Abstract
NMR structural studies of large monomeric and multimeric proteins face distinct challenges. In large monomeric proteins, the common occurrence of frequency degeneracies between residues impedes unambiguous assignment of NMR signals. To overcome this barrier, non-uniform sampling is used to measure spectra with optimal resolution within reasonable time, new correlation maps resolve previous impasses in assignment strategies, and novel selective methyl labeling schemes provide additional structural probes without cluttering NMR spectra. These advances push the limits of NMR studies of large monomeric proteins. Large multimeric and multi-domain proteins are studied by NMR when individual components can also be studied by NMR and have known structures. The structural properties of large assemblies are obtained by identifying binding surfaces, by orienting domains, and employing limited distance constraints. Segmental labeling and the combination of NMR with other methods have helped popularise NMR studies of such systems.
Introduction
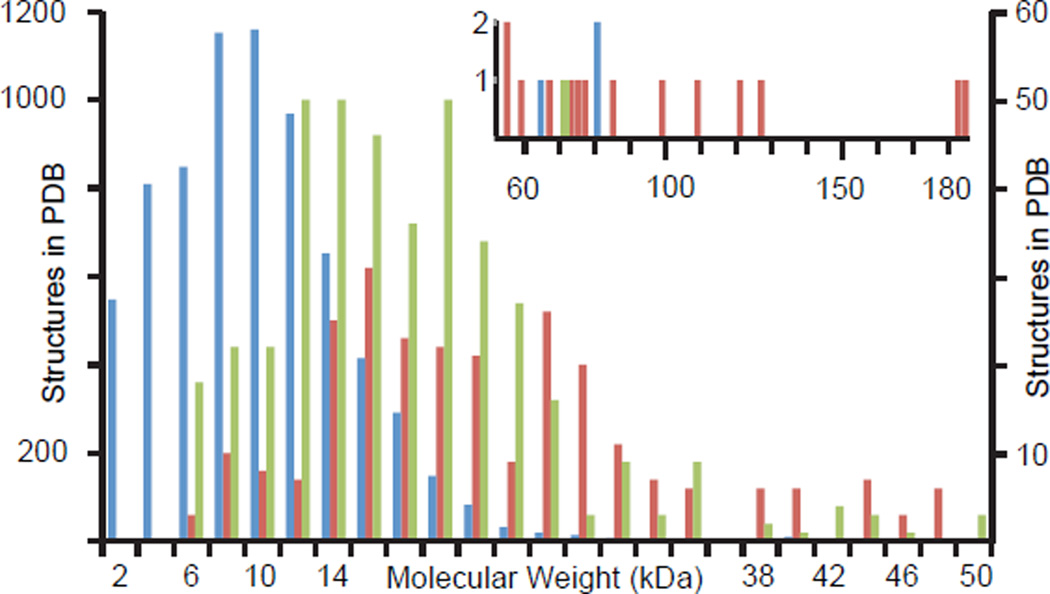
Since its inception nuclear magnetic resonance (NMR) has evolved from a technique devoted to chemical analysis to a powerful and versatile tool for biological studies. Notably, NMR provides structural models of proteins in near physiological conditions and thus offers a complementary alternative to crystallographic studies. Functional NMR studies further contribute to the popularity of the method; NMR can probe protein dynamics, kinetics, and thermodynamics all at atomic resolution. This versatility results from the power of NMR correlation maps that simultaneously report on targeted molecular properties and correlate various nuclei in the protein. Unfortunately, the method has historically been limited to proteins smaller than 25 kDa. The two major obstacles to a universal application of NMR are sensitivity losses and increased spectral complexity, both of which are pronounced in large proteins. The loss in sensitivity originates from NMR relaxation, the process by which the NMR spin systems return to their equilibrium state. In particular, transverse relaxation leads to concomitant losses in signal intensities and signal line-broadening. In a major breakthrough, so-called TROSY (transverse relaxation optimised spectroscopy) techniques have provided a means to combat these adverse effects when combined with isotope labeling [1,2]. Many NMR experiments were adapted to TROSY methods and structural studies of larger systems emerged. However, these studies have most often focused on multimeric proteins, as reflected by depositions in the PDB (Figure 1), highlighting the additional challenges to which large monomeric proteins are subject. As a consequence, this review covers NMR studies of monomeric and multimeric proteins separately, with an emphasis on NMR method development for monomeric proteins and on combining NMR and other biophysical methods for multimeric proteins.
Figure 1.
Molecular-weight distribution of NMR structures in the PDB. Blue: Monomeric proteins, scaled to left axis. Green: homomeric proteins, right axis. Red: heteromeric proteins, right axis. The number of structures in all categories drops off dramatically at molecular weights above 20 kDa reflecting difficulties associated with solving these structures. The inset graph shows the number of structures greater than 50 kDa. These structures are predominantly heteromeric, reflecting a strategy in which structures of each subunit are solved individually and combined to model a complex.
Structures of large single domain monomeric proteins
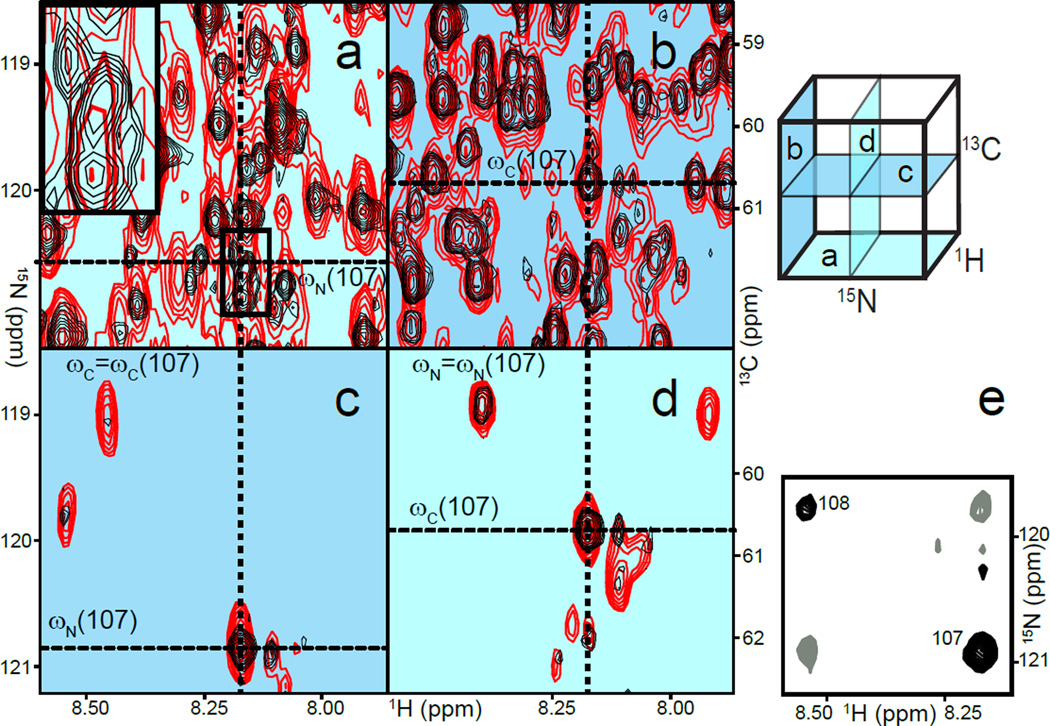
NMR structure determination necessitates successful assignment of NMR signals, which relies on resolving NMR signals in correlation maps. The abundance of NMR signals in larger proteins results in spectral crowding and higher resolution is needed to prevent signal overlap. Higher-magnetic fields and TROSY help decrease signal overlap, but the high experimental resolution needed to resolve signals requires impractical measurement times. The acquisition time needed for an N-dimensional correlation map scales as the product of the number of points in each indirect dimension. Maximal resolution for a 50 kDa protein at 900 MHz [3] necessitates 6h30 for a 2D HN-TROSY-HSQC spectrum and 28 days for a 3D HNCO (assuming a 1D spectrum takes 1 minute). Many strategies have been suggested to accelerate NMR data acquisition [4]. In particular, the measurement time can be shortened dramatically by recording a sparse subset of points in the indirect dimensions, a method called non-uniform sampling (NUS)[5]. Alternatives to Fourier transformation such as maximum entropy reconstruction (MaxEnt) or multidimensional decomposition (MDD) have been used to produce 3D and 4D spectra[6,7], but suffer from non-linearity in signal intensities or leak-through of adjacent signals in crowded spectra, respectively. Recent processing techniques provide a faithful reproduction of the spectrum and do not require human intervention for optimizing parameters. Forward Maximum Entropy[8] can reproduce crowded protein NOESY spectra without false cross-peaks and maintains linearity in signal amplitudes, but requires extensive processing time. Minimisation of l1-norm, paired with iterative soft threshold [8–11], and variations of compressed sensing [12,13] have been equally successful and benefit from fast processing time. Thus, NUS is now practical for routine acquisition of reliable multidimensional NMR spectra with optimal resolution, which alleviates partial overlap and facilitates unambiguous assignments in larger proteins as shown in Figure 2a-d.
Figure 2.
Overcoming frequency degeneracies in a 53 kDa monomeric protein. Upper right: cartoon representation of a 3D HNCA; the labels refer to panels a,b,c, and d. a) H/N projection of HNCA with uniform (red) and non-uniform sampling (NUS, black). The inset emphasises a region in which four correlations appear as a single signal in the uniform experiment (thick line) but are resolved in the NUS experiment. The cross-hairs denote the frequencies of residue 107. b) H/C projection of the same spectrum. c) H/N plane at the frequency ωC=ωC(107), as defined in panel b. Signals only seen in the uniform data result from a leak-through of adjacent planes due to poor resolution in the carbon dimension. d) The corresponding H/C plane further emphasises the advantage of an increased resolution in nitrogen since fewer signals accidentally appear to share the same nitrogen frequency. e) The corresponding H/N plane of the (H)NCA(N)H experiment allows for straightforward identification of sequential residues 107 and 108.
The multitude of atoms in larger proteins results in frequency degeneracies that lead to indistinguishable correlations along various dimensions of NMR spectra, and in these cases traditional NMR experiments fail to provide unambiguous assignment. Here, NUS can be used to design experiments that would otherwise not be feasible for larger proteins. For instance, the 3D double-TROSY (H)NCA(N)H [14] provides correlations between backbone atoms of sequential residues along all three dimensions of the spectrum, 1H,15N and 13C. Other experiments only provide sequential correlations along a single dimension. As a consequence, a tedious pairwise comparison of correlations can be replaced with visual identification of sequential cross-peaks akin to NOESY cross-peaks (Figure 2e). Here, NUS was used to rescue sensitivity by increasing the number of NMR transients accumulated whilst maintaining high-resolution in all dimensions. The application of NUS to improve sensitivity has recently been discussed in detail[15]. For 4D backbone experiments[16], NUS can overcome severe limitations in resolution that are otherwise needed to record spectra in reasonable time. Without NUS, low resolution induces overlap in each dimension and offsets the benefits of separating signals along additional dimensions.
Another impediment to determining structures of larger proteins is the scarcity of structural constraints. Recent advances have attenuated this limitation: torsion angles can be predicted from assigned backbone resonances with increased reliability [17] and novel experiments are available to measure residual dipolar couplings and provide bond orientations in larger proteins [18–20]. Distance constraints between protons are nevertheless necessary for de novo structure determinations; however, to minimise relaxation, larger proteins are predominantly deuterated, severely reducing the number of distance constraints from the outset. A protocol introduced by Kay and coworkers, which combines methyl-TROSY[2] with selective protonation of Ile (δ), Leu, and Val methyls[21], paved the way for overcoming this limitation. Other methyl groups can now be used as probes: Met [22–25], Ala [26,27], Ile (γ2) [28], and Thr methyls [29–31] can all be selectively labeled. Novel experiments have been designed to assign the methyl resonances of Ala and Ile(γ2) methyls[32]. Met and Thr methyls are assigned by using NOESY or mutagenesis and comparing the resulting spectra. Thus, all methyls that can be found in a protein can be selectively labeled to provide distance constraints.
Even with the accumulation of distance probes in selectively labeled large proteins it remains critical to maximise the assignment of nOe correlations to compensate for the loss of distance constraints when compared to protonated samples. Indeed, the accuracy and precision of NMR structures depends on the number of constraints and on their spatial distribution throughout the molecule; discarding a single constraint may have dramatic consequences for sparsely labeled samples. Non-uniform sampling can be combined with time-shared acquisition, which simultaneously provides several correlation maps with minimal impact on signal to noise[33,34], to measure both 15N and 13C edited 3D NOESY at once. In larger, selectively methyl labeled proteins, all distances involving either methyl or amide protons or both can be obtained with a single experiment that benefits from TROSY [35–38]. Likewise, a time-shared 4D experiment provides four 4D spectra involving methyl and amide protons in a single acquisition[38]. Recent progress in processing NUS data enables its implementation in time-shared NOESY experiments; together, the methods minimise the number of sacrificed constraints and increase the accuracy and precision of NMR structures of large proteins.
The methods described in this section render structure determination by NMR tractable for proteins up to 80 kDa. The exact limit depends on the protein structural and dynamic properties and on its solubility, which will affect the quality of NMR spectra. Fortunately, these effects can be identified upfront with a simple 2D HN-TROSY-HSQC and the sample design, as well as the buffer, can be modified to improve the spectral quality. In the end, recent developments should help structure determinations of large monomeric proteins become more common, although a certain degree of expertise is still required.
Structures of supra-molecular assemblies
The advent of TROSY techniques has readily provided access to studies of large homomeric proteins. The 900 kDa complex between tetradecameric GroEL and heptameric GroES was studied by amide proton and nitrogen HN-correlation maps[39] and the 670 kDa α7β7β7α7 core particle of the proteasome was studied by methyl spectra [40]. In both cases a divide and conquer approach was central in assigning the resonances. The monomeric units were assigned using conventional 3D or 4D HN-TROSY strategies and comparison with 2D correlation maps of the homomeric protein allowed transfer of the assignments. For GroEL/GroES, HN-correlation maps were used, and only resonances that did not undergo extensive shifts upon oligomerization were considered during subsequent analysis. In the proteasome studies, 3D experiments correlating methyl and beta or gamma carbons were also performed to assign signals that differ in monomer and multimer spectra; near complete assignment of the methyls in the proteasome core particle was achieved. More recent investigations of the folded states of protein substrates inside the proteasome brilliantly illustrated the biological impact of such methods [41].
Heteromeric systems can be studied with a similar divide and conquer strategy but are subject to further limitations. Symmetric homomers benefit from a signal accumulation of their individual units that heteromers lack; a 100 µM sample of a heptamer corresponds to an effective concentration of 700 µM in monomer units. In addition more resonances are likely to be affected by complex formation in heteromers when binding partners are different, thus reducing the number of resonance assignments that can be transferred from isolated monomeric units. For these reasons, the size of the systems that can be studied by NMR is somewhat smaller for heteromeric than for homomeric systems and many techniques need to be combined including non NMR methods[42]. In such cases, NMR is used to orient and position the components of the heteromer with respect to one another. Contacts between domains are identified by chemical shift perturbation[43], saturation transfer[44], or paramagnetic relaxation enhancement (PRE)[45], while domain orientations can be determined either by residual dipolar couplings[46–48] or relaxation analysis[49,50]. PRE was instrumental in determining the solution structure of a 40 kDa di-domain ubiquitin receptor associated with the proteasome, in which a flexible linker prevented traditional structure determination [51]. Likewise, in studies of protein ubiquitination, PRE could demonstrate that a ubiquitin-ligated E2 shifted to a tighter complex upon binding with its E3[52]. RDCs were used to orient two proteins and one RNA fragment of a ternary complex involved in mRNA processing, and PRE resolved the degeneracies in relative unit orientations[53]. Small (or wide) angle X-ray scattering has been particularly useful for determining quaternary models in conjunction with NMR54]. Chemical shift perturbation, RDCs, PRE, and 15N relaxation have all been used with SAXS, notably for structures with disordered regions [55–58]. CryoEM, X-ray crystallography, mass-spectrometry, and other techniques can all be combined with NMR to provide molecular models of heteromers[59]. These strategies can be extended to multi-domain proteins by using segmental labeling[60]. Each domain can be produced with a different isotopic composition and ligated subsequently either in vitro, when all domains are stable, or in-vivo with sequential induction of protein expression concomitant with alteration of growth media. The development of novel methods of ligation, e.g. by using sortases [61], should help increase the chances of successful segmental labeling for a given system.
Conclusions
The last decade has been relatively rich in NMR studies of larger proteins, and many biological studies have benefited from the unique versatility provided by the method. While the focus of this review has been on structural studies, an important observation emerges from inspecting the examples presented. NMR structural investigations of larger proteins are rarely pursued without accompanying studies exploiting other applications of NMR, such as defining ligand affinity or studying protein dynamics. This observation emphasises the rationale for utilizing NMR in studying larger proteins: the objective is not to compete with X-ray crystallography, which is much more suitable for high-resolution structures, but rather to use NMR for systems in which other functional studies are needed. NMR has the unique advantage of providing an atomic level read out. In the extreme case of supramolecular assemblies (>500 kDa), the “structural” study in fact consists in probing for molecular interactions, including structural rearrangements or modulation of dynamics upon binding events. When a molecular model of heteromers or multidomain proteins needs to be determined in solution, e.g. in presence of transient interactions, NMR is often combined with other techniques to provide a structure. Again, the structural studies are most often accompanied by other NMR studies, such as protein dynamics. The versatility of NMR has been the major driving force for developing means of overcoming challenges in studies of large proteins and the methods described in this review should help promulgate its application to heretofore inaccessible systems.
Highlights.
Large monomeric proteins suffer from spectral crowding
Non-uniform sampling of 3D and 4D data optimises spectral resolution
Selective methyl labeling and tailored NMR experiments provide distance constraints
Large multimeric proteins are studied with divide and conquer approaches
Multimeric proteins studied by combining NMR methods with other techniques
Acknowledgements
We thank Bradley Harden, Ananya Majumdar, Albert Lau, Dan Leahy, and Gerhard Wagner for careful reading of the manuscript and insightful comments. Original work from our laboratory was funded with start-up funds from the Johns Hopkins School of Medicine. We acknowledge current funding from NIH (R01GM104257).
Abbreviations
- CryoEM
Cryo-electron microscopy
- HNCO
a 3D experiment correlating amide protons and nitrogens with carbonyl carbons
- HSQC
Heteronuclear Single-Quantum Coherence
- MaxEnt
A particular implementation of maximum entropy reconstruction with a modified entropy function used for processing NUS data.
- MDD
Multi-Dimensional Decomposition
- NOESY
Nuclear Overhauser Effect (nOe) Spectroscopy
- NUS
Non-uniform sampling
- PDB
Protein Data Bank
- PRE
Paramagnetic Relaxation Enhancement
- RDC
Residual Dipolar Couplings
- SAXS
Small Angle X-ray Scattering
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. Cross-correlated relaxation enhanced 1H[bond]13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc. 2003;125:10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]
- 3.Rovnyak D, Hoch JC, Stern AS, Wagner G. Resolution and sensitivity of high field nuclear magnetic resonance spectroscopy. J Biomol NMR. 2004;30:1–10. doi: 10.1023/B:JNMR.0000042946.04002.19. [DOI] [PubMed] [Google Scholar]
- 4.Isabella C, Felli BB. Recent Advances in Solution NMR: Fast Methods and Heteronuclear Direct Detection. Chemphyschem. 2009;9999 doi: 10.1002/cphc.200900133. NA. [DOI] [PubMed] [Google Scholar]
- 5.Barna JCJ, Laue ED, Mayger MR, Skilling J, Worrall SJP. Exponential sampling, an alternative method for sampling in two-dimensional NMR experiments. J Magn Reson. 1987;73:69–77. [Google Scholar]
- 6.Luan T, Jaravine V, Yee A, Arrowsmith CH, Orekhov VY. Optimization of resolution and sensitivity of 4D NOESY using multi-dimensional decomposition. J Biomol NMR. 2005;33:1–14. doi: 10.1007/s10858-005-1363-6. [DOI] [PubMed] [Google Scholar]
- 7.Rovnyak D, Frueh DP, Sastry M, Sun ZY, Stern AS, Hoch JC, Wagner G. Accelerated acquisition of high resolution triple-resonance spectra using non-uniform sampling and maximum entropy reconstruction. J Magn Reson. 2004;170:15–21. doi: 10.1016/j.jmr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 8.Hyberts SG, Frueh DP, Arthanari H, Wagner G. FM reconstruction of non-uniformly sampled protein NMR data at higher dimensions and optimization by distillation. J Biomol NMR. 2009;45:283–294. doi: 10.1007/s10858-009-9368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stern AS, Donoho DL, Hoch JC. NMR data processing using iterative thresholding and minimum l(1)-norm reconstruction. J. Magn. Reson. 2007;188:295–300. doi: 10.1016/j.jmr.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hyberts S, Arthanari H, Wagner G. Topics in Current Chemistry. Springer Berlin: Heidelberg; 2011. Applications of Non-Uniform Sampling and Processing; pp. 1–24. •This review provides an introduction to non-uniform sampling and analyses various factors influencing the reconstructed spectra. Forward Maximum Entropy is used as a read-out to evaluate various sampling strategies and analyse the effect of noise.
- 11.Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, Wagner G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR. 2012;52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holland DJ, Bostock MJ, Gladden LF, Nietlispach D. Fast multidimensional NMR spectroscopy using compressed sensing. Angew. Chem. 2011;50:6548–6551. doi: 10.1002/anie.201100440. •The first application of compressed sensing, together with ref 13, applied here to a 3D backbone experiment.
- 13. Kazimierczuk K, Orekhov VY. Accelerated NMR spectroscopy by using compressed sensing. Angew. Chem. 2011;50:5556–5559. doi: 10.1002/anie.201100370. •The first application of compressed sensing, together with ref 11, demonstrated on crowded NOESY spectra.
- 14.Frueh DP, Arthanari H, Koglin A, Walsh CT, Wagner G. A double TROSY hNCAnH experiment for efficient assignment of large and challenging proteins. J Am Chem Soc. 2009;131:12880–12881. doi: 10.1021/ja9046685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyberts SG, Robson SA, Wagner G. Exploring signal-to-noise ratio and sensitivity in non-uniformly sampled multi-dimensional NMR spectra. J. Biomol. NMR. 2013;55:167–178. doi: 10.1007/s10858-012-9698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tugarinov V, Hwang PM, Kay LE. Nuclear magnetic resonance spectroscopy of high-molecular-weight proteins. Annu Rev Biochem. 2004;73:107–146. doi: 10.1146/annurev.biochem.73.011303.074004. [DOI] [PubMed] [Google Scholar]
- 17.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharya A, Revington M, Zuiderweg ERP. Measurement and interpretation of 15N-1H residual dipolar couplings in larger proteins. J. Magn. Reson. 2010;203:11–28. doi: 10.1016/j.jmr.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arbogast L, Majumdar A, Tolman J. HNCO-based measurement of one-bond amide 15N-1H couplings with optimized precision. J. Biomol. NMR. 2010;46:175–189. doi: 10.1007/s10858-009-9391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Prestegard JH. Measurement of one and two bond N–C couplings in large proteins by TROSY-based J-modulation experiments. J. Magn. Reson. 2009;200:109–118. doi: 10.1016/j.jmr.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosen MK, Gardner KH, Willis RC, Parris WE, Pawson T, Kay LE. Selective Methyl Group Protonation of Perdeuterated Proteins. J. Mol. Biol. 1996;263:627–636. doi: 10.1006/jmbi.1996.0603. [DOI] [PubMed] [Google Scholar]
- 22.Fischer M, Kloiber K, Häusler J, Ledolter K, Konrat R, Schmid W. Synthesis of a 13C-Methyl-Group-Labeled Methionine Precursor as a Useful Tool for Simplifying Protein Structural Analysis by NMR Spectroscopy. ChemBioChem. 2007;8:610–612. doi: 10.1002/cbic.200600551. [DOI] [PubMed] [Google Scholar]
- 23.Stoffregen Mira C, Schwer Matthias M, Renschler Fabian A, Wiesner S. Methionine Scanning as an NMR Tool for Detecting and Analyzing Biomolecular Interaction Surfaces. Structure. 2012;20:573–581. doi: 10.1016/j.str.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Gelis I, Bonvin AMJJ, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG. Structural Basis for Signal-Sequence Recognition by the Translocase Motor SecA as Determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weininger U, Liu Z, McIntyre DD, Vogel HJ, Akke M. Specific 12CβD212CγD2S13CεHD2 Isotopomer Labeling of Methionine To Characterize Protein Dynamics by 1H and 13C NMR Relaxation Dispersion. J. Am. Chem. Soc. 2012;134:18562–18565. doi: 10.1021/ja309294u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isaacson RL, Simpson PJ, Liu M, Cota E, Zhang X, Freemont P, Matthews S. A New Labeling Method for Methyl Transverse Relaxation-Optimized Spectroscopy NMR Spectra of Alanine Residues. J. Am. Chem. Soc. 2007;129:15428–15429. doi: 10.1021/ja0761784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayala I, Sounier R, Usé N, Gans P, Boisbouvier J. An efficient protocol for the complete incorporation of methyl-protonated alanine in perdeuterated protein. J. Biomol. NMR. 2009;43:111–119. doi: 10.1007/s10858-008-9294-7. [DOI] [PubMed] [Google Scholar]
- 28.Ruschak AM, Velyvis A, Kay LE. A simple strategy for (1)(3)C, (1)H labeling at the Ile-gamma2 methyl position in highly deuterated proteins. J. Biomol. NMR. 2010;48:129–135. doi: 10.1007/s10858-010-9449-1. [DOI] [PubMed] [Google Scholar]
- 29.Takeda M, Jee J, Ono AM, Terauchi T, Kainosho M. Hydrogen Exchange Study on the Hydroxyl Groups of Serine and Threonine Residues in Proteins and Structure Refinement Using NOE Restraints with Polar Side-Chain Groups. J. Am. Chem. Soc. 2011;133:17420–17427. doi: 10.1021/ja206799v. [DOI] [PubMed] [Google Scholar]
- 30.Sinha K, Jen-Jacobson L, Rule GS. Specific Labeling of Threonine Methyl Groups for NMR Studies of Protein–Nucleic Acid Complexes. Biochemistry. 2011;50:10189–10191. doi: 10.1021/bi201496d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velyvis A, Ruschak AM, Kay LE. An Economical Method for Production of 2H,13CH3--Threonine for Solution NMR Studies of Large Protein Complexes: Application to the 670 kDa Proteasome. PLoS ONE. 2012;7:e43725. doi: 10.1371/journal.pone.0043725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheppard D, Guo C, Tugarinov V. Methyl-detected ‘out-and-back’ NMR experiments for simultaneous assignments of Alaβ and Ileγ2 methyl groups in large proteins. J. Biomol. NMR. 2009;43:229–238. doi: 10.1007/s10858-009-9305-3. [DOI] [PubMed] [Google Scholar]
- 33.Farmer B., II Simultaneous [13C,15N]-HMQC, A Pseudo-Triple Resonance Experiment. J Magn Reson. 1991;93:635–641. [Google Scholar]
- 34.Sattler M, Maurer M, Schleucher J, Griesinger C. A simultaneous 15N.1H and 13C,1H-HSQC with sensitivity enhancement and a heteronuclear gradient echo. J. Biomol. NMR. 1995;5:97–102. doi: 10.1007/BF00227475. [DOI] [PubMed] [Google Scholar]
- 35.Guo C, Tugarinov V. Identification of HN-methyl NOEs in large proteins using simultaneous amide-methyl TROSY-based detection. J Biomol NMR. 2009;43:21–30. doi: 10.1007/s10858-008-9285-8. [DOI] [PubMed] [Google Scholar]
- 36.Frueh DP, Leed A, Arthanari H, Koglin A, Walsh CT, Wagner G. Time-shared HSQC-NOESY for accurate distance constraints measured at high-field in (15)N-(13)C-ILV methyl labeled proteins. J Biomol NMR. 2009;45:311–318. doi: 10.1007/s10858-009-9372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wurtz P, Aitio O, Hellman M, Permi P. Simultaneous detection of amide and methyl correlations using a time shared NMR experiment: application to binding epitope mapping. J Biomol NMR. 2007;39:97–105. doi: 10.1007/s10858-007-9178-2. [DOI] [PubMed] [Google Scholar]
- 38.Frueh DP, Vosburg DA, Walsh CT, Wagner G. Determination of all nOes in 1H-13C-Me-ILV-U-2H-15N proteins with two time-shared experiments. J Biomol NMR. 2006;34:31–40. doi: 10.1007/s10858-005-5338-4. [DOI] [PubMed] [Google Scholar]
- 39.Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K. NMR analysis of a 900K GroEL GroES complex. Nature. 2002;418:207–211. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- 40.Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 41. Ruschak AM, Religa TL, Breuer S, Witt S, Kay LE. The proteasome antechamber maintains substrates in an unfolded state. Nature. 2010;467:868–871. doi: 10.1038/nature09444. •• A demonstration of both the power of methyl TROSY techniques and the impact of NMR in biological studies. The authors study how three protein substrates of the proteasome, with representative alpha helical or beta-sheet secondary structures, interact with the proteaseome chamber and are maintained in an unfolded state facilitating proteolysis.
- 42.Wang X, Lee H-W, Liu Y, Prestegard JH. Structural NMR of protein oligomers using hybrid methods. J Struct Biol. 2011;173:515–529. doi: 10.1016/j.jsb.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuiderweg ER. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 44.Nakanishi T, Miyazawa M, Sakakura M, Terasawa H, Takahashi H, Shimada I. Determination of the interface of a large protein complex by transferred cross-saturation measurements. J Mol Biol. 2002;318:245–249. doi: 10.1016/S0022-2836(02)00018-9. [DOI] [PubMed] [Google Scholar]
- 45.Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-oriented proteins: information for structure determination in solution. Proc Natl Acad Sci U S A. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 48.Fischer MW, Losonczi JA, Weaver JL, Prestegard JH. Domain orientation and dynamics in multidomain proteins from residual dipolar couplings. Biochemistry. 1999;38:9013–9022. doi: 10.1021/bi9905213. [DOI] [PubMed] [Google Scholar]
- 49.Bruschweiler R, Liao X, Wright PE. Long-range motional restrictions in a multidomain zinc-finger protein from anisotropic tumbling. Science. 1995;268:886–889. doi: 10.1126/science.7754375. [DOI] [PubMed] [Google Scholar]
- 50.Fushman D, Varadan R, Assfalg M, Walker O. Determining domain orientation in macromolecules by using spin-relaxation and residual dipolar coupling measurements. Progress in NMR Spectroscopy. 2004;44:189–214. [Google Scholar]
- 51.Chen X, Lee BH, Finley D, Walters KJ. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol. Cell. 2010;38:404–415. doi: 10.1016/j.molcel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. •Using paramagnetic relaxation enhancement, the authors showed that an E2 tethered to a ubiquitin shifts into a conformation in which E2 and ubiquitin are closer upon E3 binding to E2. They then show that mutations predicted to disrupt this interaction also impede transfer of ubiquitin.
- 53.Leeper TC, Qu X, Lu C, Moore C, Varani G. Novel protein-protein contacts facilitate mRNA 3'-processing signal recognition by Rna15 and Hrp1. J. Mol. Biol. 2010;401:334–349. doi: 10.1016/j.jmb.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Madl T, Gabel F, Sattler M. NMR and small-angle scattering-based structural analysis of protein complexes in solution. J Struct Biol. 2011;173:472–482. doi: 10.1016/j.jsb.2010.11.004. •An excelent review discussing the combination of SAXS and NMR data in structural investigations of biological systems.
- 55. Bernado P, Modig K, Grela P, Svergun DI, Tchorzewski M, Pons M, Akke M. Structure and Dynamics of Ribosomal Protein L12: An Ensemble Model Based on SAXS and NMR Relaxation. Biophys. J. 2010;98:2374–2382. doi: 10.1016/j.bpj.2010.02.012. ••While not applied to a very large system, a rare and excellent example of combining NMR relaxation data with SAXS. Ensembles models of the ribosomal protein L12 (18 kDa) were generated from 15N-relaxation data, which were further optimised based on agreement with SAXS data. The optimised ensembles were used to model domain motions.
- 56. Schwieters CD, Suh JY, Grishaev A, Ghirlando R, Takayama Y, Clore GM. Solution structure of the 128 kDa enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering. J. Am. Chem. Soc. 2010;132:13026–13045. doi: 10.1021/ja105485b. ••The solution structure of a 146 kDa dimer of heterodimers, (EI:HPr)2 is determined with a combination of NMR and SAXS. Previously available NMR- and X-ray-structures of individual domains were oriented with residual dipolar couplings, quaternary shape information was obtained from SAXS/WAXS data, and the oligomeric state was validated with analytical ultracentrifugation.
- 57.Dancheck B, Ragusa MJ, Allaire M, Nairn AC, Page R, Peti W. Molecular investigations of the structure and function of the protein phosphatase 1-spinophilin-inhibitor 2 heterotrimeric complex. Biochemistry. 2011;50:1238–1246. doi: 10.1021/bi101774g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mittag T, Marsh J, Grishaev A, Orlicky S, Lin H, Sicheri F, Tyers M, Forman-Kay JD. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure. 2010;18:494–506. doi: 10.1016/j.str.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Armache J-P, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. Proc Natl Acad Sci USA. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat. Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 61.Refaei M, Combs A, Kojetin D, Cavanagh J, Caperelli C, Rance M, Sapitro J, Tsang P. Observing selected domains in multi-domain proteins via sortase-mediated ligation and NMR spectroscopy. J. Biomol. NMR. 2011;49:3–7. doi: 10.1007/s10858-010-9464-2. [DOI] [PMC free article] [PubMed] [Google Scholar]