito M. Age-Related Decrease in Brown Adipose Tissue and Obesity in Humans. Obesity. 2011;19:S79–S79. doi: 10.1038/oby.2011.125. [DOI] [PubMed] [Google Scholar]
- [28].Yoneshiro T, Aita S, Matsushita M, Kameya T, Nakada K, Kawai Y, Saito M. Brown Adipose Tissue, Whole-Body Energy Expenditure, and Thermogenesis in Healthy Adult Men. Obesity. 2011;19:13–16. doi: 10.1038/oby.2010.105. [DOI] [PubMed] [Google Scholar]
- [29].Cypess AM, Chen Y-C, Sze C, Wang K, English J, Chan O, Holman AR, Tal I, Palmer MR, Kolodny GM, Kahn CR. Cold but not sympathomimetics activates human brown adipose tissue in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:10001–10005. doi: 10.1073/pnas.1207911109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang Q, Zhang M, Ning G, Gu W, Su T, Xu M, Li B, Wang W. Brown Adipose Tissue in Humans Is Activated by Elevated Plasma Catecholamines Levels and Is Inversely Related to Central Obesity. Plos One. 2011;6 doi: 10.1371/journal.pone.0021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ouellet V, Labbe SM, Blondin DP, Phoenix S, Guerin B, Haman F, Turcotte EE, Richard D, Carpentier AC. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. Journal of Clinical Investigation. 2012;122:545–552. doi: 10.1172/JCI60433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vijgen GHEJ, Bouvy ND, Teule GJJ, Brans B, Hoeks J, Schrauwen P, Lichtenbelt W.D.v.M. Increase in Brown Adipose Tissue Activity after Weight Loss in Morbidly Obese Subjects. Journal of Clinical Endocrinology & Metabolism. 2012;97:E1229–E1233. doi: 10.1210/jc.2012-1289. [DOI] [PubMed] [Google Scholar]
- [33].Vijgen GHEJ, Bouvy ND, Teule GJJ, Brans B, Schrauwen P, Lichtenbelt W.D.v.M. Brown Adipose Tissue in Morbidly Obese Subjects. Plos One. 2011;6 doi: 10.1371/journal.pone.0017247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gelfand MJ, O’Hara SM, Curtwright LA, MacLean JR. Pre-medication to block F-18 FDG uptake in the brown adipose tissue of pediatric and adolescent patients. Pediatric Radiology. 2005;35:984–990. doi: 10.1007/s00247-005-1505-8. [DOI] [PubMed] [Google Scholar]
- [35].Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic Peroxisome Proliferator-activated Receptor gamma (PPAR gamma) Activation of Epididymally Derived White Adipocyte Cultures Reveals a Population of Thermogenically Competent, UCP1-containing Adipocytes Molecularly Distinct from Classic Brown Adipocytes. Journal of Biological Chemistry. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ishibashi J, Seale P. Medicine. Beige can be slimming. Science. 2010;328:1113–1114. doi: 10.1126/science.1190816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, Schrier D, Falb D, Kirkland JL, Wagers AJ, Tseng Y-H. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Young P, Arch JRS, Ashwell M. Brown adipose-tissue in the parametrial fat pad of the mouse. Febs Letters. 1984;167:10–14. doi: 10.1016/0014-5793(84)80822-4. [DOI] [PubMed] [Google Scholar]
- [39].Cannon B, Nedergaard J. CELL BIOLOGY Neither brown nor white. Nature. 2012;488:286–287. doi: 10.1038/488286a. [DOI] [PubMed] [Google Scholar]
- [40].Cinti S. Between brown and white: Novel aspects of adipocyte differentiation. Annals of Medicine. 2011;43:104–115. doi: 10.3109/07853890.2010.535557. [DOI] [PubMed] [Google Scholar]
- [41].Cinti S. Transdifferentiation properties of adipocytes in the adipose organ. American Journal of Physiology-Endocrinology and Metabolism. 2009;297:E977–E986. doi: 10.1152/ajpendo.00183.2009. [DOI] [PubMed] [Google Scholar]
- [42].Nedergaard J, Bengtson T, Cannon B. Three years with adult human brown adipose tissue. In: Powers AC, Ahima RS, editors. Year in Diabetes and Obesity. Vol. 1212. 2010. pp. E20–E36. [DOI] [PubMed] [Google Scholar]
- [43].Hofmann WE, Liu XT, Bearden CM, Harper ME, Kozak LP. Effects of genetic background on thermoregulation and fatty acid-induced uncoupling of mitochondria in UCP1-deficient mice. Journal of Biological Chemistry. 2001;276:12460–12465. doi: 10.1074/jbc.M100466200. [DOI] [PubMed] [Google Scholar]
- [44].Vitali A, Murano I, Zingaretti MC, Frontini A, Ricquier D, Cinti S. The adipose organ of obesity-prone C57BL/6J mice is composed of mixed white and brown adipocytes. Journal of Lipid Research. 2012;53:619–629. doi: 10.1194/jlr.M018846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Xue B, Rim J-S, Hogan JC, Coulter AA, Koza RA, Kozak LP. Genetic variability affects the development of brown adipocytes in white fat but not in interscapular brown fat. Journal of Lipid Research. 2007;48:41–51. doi: 10.1194/jlr.M600287-JLR200. [DOI] [PubMed] [Google Scholar]
- [46].Xue BZ, Coulter A, Rim JS, Koza RA, Kozak LP. Transcriptional synergy and the regulation of Ucp1 during brown adipocyte induction in white fat depots. Molecular and Cellular Biology. 2005;25:8311–8322. doi: 10.1128/MCB.25.18.8311-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Coulter AA, Bearden CM, Liu XT, Koza RA, Kozak LP. Dietary fat interacts with QTLs controlling induction of Pgc-1 alpha and Ucp1 during conversion of white to brown fat. Physiological Genomics. 2003;14:139–147. doi: 10.1152/physiolgenomics.00057.2003. [DOI] [PubMed] [Google Scholar]
- [48].Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang A-H, Khandekar M, Virtanen KA, Nuutila P, Schaart G, Huang K, Tu H, Lichtenbelt W.D.v.M., Hoeks J, Enerbaeck S, Schrauwen P, Spiegelman BM. Beige Adipocytes Are a Distinct Type of Thermogenic Fat Cell in Mouse and Human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sharp LZ, Shinoda K, Ohno H, Scheel DW, Tomoda E, Ruiz L, Hu H, Wang L, Pavlova Z, Gilsanz V, Kajimura S. Human BAT Possesses Molecular Signatures That Resemble Beige/Brite Cells. PloS one. 2012;7:e49452–e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lee Y-H, Petkova AP, Mottillo EP, Granneman JG. In Vivo Identification of Bipotential Adipocyte Progenitors Recruited by beta 3-Adrenoceptor Activation and High-Fat Feeding. Cell Metabolism. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rosenwald M, Perdikari A, Rulicke T, Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nat Cell Biol. 2013 doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- [52].Nedergaard J, Cannon B. UCP1 mRNA does not produce heat. Biochimica et biophysica acta. 2013 doi: 10.1016/j.bbalip.2013.01.009. [DOI] [PubMed] [Google Scholar]
- [53].Cousin B, Cinti S, Morroni M, Raimbault S, Ricquier D, Penicaud L, Casteilla L. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J Cell Sci. 1992;103(Pt 4):931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- [54].Lidell ME, Betz MJ, Leinhard OD, Heglind M, Elander L, Slawik M, Mussack T, Nilsson D, Romu T, Nuutila P, Virtanen KA, Beuschlein F, Persson A, Borga M, Enerback S. Evidence for two types of brown adipose tissue in humans. Nat Med. 2013 doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- [55].Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, Huang TL, Roberts-Toler C, Weiner LS, Sze C, Chacko AT, Deschamps LN, Herder LM, Truchan N, Glasgow AL, Holman AR, Gavrila A, Hasselgren PO, Mori MA, Molla M, Tseng YH. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat Med. 2013 doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ahfeldt T, Schinzel RT, Lee Y-K, Hendrickson D, Kaplan A, Lum DH, Camahort R, Xia F, Shay J, Rhee EP, Clish CB, Deo RC, Shen T, Lau FH, Cowley A, Mowrer G, Al-Siddiqi H, Nahrendorf M, Musunuru K, Gerszten RE, Rinn JL, Cowan CA. Programming human pluripotent stem cells into white and brown adipocytes. Nature Cell Biology. 2012;14:209–219. doi: 10.1038/ncb2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nishio M, Yoneshiro T, Nakahara M, Suzuki S, Saeki K, Hasegawa M, Kawai Y, Akutsu H, Umezawa A, Yasuda K, Tobe K, Yuo A, Kubota K, Saito M, Saeki K. Production of Functional Classical Brown Adipocytes from Human Pluripotent Stem Cells using Specific Hemopoietin Cocktail without Gene Transfer. Cell Metabolism. 2012;16:394–406. doi: 10.1016/j.cmet.2012.08.001. [DOI] [PubMed] [Google Scholar]
- [58].Hondares E, Rosell M, Gonzalez FJ, Giralt M, Iglesias R, Villarroya F. Hepatic FGF21 Expression Is Induced at Birth via PPAR alpha in Response to Milk Intake and Contributes to Thermogenic Activation of Neonatal Brown Fat. Cell Metabolism. 2010;11:206–212. doi: 10.1016/j.cmet.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Fisher FM, Kleiner S, Douris N, Fox EC, Mepani RJ, Verdeguer F, Wu J, Kharitonenkov A, Flier JS, Maratos-Flier E, Spiegelman BM. FGF21 regulates PGC-1 alpha and browning of white adipose tissues in adaptive thermogenesis. Genes & Development. 2012;26:271–281. doi: 10.1101/gad.177857.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Edens NK, Fried SK, Kral JG, Hirsch J, Leibel RL. In-vitro lipid-synthesis in human adipose-tissue from 3 abdominal sites. American Journal of Physiology. 1993;265:E374–E379. doi: 10.1152/ajpendo.1993.265.3.E374. [DOI] [PubMed] [Google Scholar]
- [61].Tchkonia T, Lenburg M, Thomou T, Giorgadze N, Frampton G, Pirtskhalava T, Cartwright A, Cartwright M, Flanagan J, Karagiannides I, Gerry N, Forse RA, Tchoukalova Y, Jensen MD, Pothoulakis C, Kirkland JL. Identification of depot-specific human fat cell progenitors through distinct expression profiles and developmental gene patterns. American Journal of Physiology-Endocrinology and Metabolism. 2007;292:E298–E307. doi: 10.1152/ajpendo.00202.2006. [DOI] [PubMed] [Google Scholar]
- [62].Tchkonia T, Tchoukalova YD, Giorgadze N, Pirtskhalava T, Karagiannides I, Forse RA, Koo A, Stevenson M, Chinnappan D, Cartwright A, Jensen MD, Kirkland JL. Abundance of two human preadipocyte subtypes with distinct capacities for replication, adipogenesis, and apoptosis varies among fat depots. American Journal of Physiology-Endocrinology and Metabolism. 2005;288:E267–E277. doi: 10.1152/ajpendo.00265.2004. [DOI] [PubMed] [Google Scholar]
- [63].Rehrer CW, Karimpour-Fard A, Hernandez TL, Law CK, Stob NR, Hunter LE, Eckel RH. Regional Differences in Subcutaneous Adipose Tissue Gene Expression. Obesity. 2012;20:2168–2173. doi: 10.1038/oby.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gesta S, Bluher M, Yamamoto Y, Norris AW, Berndt J, Kralisch S, Boucher J, Lewis C, Kahn CR. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Vohl MC, Sladek R, Robitaille J, Gurd S, Marceau P, Richard D, Hudson TJ, Tchernof A. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obesity Research. 2004;12:1217–1222. doi: 10.1038/oby.2004.153. [DOI] [PubMed] [Google Scholar]
- [66].Yamamoto Y, Gesta S, Lee KY, Tran TT, Saadatirad P, Kahn CR. Adipose Depots Possess Unique Developmental Gene Signatures. Obesity. 2010;18:872–878. doi: 10.1038/oby.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sackmann-Sala L, Berryman DE, Munn RD, Lubbers ER, Kopchick JJ. Heterogeneity Among White Adipose Tissue Depots in Male C57BL/6J Mice. Obesity. 2012;20:101–111. doi: 10.1038/oby.2011.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bluher M, Michael MD, Peroni OD, Ueki K, Carter N, Kahn BB, Kahn CR. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Developmental Cell. 2002;3:25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- [69].Bluher M, Patti ME, Gesta S, Kahn BB, Kahn CR. Intrinsic heterogeneity in adipose tissue of fat-specific insulin receptor knock-out mice is associated with differences in patterns of gene expression. Journal of Biological Chemistry. 2004;279:31891–31901. doi: 10.1074/jbc.M404569200. [DOI] [PubMed] [Google Scholar]
- [70].de Souza CJ, Eckhardt M, Gagen K, Dong M, Chen W, Laurent D, Burkey BF. Effects of pioglitazone on adipose tissue remodeling within the setting of obesity and insulin resistance. Diabetes. 2001;50:1863–1871. doi: 10.2337/diabetes.50.8.1863. [DOI] [PubMed] [Google Scholar]
- [71].Fortier M, Soni K, Laurin N, Wang SP, Mauriege P, Jirik FR, Mitchell GA. Human hormone-sensitive lipase (HSL): expression in white fat corrects the white adipose phenotype of HSL-deficient mice. Journal of Lipid Research. 2005;46:1860–1867. doi: 10.1194/jlr.M500081-JLR200. [DOI] [PubMed] [Google Scholar]
- [72].Jernas M, Palming J, Sjoholm K, Jennische E, Svensson P-A, Gabrielsson BG, Levin M, Sjogren A, Rudemo M, Lystig TC, Carlsson B, Carlsson LMS, Lonn M. Separation of human adipocytes by size: hypertrophic fat cells display distinct gene expression. Faseb Journal. 2006;20:1540–+. doi: 10.1096/fj.05-5678fje. [DOI] [PubMed] [Google Scholar]
- [73].Casteilla L, Planat-Benard V, Laharrague P, Cousin B. Adipose-derived stromal cells: Their identity and uses in clinical trials, an update. World journal of stem cells. 2011;3:25–33. doi: 10.4252/wjsc.v3.i4.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tran TT, Kahn CR. Transplantation of adipose tissue and stem cells: role in metabolism and disease. Nature reviews. Endocrinology. 2010;6:195–213. doi: 10.1038/nrendo.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Lemonnier D. Effect of age, sex, and site on cellularity of adipose-tissue in mice and rats rendered obese by a high-fat diet. Journal of Clinical Investigation. 1972;51:2907–&. doi: 10.1172/JCI107115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Klyde BJ, Hirsch J. Increased cellular proliferation in adipose-tissue of adult-rats fed a high-fat diet. Journal of Lipid Research. 1979;20:705–715. [PubMed] [Google Scholar]
- [77].Hausman DB, DiGirolamo M, Bartness TJ, Hausman GJ, Martin RJ. The biology of white adipocyte proliferation. Obesity reviews: an official journal of the International Association for the Study of Obesity. 2001;2:239–254. doi: 10.1046/j.1467-789x.2001.00042.x. [DOI] [PubMed] [Google Scholar]
- [78].Tchoukalova YD, Votruba SB, Tchkonia T, Giorgadze N, Kirkland JL, Jensen MD. Regional differences in cellular mechanisms of adipose tissue gain with overfeeding. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18226–18231. doi: 10.1073/pnas.1005259107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, Bergmann O, Blomqvist L, Hoffstedt J, Naslund E, Britton T, Concha H, Hassan M, Ryden M, Frisen J, Arner P. Dynamics of fat cell turnover in humans. Nature. 2008;453:783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- [80].Rodeheffer MS, Birsoy K, Friedman JM. Identification of White Adipocyte Progenitor Cells In Vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- [81].Joe AWB, Yi L, Even Y, Vogl AW, Rossi FMV. Depot-Specific Differences in Adipogenic Progenitor Abundance and Proliferative Response to High-Fat Diet. Stem Cells. 2009;27:2563–2570. doi: 10.1002/stem.190. [DOI] [PubMed] [Google Scholar]
- [82].Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Sanchez-Gurmaches J, Hung C-M, Sparks CA, Tang Y, Li H, Guertin DA. PTEN Loss in the Myf5 Lineage Redistributes Body Fat and Reveals Subsets of White Adipocytes that Arise from Myf5 Precursors. Cell Metabolism. 2012;16:348–362. doi: 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Joe AWB, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FMV. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nature Cell Biology. 2010;12:153–U144. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Macotela Y, Emanuelli B, Mori MA, Gesta S, Schulz TJ, Tseng Y-H, Kahn CR. Intrinsic Differences in Adipocyte Precursor Cells From Different White Fat Depots. Diabetes. 2012;61:1691–1699. doi: 10.2337/db11-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Prunet-Marcassus B, Cousin B, Caton D, Andre M, Penicaud L, Casteilla L. From heterogeneity to plasticity in adipose tissues: Site-specific differences. Experimental Cell Research. 2006;312:727–736. doi: 10.1016/j.yexcr.2005.11.021. [DOI] [PubMed] [Google Scholar]
- [87].Baglioni S, Cantini G, Poli G, Francalanci M, Squecco R, Di Franco A, Borgogni E, Frontera S, Nesi G, Liotta F, Lucchese M, Perigli G, Francini F, Forti G, Serio M, Luconi M. Functional Differences in Visceral and Subcutaneous Fat Pads Originate from Differences in the Adipose Stem Cell. Plos One. 2012;7 doi: 10.1371/journal.pone.0036569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619–U187. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Billon N, Dani C. Developmental Origins of the Adipocyte Lineage: New Insights from Genetics and Genomics Studies. Stem Cell Reviews and Reports. 2012;8:55–66. doi: 10.1007/s12015-011-9242-x. [DOI] [PubMed] [Google Scholar]
- [90].Majka SM, Barak Y, Klemm DJ. Concise review: adipocyte origins: weighing the possibilities. Stem Cells. 2011;29:1034–1040. doi: 10.1002/stem.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cinti S, Cigolini M, Bosello O, Bjorntorp P. A morphological-study of the adipocyte precursor. Journal of Submicroscopic Cytology and Pathology. 1984;16:243–251. [PubMed] [Google Scholar]
- [93].Barnard T. Ultrastructural differentiation of brown adipose tissue in rat. Journal of Ultrastructure Research. 1969;29:311–&. doi: 10.1016/s0022-5320(69)90109-9. [DOI] [PubMed] [Google Scholar]
- [94].Napolitano L. The differentiation of white adipose cells. An electron microscope study. The Journal of cell biology. 1963;18:663–679. doi: 10.1083/jcb.18.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Gupta RK, Mepani RJ, Kleiner S, Lo JC, Khandekar MJ, Cohen P, Frontini A, Bhowmick DC, Ye L, Cinti S, Spiegelman BM. Zfp423 Expression Identifies Committed Preadipocytes and Localizes to Adipose Endothelial and Perivascular Cells. Cell Metabolism. 2012;15:230–239. doi: 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Alva JA, Zovein AC, Monvoisin A, Murphy T, Salazar A, Harvey NL, Carmeliet P, Iruela-Arispe ML. VE-cadherin-Cre-recombinase transgenic mouse: A tool for lineage analysis and gene deletion in endothelial cells. Developmental Dynamics. 2006;235:759–767. doi: 10.1002/dvdy.20643. [DOI] [PubMed] [Google Scholar]
- [97].Monvoisin A, Alva JA, Hofmann JJ, Zovein AC, Lane TF, Iruela-Arispe ML. VE-cadherin-CreER(T2) transgenic mouse: A model for inducible recombination in the endothelium. Developmental Dynamics. 2006;235:3413–3422. doi: 10.1002/dvdy.20982. [DOI] [PubMed] [Google Scholar]
- [98].Khanh-Van T, Gealekman O, Frontini A, Zingaretti MC, Morroni M, Giordano A, Smorlesi A, Perugini J, De Matteis R, Sbarbati A, Corvera S, Cinti S. The Vascular Endothelium of the Adipose Tissue Gives Rise to Both White and Brown Fat Cells. Cell Metabolism. 2012;15:222–229. doi: 10.1016/j.cmet.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Kanda T, Brown JD, Orasanu G, Vogel S, Gonzalez FJ, Sartoretto J, Michel T, Plutzky J. PPARgamma in the endothelium regulates metabolic responses to high-fat diet in mice. The Journal of clinical investigation. 2009;119:110–124. doi: 10.1172/JCI36233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nat Cell Biol. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Kamal D, Breton P, Bouletreau P. Congenital infiltrating lipomatosis of the face: Report of three cases and review of the literature. Journal of Cranio-Maxillofacial Surgery. 2010;38:610–614. doi: 10.1016/j.jcms.2010.02.014. [DOI] [PubMed] [Google Scholar]
- [102].Slavin SA, Baker DC, McCarthy JG, Mufarrij A. Congenital infiltrating lipomatosis of the face - Clinicopathologic evaluation and treatment. Plastic and Reconstructive Surgery. 1983;72:158–164. doi: 10.1097/00006534-198308000-00006. [DOI] [PubMed] [Google Scholar]
- [103].Billon N, Iannarelli P, Monteiro MC, Glavieux-Pardanaud C, Richardson WD, Kessaris N, Dani C, Dupin E. The generation of adipocytes by the neural crest. Development. 2007;134:2283–2292. doi: 10.1242/dev.002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Lemos DR, Paylor B, Chang C, Sampaio A, Underhill TM, Rossi FMV. Functionally Convergent White Adipogenic Progenitors of Different Lineages Participate in a Diffused System Supporting Tissue Regeneration. Stem Cells. 2012;30:1152–1162. doi: 10.1002/stem.1082. [DOI] [PubMed] [Google Scholar]
- [105].Bronnerfraser M. Neural crest cell-formation and migration in the developing embryo. Faseb Journal. 1994;8:699–706. doi: 10.1096/fasebj.8.10.8050668. [DOI] [PubMed] [Google Scholar]
- [106].Crossno JT, Jr., Majka SM, Grazia T, Gill RG, Klemm DJ. Rosiglitazone promotes development of a novel adipocyte population from bone marrow-derived circulating progenitor cells. The Journal of clinical investigation. 2006;116:3220–3228. doi: 10.1172/JCI28510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Tomiyama K, Murase N, Stolz DB, Toyokawa H, O’Donnell DR, Smith DM, Dudas JR, Rubin JP, Marra KG. Characterization of transplanted green fluorescent protein+ bone marrow cells into adipose tissue. Stem Cells. 2008;26:330–338. doi: 10.1634/stemcells.2007-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Sera Y, LaRue AC, Moussa O, Mehrotra M, Duncan JD, Williams CR, Nishimoto E, Schulte BA, Watson PM, Watson DK, Ogawa M. Hematopoietic stem cell origin of adipocytes. Experimental hematology. 2009;37:1108–1120. 1120 e1101–1104. doi: 10.1016/j.exphem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Camargo FD, Green R, Capetanaki Y, Jackson KA, Goodell MA. Single hematopoietic stem cells generate skeletal muscle through myeloid intermediates. Nat Med. 2003;9:1520–1527. doi: 10.1038/nm963. [DOI] [PubMed] [Google Scholar]
- [110].Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic research. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- [111].Majka SM, Fox KE, Psilas JC, Helm KM, Childs CR, Acosta AS, Janssen RC, Friedman JE, Woessner BT, Shade TR, Varella-Garcia M, Klemm DJ. De novo generation of white adipocytes from the myeloid lineage via mesenchymal intermediates is age, adipose depot, and gender specific. Proc Natl Acad Sci U S A. 2010;107:14781–14786. doi: 10.1073/pnas.1003512107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. European journal of immunology. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- [113].Sgaier SK, Millet S, Villanueva MP, Berenshteyn F, Song C, Joyner AL. Morphogenetic and cellular movements that shape the mouse cerebellum: Insights from genetic fate mapping. Neuron. 2005;45:27–40. doi: 10.1016/j.neuron.2004.12.021. [DOI] [PubMed] [Google Scholar]
- [114].Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Developmental Biology. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- [115].Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, Nedergaard J, Cannon B. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–U927. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Tallquist MD, Weismann KE, Hellstrom M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- [118].Komatsu N, Oda T, Muramatsu T. Involvement of both caspase-like proteases and serine proteases in apoptotic cell death induced by ricin, modeccin, diphtheria toxin, and Pseudomonas toxin. Journal of Biochemistry. 1998;124:1038–1044. doi: 10.1093/oxfordjournals.jbchem.a022197. [DOI] [PubMed] [Google Scholar]
- [119].Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development. 2008;135:1597–1604. doi: 10.1242/dev.019331. [DOI] [PubMed] [Google Scholar]
- [120].Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nature Reviews Molecular Cell Biology. 2011;12:349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- [121].Lepper C, Conway SJ, Fan C-M. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–U694. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Lepper C, Fan C-M. Inducible Lineage Tracing of Pax7-Descendant Cells Reveals Embryonic Origin of Adult Satellite Cells. Genesis. 2010;48:424–436. doi: 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Lang D, Lu MM, Huang L, Engleka KA, Zhang MZ, Chu EY, Lipner S, Skoultchi A, Millar SE, Epstein JA. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433:884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- [124].Liu W, Liu Y, Lai X, Kuang S. Intramuscular adipose is derived from a non-Pax3 lineage and required for efficient regeneration of skeletal muscles. Developmental Biology. 2012;361:27–38. doi: 10.1016/j.ydbio.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Carracedo A, Alimonti A, Pandolfi PP. PTEN Level in Tumor Suppression: How Much Is Too Little? Cancer Research. 2011;71:629–633. doi: 10.1158/0008-5472.CAN-10-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Jinno H, Morozova O, Jones KL, Biernaskie JA, Paris M, Hosokawa R, Rudnicki MA, Chai Y, Rossi F, Marra MA, Miller FD. Convergent Genesis of an Adult Neural Crest-Like Dermal Stem Cell from Distinct Developmental Origins. Stem Cells. 2010;28:2027–2040. doi: 10.1002/stem.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Haldar M, Karan G, Tvrdik P, Capecchi MR. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev Cell. 2008;14:437–445. doi: 10.1016/j.devcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Daubas P, Tajbakhsh S, Hadchouel J, Primig M, Buckingham M. Myf5 is a novel early axonal marker in the mouse brain and is subjected to post-transcriptional regulation in neurons. Development. 2000;127:319–331. doi: 10.1242/dev.127.2.319. [DOI] [PubMed] [Google Scholar]
- [129].Misra A, Peethambaram A, Garg A. Clinical features and metabolic and autoimmune derangements in acquired partial lipodystrophy: report of 35 cases and review of the literature. Medicine. 2004;83:18–34. doi: 10.1097/01.md.0000111061.69212.59. [DOI] [PubMed] [Google Scholar]
- [130].Mallewa JE, Wilkins E, Vilar J, Mallewa M, Doran D, Back D, Pirmohamed M. HIV-associated lipodystrophy: a review of underlying mechanisms and therapeutic options. The Journal of antimicrobial chemotherapy. 2008;62:648–660. doi: 10.1093/jac/dkn251. [DOI] [PubMed] [Google Scholar]
- [131].Vegiopoulos A, Muller-Decker K, Strzoda D, Schmitt I, Chichelnitskiy E, Ostertag A, Berriel Diaz M, Rozman J, Hrabe de Angelis M, Nusing RM, Meyer CW, Wahli W, Klingenspor M, Herzig S. Cyclooxygenase-2 controls energy homeostasis in mice by de novo recruitment of brown adipocytes. Science. 2010;328:1158–1161. doi: 10.1126/science.1186034. [DOI] [PubMed] [Google Scholar]
- [132].Artemenko Y, Gagnon A, Aubin D, Sorisky A. Anti-adipogenic effect of PDGF is reversed by PKC inhibition. Journal of cellular physiology. 2005;204:646–653. doi: 10.1002/jcp.20314. [DOI] [PubMed] [Google Scholar]
- [133].Olson LE, Soriano P. PDGFRbeta signaling regulates mural cell plasticity and inhibits fat development. Dev Cell. 2011;20:815–826. doi: 10.1016/j.devcel.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Goodpaster BH, Wolf D. Skeletal muscle lipid accumulation in obesity, insulin resistance, and type 2 diabetes. Pediatric Diabetes. 2004;5:219–226. doi: 10.1111/j.1399-543X.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- [135].Greco AV, Mingrone G, Giancaterini A, Manco M, Morroni M, Cinti S, Granzotto M, Vettor R, Camastra S, Ferrannini E. Insulin resistance in morbid obesity - Reversal with intramyocellular fat depletion. Diabetes. 2002;51:144–151. doi: 10.2337/diabetes.51.1.144. [DOI] [PubMed] [Google Scholar]
- [136].Uezumi A, Fukada S.-i., Yamamoto N, Takeda S.i., Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nature Cell Biology. 2010;12:143–U116. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- [137].Almind K, Manieri M, Sivitz WI, Cinti S, Kahn CR. Ectopic brown adipose tissue in muscle provides a mechanism for differences in risk of metabolic syndrome in mice. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2366–2371. doi: 10.1073/pnas.0610416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Birsoy K, Berry R, Wang T, Ceyhan O, Tavazoie S, Friedman JM, Rodeheffer MS. Analysis of gene networks in white adipose tissue development reveals a role for ETS2 in adipogenesis. Development. 2011;138:4709–4719. doi: 10.1242/dev.067710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Matsuoka T, Ahlberg PE, Kessaris N, Iannarelli P, Dennehy U, Richardson WD, McMahon AP, Koentges G. Neural crest origins of the neck and shoulder. Nature. 2005;436:347–355. doi: 10.1038/nature03837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Foo SS, Turner CJ, Adams S, Compagni A, Aubyn D, Kogata N, Lindblom P, Shani M, Zicha D, Adams RH. Ephrin-B2 controls cell motility and adhesion during blood-vessel-wall assembly. Cell. 2006;124:161–173. doi: 10.1016/j.cell.2005.10.034. [DOI] [PubMed] [Google Scholar]
- [141].Chai Y, Jiang XB, Ito Y, Bringas P, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- [142].Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- [143].Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL. The transcriptome of retinal Muller glial cells. The Journal of comparative neurology. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 2000;14:1377–1389. [PMC free article] [PubMed] [Google Scholar]
- [145].Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nature Neuroscience. 2008;11:1392–1401. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]