Abstract
We examined gene expression in the blood of six females with anorexia nervosa (AN) before and after weight restoration using RNAseq. AN cases (aged 19-39) completed clinical assessments and had blood drawn for RNA at hospital admission (T1, < ~75% ideal body weight, IBW) and again at discharge (T2, ≥ ~85% IBW). To examine the relationship between weight restoration and differential gene expression, normalized gene expression levels were analyzed using a paired design. We found 564 genes whose expression was nominally significantly different following weight restoration (p < 0.01, 231 increased and 333 decreased). With a more stringent significance threshold (false discovery rate q < 0.05), 67 genes met criteria for differential expression. Of the top 20 genes, CYP11A1, C16orf11, LINC00235, and CPA3 were down-regulated more than two-fold after weight restoration while multiple olfactory receptor genes (OR52J3, OR51L1, OR51A4, OR51A2) were up-regulated more than two-fold after weight restoration. Pathway analysis revealed up-regulation of two broad pathways with largely overlapping genes, one related to protein secretion and signaling and the other associated with defense response to bacterial regulation. Although results are preliminary secondary to a small sample size, these data provide initial evidence of transcriptional alterations during weight restoration in AN.
Keywords: biomarkers, renourishment, transcriptome, eating disorder, genetic, genome
1. Introduction
Anorexia nervosa (AN), a severe psychiatric illness marked by extremely low body weight, fear of weight gain, and inability to recognize the seriousness of the low weight, carries the highest mortality rate of any psychiatric disorder (Sullivan 1995; Zipfel et al., 2000; Birmingham et al., 2005; Millar et al., 2005; Papadopoulos et al., 2009). The DSM-IV-TR (American Psychiatric Association, 2000) differentiates two types of AN; the restricting type (AN-R) and the binge-purge type (AN-BP). Unlike individuals with AN-R, individuals with AN-BP regularly engage in binge eating and/or compensatory behaviors. As in other forms of starvation, AN is associated with biochemical, metabolic, immunologic, and sensory abnormalities (Mira et al., 1987; Umeki 1988; Nova et al., 2002; Mont and Castro, 2003; Millar et al., 2005; Ulger et al., 2006). Individuals with AN who are less than 75% ideal body weight (IBW) are typically hospitalized for medically supervised weight restoration (American Psychiatric Association, 2000). Inpatient treatment is costly (Krauth et al., 2002) and relapse is common (Carte et al., 2004). There are no robust biological indices of risk, illness severity, or treatment response, and the identification of such biomarkers is an urgent area of inquiry.
To be clinically useful, AN biomarkers must distinguish indices of starvation from indices of disease. Indeed, a number of metabolic, endocrine, and neural biomarkers attributed to malnutrition have been found in AN (e.g., alterations in neuropeptide Y, leptin, ghrelin, orexin A, corticotrophin-releasing hormone, cholecystokinin, pancreatic polypeptide beta-endorphin, and brain derived neurotrophic factor; Lob et al., 2003; Nakazato et al., 2003; Connan et al., 2007; Støving et al., 2009; Bronsky et al., 2011). Other biomarkers that have been reported to persist following renourishment in AN, and thus could represent an index of disease status, include abnormalities of the dopamine (Kaye et al., 1999; Bergen et al., 2005) and serotonin (Bailer et al., 2005; Galusca et al., 2008) neurotransmitter systems. The majority of such studies, however, tested a single candidate biomarker that is a part of a metabolic or neurochemical system believed to be related to the etiology of AN. This candidate approach has been unsuccessful in the vast majority of complex biomedical traits, suggesting that a more global and unbiased search is warranted.
Although the number of studies is limited, changes in the expression of candidate genes have been reported in individuals with AN (Frieling et al., 2008; Ehrlich et al., 2010). Ehrlich et al. (2010) used quantitative polymerase chain reaction (PCR) to examine proopiomelanocortin (POMC) splice variant levels in peripheral blood ribonucleic acid (RNA). The long POMC splice variant was higher in underweight women with AN than weight-recovered individuals and healthy controls. Alterations in POMC expression were interpreted to be the result of malnutrition, rather than a persisting trait marker of AN. Similarly, Kahl et al. (2004) examined expression of tumor necrosis factor-α (TNF-α), interferon-γ (INF-γ), interleukin-6 (IL-6), and interleukin-10 (IL-10). An increase in TNF-α and IL-6 expression was found in individuals with AN at hospital admission compared with controls, and the expression of TNF-α remained significantly higher in those with AN who were weight restored, while IL-6 expression decreased. The authors concluded that TNF-α may contribute to metabolic abnormalities in AN even following weight restoration. Janas-Kozik et al. (2008) found that a leptin receptor transcript showed statistically significant differences between individuals with AN and controls. Several other investigations compared expression of P-glycoprotein (Storch et al., 2008) and prohormone preproenkephalin (Weiss et al., 2010), failed to find significant differences. Although limited in number, these studies have provided some preliminary information regarding transcriptomic profiling in AN. However, these investigations have been hampered by the study of small numbers of candidate genes and a lack of within-subjects longitudinal data.
Whole transcriptome expression profiling is a powerful, unbiased method capable of identifying genes and biological pathways correlated with a phenotypic trait or environmental perturbation. As such, gene expression levels can serve as a biomarker for a biomedically relevant state. Whole blood is an attractive tissue source for the identification of gene expression biomarkers related to AN for two main reasons. First, blood is easily accessible whereas the target organ of interest (brain) is not. Second, genes expression levels in peripheral blood are well correlated with multiple central nervous system tissues (median non-parametric correlation of 0.5; Sullivan et al., 2006), suggesting cross-tissue relevance for expression changes.
An important next step towards generating candidate biomarkers for further exploration in AN is to determine how transcriptome expression patterns change as patients gain weight. Given the profound physiological changes that occur with starvation and renourishment, as well as numerous reports from the animal literature citing transcriptomic changes secondary to starvation and caloric restriction (Narnaware and Peter, 2001; Drew et al., 2008; Dhahbi et al., 2012; Mitchell et al., 2012; Plank et al., 2012), we expected to see transcriptomic changes during renourishment in AN. Although weight gain is not the only index of recovery, it is an essential first step in treatment, and identifying differential expression between the acutely underweight and weight restored state is an essential component of our ultimate goal to differentiate biomarkers of starvation from biomarkers of disease. Thus, the purpose of this proof-of-concept investigation was to determine whether RNA-sequencing (RNAseq) interrogation of the peripheral blood transcriptome in AN can identify genes that differ before and after weight restoration. Specifically, this investigation tested for differential gene expression in the same individuals with AN at T1 (<~75% IBW) and T2 (≥ ~85% IBW).
2. Methods
2.1 Participants
Full methods and participant information are included in the Supplementary Material (Table S1). Briefly, the participants were six females ages 19-39 who met DSM-IV-TR (American Psychiatric Association, 2000) criteria for AN and were admitted for inpatient treatment. Diagnosis was verified with the Structured Clinical Interview for DSM-IV (SCID-I/P; First et al., 2002). Five participants met criteria for AN-R type and one participant met criteria for AN-BP type. This study was approved by the Biomedical Institutional Review Board at the University of North Carolina at Chapel Hill and all participants provided written informed consent.
2.2 Body Composition
Height (assessed at T1 only) and weight were assessed using a stadiometer and a calibrated digital scale. Body mass index (BMI) was calculated as the standard Quetelet index. Body fat percent was determined by dual x-ray absorptiometry (DXA). IBW and BMI varied somewhat across participants due to the complexities associated with hospital admission (meeting medical necessity criteria for inpatient) and discharge (variation due to insurance coverage and availability of step-down treatment options).
2.3 Laboratory
Venous blood samples were drawn twice: within one week of admission (T1, <~75% IBW) and at discharge (T2, ≥~85% IBW). The sample was obtained from each participant 2-3 hours following a morning meal. The timing of the blood draw was based on when it was feasible for the patients to leave the inpatient unit to participate in the study. One tube of blood was used to generate both a complete blood count (CBC) and measure a panel of metabolic analytes. A second tube of blood was drawn into PAXgene Blood RNA (PreAnalytiX, Qiagen BD, Valencia, CA) tubes to stabilize RNA and then frozen at −80°C until RNA extraction. RNA concentration was measured by fluorometry (Qubit 2.0 Fluorometer, Life Technologies Corp., Carlsbad, CA) and RNA quality was verified using a microfluidics platform (Bioanalyzer, Agilent Technologies, Santa Clara, CA).
2.4 RNA sequencing
Twelve RNA samples were sequenced (6 subjects × 2 time points/subject), using one microgram of total RNA as input. The 12 samples were randomized and sequencing libraries were prepared using the Illumina (San Diego, CA) TruSeq RNA Sample Preparation Kit v2 with 12 unique indexed adapters (AD001-AD012). Libraries were quantitated using fluorometry and all 12 samples were pooled at equimolar concentrations prior to sequencing, which helps to control for lane and batch effects (Leek et al., 2010). Two lanes of an Illumina HiSeq 2000 instrument were used to generate a total of 381 million 100 bp paired-end reads.
Following extensive quality control, we used a standard RNAseq alignment and analysis pipeline to examine differential expression (Oshlack et al., 2010). First, sequencing reads were mapped to the UCSC hg19 reference genome using Tophat2 (Trapnell et al., 2009). Second, mapped reads for each sample were summarized into gene-level expression counts. Third, the summarized data were normalized, and analyzed to detect differential gene expression. To test for differential expression, we summarized and aggregated reads at the gene-level using Rsamtools (http://bioconductor.org) by counting the number of reads overlapping each gene. Gene locations were defined using GenomeFeatures (http://www.bioconductor.org) and refGene information from UCSC genome browser (downloaded May, 2012).
Our study design had two explanatory factors: participants (six levels) and time (two levels per participant). Therefore, the study was a paired design in which participants were used as a blocking factor. Our main goal was to identify genes that were differentially expressed between the two time points. Finally, we performed pathway analysis to gain biological insight from the list of differentially expressed genes. In order to find pathways or functional categories that were over represented among differentially expressed genes, we examined the top 100 differentially expressed genes using the Database for Annotation, Visualization and Integrated Discovery (DAVID; Dennis et al., 2003).
3. Results
3.1 Laboratory
Results for the CBC and comprehensive metabolic panel for each participant at T1 and T2 can be found in Table S2. Laboratory values revealed limited abnormalities with no clear pattern across T1 and T2. Three individuals had a low white blood cell (WBC) count at T1 and two of these individuals had a low WBC count at T2. Four and three individuals had low absolute lymphocytes at T1 and T2, respectively. Only one individual had an elevated erythrocyte sedimentation rate (ESR) at T1 and her ESR was not available at T2. One individual had elevated Asparatate Amino Transferase (AST) at T1 but not at T2. Two individuals had elevated Alanine Amino Transferase (ALT) values at T1 but not at T2. No individuals had an abnormal total bilirubin value. Four individuals had abnormal lactate dehydrogenase (LDH) at T1, and one of these individuals had elevated LDH at T2.
3.2 Body Composition
BMI and adiposity (determined by DXA) for each participant at T1 and T2 are reported in Table S1. Weight restoration was verified in all six participants, with a mean increase of 3.2 BMI units (range 1.9 – 5.1).
3.2 Quality control of RNAseq data
We used multiple approaches to evaluate sequence quality and to confirm subject identify. We used FastQC to verify that the raw fastq files had no major issues (e.g., no prominent adaptor overrepresentation and similar read counts for samples between lanes). The alignment results showed that 77-93% of reads from each sample were properly mapped to the human genome (Figure S1). After mapping, we removed reads when the mate (paired-end) sequence was unmapped as well as PCR duplicates. To ensure that samples were correctly identified, we called SNPs from the RNAseq data and verified the consistent identity of all subjects across lanes and across time. Read counts for chromosome Y transcripts verified that all subjects were female. High-level descriptions of gene expression using hierarchical clustering (Figure S2) demonstrated that gene expression for the same individual at two time points clustered well.
3.3 Differential gene expression before and after renourishment
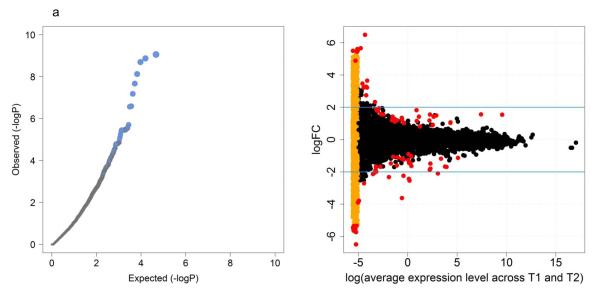
Using a paired design, we tested for differential expression before and after weight restoration, in a manner similar to computing a paired t-test for each gene but adapted for count data. Quantile-quantile plots of the p values (Figure 1a) indicated that a small subset of genes was differentially expressed. A total of 564 genes had unadjusted p-values < 0.01 between the two time points, and 67 had false discovery rate-adjusted q-values < 0.05. Table S3 summarizes the top 20 genes. The raw read counts for the top nine genes before and after weight restoration are shown in Figure S3. Four genes (CYP11A1, C16orf11, LINC00235, CPA3) were down-regulated more than two-fold after weight restoration, and four olfactory receptor genes (OR52J3, OR51L1, OR51A4, OR51A2) were up-regulated more than two-fold after renourishment. Figure 1b depicts, for every gene, the relationship between average expression level and fold change between T2 and T1. Transcripts with read counts of zero at T1 or T2 showed large fold-changes and statistical significance, but are likely to be false positives. To avoid overlooking biologically interesting genes, we manually checked their raw counts and annotation before removing them from the list of differentially expressed genes.
Figure 1.
QQ plot and visualization of differentially expressed genes.
Left panel (a): It shows a quantile-quantile (QQ) plot in which the expected distribution of −log10(p-value) is compared to the observed distribution. The gray area denotes 95% confidence bands for the expected distribution. The observed distribution deviates sharply from the expected, consistent with a small number of genes with differential expression. Right panel (b): The top 100 differentially expressed genes identified using gene-wise dispersion are highlighted in red. The orange points on the left represent transcripts with counts of zero at either T1 or T2. Blue lines denote log2FC of ±2. (FC is fold change.)
3.4 Correlation between gene expression and weight gain
In order to evaluate whether differential gene expression was secondary to increased body weight at T2, we examined the correlation between weight change and expression change across subjects. This analysis included each of the 231 up-regulated and 333 down-regulated genes (unadjusted p-value < 0.01) between T1 and T2. Only four up-regulated genes yielded correlations greater than 0.8, with the majority of genes showing no significant relationship (Figure S4). The majority of the 333 down-regulated genes also showed weak relationships between weight change and expression change (Figure S5) with, again, four genes yielding correlations greater than 0.8. The correlation between BMI change and expression change showed a similar pattern (data not shown).
3.5 Pathway analysis
To interpret the observed pattern of differential gene expression for biological relevance, we selected the top 100 most significant genes (48 down, 52 up) for functional clustering analysis via DAVID. We found modest evidence of enrichment in two broad pathways (Table 1). For these particular pathways, genes tended to be up-regulated with weight restoration.
Table 1.
Top two functional clusters from gene-set enrichment analysis of top 100 genes using DAVID.
| Annotation Cluster 1 | Enrichment Score: 4.62 | ||||||
|---|---|---|---|---|---|---|---|
| Category | Term | Count€ | PValue | Pop Hits |
Fold Enrichment |
FDR | Up vs. Down£ |
| SP_PIR_KEYWORDS | disulfide bond | 26 | 5.86E-08 | 2924 | 3.10975 | 7.01E-05 | 17/2 |
| UP_SEQ_FEATURE | disulfide bond | 25 | 1.57E-07 | 2819 | 3.081847 | 2.00E-04 | 17/8 |
| SP_PIR_KEYWORDS | signal | 22 | 9.75E-05 | 3250 | 2.367385 | 0.116642 | 14/8 |
| UP_SEQ_FEATURE | signal peptide | 22 | 1.07E-04 | 3250 | 2.352369 | 0.136868 | 14/8 |
| GOTERM_CC_FAT | GO:0005576~extracellular region |
16 | 9.13E-04 | 2010 | 2.422554 | 1.016557 | 13/3 |
| SP_PIR_KEYWORDS | Secreted | 13 | 0.002134 | 1689 | 2.691803 | 2.524341 | 10/3 |
| Annotation Cluster 2 | Enrichment Score: 2.58 | ||||||
| Category | Term | Count | PValue | Pop Hits |
Fold Enrichment |
FDR | Up vs. Down |
| GOTERM_BP_FAT | GO:0006952~defense response |
9 | 8.46E-04 | 615 | 4.303712 | 1.271136 | 7/2 |
| SP_PIR_KEYWORDS | antibiotic | 4 | 9.21E-04 | 68 | 20.57219 | 1.097168 | 4/0 |
| SP_PIR_KEYWORDS | Antimicrobial | 4 | 0.001044 | 71 | 19.70294 | 1.243026 | 4/0 |
| GOTERM_BP_FAT | GO:0042742~defense response to bacterium |
4 | 0.006089 | 112 | 10.50311 | 8.819647 | 4/0 |
| GOTERM_BP_FAT | GO:0009617~response to bacterium |
4 | 0.026167 | 193 | 6.095066 | 33.02563 | 4/0 |
number of genes in each functional category
Number of up-regulated genes versus down-regulated genes
3.6 Chromosome × inactivation skewing
We also used the RNAseq data to evaluate chromosome × inactivation skewing (Gribnau and Grootegoed 2012). Although the expectations are that the maternally and paternally inherited × chromosomes will be inactivated randomly, marked deviation from the expected 50:50 ratio can be observed and could conceivably track with weight restoration. We focused on chromosome × transcripts with read counts > 10 and with good evidence for the presence of heterozygous SNPs. Although we could not phase the alleles or differentiate maternal from paternal inheritance, we could track consistency over time. The expression levels of alternative alleles were similar between T1 and T2 in all samples combined (Figure S6) and for each participant (Figure S7). These results are consistent with random × inactivation and the absence of skewing during short-term weight restoration.
4. Discussion
Eight genes were up or down-regulated more than two-fold after renourishment in these patients with AN. Laboratory results revealed limited abnormalities that likely had minimal influence on differences in gene expression between T1 and T2. Expression of both up and down-regulated genes was only weakly correlated with changes in weight status, suggesting that subsequent studies with larger samples sizes may have the ability to detect biomarkers of illness rather than biomarkers that simply reflect changes in weight status. However, starvation and subsequent renourishment can impact every major organ system in the body (Fairburn and Harrison, 2003), leading to hormonal aberrations and dramatic effects on normal growth, bone turnover, and nutrient metabolism (Usdan et al., 2008). In addition, although normalization of biological parameters and improvement in health may be associated with renourishment, the timing of these improvements may not precisely coincide with improved weight status. In fact, studies have shown that normalization of many biological parameters including metabolism, bone mineral density, and reproductive function (Kaye et al., 1986; Delvenne et al., 1996; Brambilla et al., 2003; Mika et al., 2007) can take months or even years following weight restoration.
Although little is known regarding the function of the significantly down-regulated genes C16orf11, LINC00235 and CPA3, CPY11A1 encodes for cholesterol side-chain cleavage enzyme (CYPP450scc), a member of the cytochrome P450 (CYP450) super family of enzymes and the first and rate-limiting step for the synthesis of steroids (Tuckey, 2005). The xenobiotic-metabolizing P450 enzymes, such as CYP11A1, play a central role in the oxidative metabolism of a wide variety of endogenous and exogenous compounds including environmental carcinogens, plant toxins, and a wide range of pharmaceuticals (Gonzalez and Gelboin, 1994; Gut et al., 2000; Huang et al., 2000). CYP11A1 mRNA and protein have been found in several regions of the nervous system including the cerebellum, retina, and hippocampus; however, the transcriptional regulation and cellular and anatomical distribution of CYP11A1 in the brain have not been clearly determined (Wu et al., 2007; Chiang et al., 2011).
There are several possible explanations for the down-regulation of CYP11A1 following renourishment in AN. The expression of CYP450 enzymes is influenced by a variety of factors including drugs, hormones, and diet. There is some evidence to suggest that cytokines, which are involved in inflammation and relay information among cells of the immune system to other major systems (Raymond et al., 2000), may impact the transcriptional activation of the CYP11A1 gene. For example, many studies have shown down-regulation of various P450 mRNAs with inflammation (Turecki et al., 1997). During renourishment, weight gain is secondary to a disproportionate increase in adipose tissue (Morgan et al., 1999; Scalfi et al., 2002), which secretes numerous bioactive substances, including several proinflammatory cytokines, such as TNF-α and IL-6 (Hotamisligil, 2006; Tilg and Moschen, 2006). Thus, one plausible hypothesis is that there may be an association between an increase in proinflammatory cytokines, secondary to an increase in adipose tissue during renourishment, and the down-regulation of CYP11A1.
Another interesting finding was that four olfactory receptor genes (OR52J3, OR51L1, OR51A4, OR51A2), which are involved in the perception of smell, were up-regulated after renourishment. Genes encoding olfactory receptors represent the largest family of genes in the human genome, comprising approximately 3% (Choquette et al., 2012). Further, olfactory gene receptors have been found to have additional functions in the brain, associated with their direct anatomical connection to the limbic system (Ortega-Hernandez et al., 2009). Genetic variants in olfactory receptors have been associated with adiposity levels, cognitive dietary restraint, and susceptibility to hunger (Choquette et al., 2012). Interestingly, although findings have not been entirely consistent (Goldzak-Kunik et al., 2012; Schecklmann et al., 2012), several studies have also found olfactory deficits in individuals with AN (Fedoroff et al., 1995; Aschenbrenner et al., 2009; Rapps et al., 2010) and have proposed that these deficits may be associated with weight loss and food aversion observed in AN (Schecklmann et al., 2012). Further, several of these studies also found a significant positive correlation between olfactory sensitivities and BMI in AN (Aschenbrenner et al., 2009; Rapps et al., 2010). Thus, one hypothesis is that olfactory dysfunction in AN may improve with weight restoration and may be associated with up-regulation of olfactory genes, as seen in this investigation.
Up-regulation of two broad pathways, one related to protein secretion and signaling and the other associated with defense response to bacterial regulation, was also identified. One hypothesis is that these pathways may represent a return to improved immune function following weight restoration. This hypothesis is consistent with robust literature suggesting that inadequate nutrition can severely limit the immune system’s ability to fight infection, illness and disease (Marcos et al., 2003). However, individuals with AN are less prone to infection than individuals experiencing other forms of malnutrition (Silber and Chan, 1996) and studies on the impact of malnutrition secondary to AN on immune function have yielded conflicting findings (Nova et al., 2002; Brown et al., 2005). This hypothesis is also consistent with the finding in this investigation that CYP11A1, which may be related to inflammation and the immune response, is down-regulated following renourishment.
Limitations of this study should be considered. First, the sample size in this investigation was small. Thus, results should be considered preliminary pending additional investigations with larger samples. Second, this investigation included only Caucasian women between the ages of 19 and 39 and results might differ for men or for different ancestry and age groups. Third, this study did not take into account AN type. Although all participants, except one, met criteria for AN-R type, it is plausible that transcriptomic changes during renourishment may differ between AN types. Fourth, although every effort was made to control extraneous variables, due to the constraints of working with individuals undergoing inpatient treatment, all patients in this investigation did not experience the same time lapse or weight gain between T1 and T2, which may have impacted findings. Fifth, this investigation did not include healthy control participants, which would have provided important control information about transcriptomic changes overtime in unaffected individuals.
Despite these limitations, the genes and genetic pathways identified in this investigation warrant further exploration as potential candidates involved in recovery from AN. Identifying alterations in transcriptome expression before and after weight restoration in AN is of clinical and scientific importance. The long-term goal will be to identify abnormalities in the transcriptome profile of individuals with AN that are not a direct result of starvation, but rather indices of disease. Understanding of the biological processes that occur during acute weight restoration may also help elucidate biologically mediated aspects of the patients’ experience and provide novel information to inform how to improve the experience of renourishment which is both psychologically difficult and physically uncomfortable for patients with AN. For example, understanding transcriptomic changes of CYP11A1 during renourishment could have important implications for facilitating pharmacotherapy treatment and dosage in individuals acutely ill with AN. Similarly, understanding transcriptomic changes in olfactory genes during renourishment may generate ideas about buffering patients from “olfactory rebound” reducing their reactivity to smell cues when undergoing renourishment. An obvious empirical next step would be to evaluate actual changes in olfactory sensitivity over the course of renourishment.
Although preliminary, these data provide initial evidence of transcriptional alterations during weight restoration in AN and support further investigations with larger samples.
Supplementary Material
Acknowledgements
This project was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Award Number UL1TR000083. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Additional funding was from the Foundation of Hope, Raleigh, NC. Dr. Trace was supported by National Institute of Health grant T32MH076694 (PI: Bulik) and 2012 – 2015 Hilda and Preston Davis Foundation Postdoctoral Fellowship Program in Eating Disorders Research Award.
Footnotes
Financial Disclosures. Dr. Sullivan was on the SAB of Expression Analysis (Durham, NC). The other authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. 4th ed. Text Revision American Psychiatric Press; Washington, D.C.: 2000. [Google Scholar]
- Aschenbrenner K, Scholze N, Joraschky P, Hummel T. Gustatory and olfactory sensitivity in patients with anorexia and bulimia in the course of treatment. Journal of Psychiatric Research. 2008;43:129–137. doi: 10.1016/j.jpsychires.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Bailer UF, Frank GK, Henry SE, Price JC, Meltzer CC, Weissfeld L, Mathis CA, Drevets WC, Wagner A, Hoge J, Ziolko SK. Altered brain serotioni 5-HT1A receptor binding after recovery from anorexia nervosa measured by positron emission tomography and [carbonyl11C]WAY-100635. Archives of General Psychiatry. 2005;62:1032–1041. doi: 10.1001/archpsyc.62.9.1032. [DOI] [PubMed] [Google Scholar]
- Bergen AW, Yeager M, Welch RA, Haque K, Ganjei JK, van den Bree MB, Mazzanti C, Nardi I, Fichter MM, Halmi KA, Kaplan AS, Strober M, Treasure J, Woodside DB, Bulik CM, Bacanu SA, Devlin B, Berrettini WH, Goldman D, Kaye WH. Association of multiple DRD2 polymorphisms with anorexia nervosa. Neuropsychopharmacology. 2005;30:1703–1710. doi: 10.1038/sj.npp.1300719. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Su J, Hlynsky JA, Goldner EM, Gao M. The mortality rate from anorexia nervosa. International Journal of Eating Disorders. 2005;38:143–146. doi: 10.1002/eat.20164. [DOI] [PubMed] [Google Scholar]
- Brambilla F, Monteleone P, Bortolotti F, Dalle Grave R, Todisco P, Favaro A, Santonastaso P, Ramacciotti C, Paoli, Maj M. Persistent amenorrhoea in weight-recovered anorexics: psychological and biological aspects. Psychiatry Research. 2003;118:249–257. doi: 10.1016/s0165-1781(03)00074-x. [DOI] [PubMed] [Google Scholar]
- Bronsky J, Nedvidkova J, Krasnicanova H, Vesela M, Schmidtova J, Koutek J, Kellermayer R, Chada M, Kabelka Z, Hrdlicka M, Nevoral J, Prusa R. Changes of orexin A plasma levels in girls with anorexia nervosa during eight weeks of realimentation. International Journal of Eating Disorders. 2011;44:547–552. doi: 10.1002/eat.20857. [DOI] [PubMed] [Google Scholar]
- Brown RF, Bartrop R, Beumont P, Birmingham CL. Bacterial infections in anorexia nervosa: delayed recognition increases complications. International Journal of Eating Disorders. 2005;37:261–261. doi: 10.1002/eat.20135. [DOI] [PubMed] [Google Scholar]
- Carter JC, Blackmore E, Sutandar-Pinnock K, Woodside DB. Relapse in anorexia nervosa: a survival analysis. Psychological Medicine. 2004;34:671–619. doi: 10.1017/S0033291703001168. [DOI] [PubMed] [Google Scholar]
- Chiang YF, Lin HT, Hu JW, Tai YC, Lin YC, Hu MC. Differential regulation of the human CYP11A1 promoter in mouse brain and adrenals. Journal of Cellular Physiology. 2011;226:1998–2005. doi: 10.1002/jcp.22534. [DOI] [PubMed] [Google Scholar]
- Choquette AC, Bouchard L, Drapeau V, Lemieux S, Tremblay A, Bouchard C, Vohl MC, Pérusse L. Association between olfactory receptor genes, eating behavior and adiposity: results from the Quebec Family Study. Physiology and Behavior. 2012;105:772–776. doi: 10.1016/j.physbeh.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Connan F, Lightman SL, Landau S, Wheeler M, Treasure J, Campbell IC. An investigation of hypothalamic-pituitary-adrenal axis hyperactivity in anorexia nervosa: The role of CRH and AVP. Journal of Psychiatric Research. 2007;41:131–43. doi: 10.1016/j.jpsychires.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Delvenne V, Goldman S, De Maertelaer V, Simon Y, Luxen A, Lotstra F. Brain hypometabolism of glucose in anorexia nervosa: normalizaiton after weight gain. Biological Psychiatry. 1996;40:761–768. doi: 10.1016/0006-3223(95)00522-6. [DOI] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane LC, Lempicki RA. Database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4:3. [PubMed] [Google Scholar]
- Dhahbi JM, Atamna H, Boffelli D, Martin DI, Spindler SR. mRNA-Seq reveals complex patterns of gene regulation and expression in the mouse skeletal muscle transcriptome associated with calorie restriction. Physiological Genomics. 2012;44:331–44. doi: 10.1152/physiolgenomics.00129.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew RE, Rodnick KJ, Settles M, Wacyk J, Churchill E, Powell MS, Hardy RW, Murdoch GK, Hill RA, Robison BD. Effect of starvation on transcriptomes of brain and liver in adult female zebrafish (Danio rerio) Physiological Genomics. 2008;35:283–295. doi: 10.1152/physiolgenomics.90213.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S, Weiss D, Burghardt R, Infante-Duarte C, Brockhaus S, Muschler MA, Bleich S, Lehmkuhl U, Frieling H. Promoter specific DNA methylation and gene expression of POMC in acutely underweight and recovered patients with anorexia nervosa. Journal of Psychiatric Research. 2010;44:827–833. doi: 10.1016/j.jpsychires.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Fedoroff IC, Stoner SA, Andersen AE, Doty RL, Rolls BJ. Olfactory dysfunction in anorexia and bulimia. International Journal of Eating Disorders. 1995;18:71–77. doi: 10.1002/1098-108x(199507)18:1<71::aid-eat2260180108>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Fairburn CG, Harrison PJ. Eating disorders. Lancet. 2003;361:407–416. doi: 10.1016/S0140-6736(03)12378-1. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer R, Gibbon M, Williams JB. Structured Clinical Interview for DSM-IV-TR Axis I Disorders (SCID-I) Clinican Version American Psychiatric Press; Washington, D.C.: 2002. [Google Scholar]
- Frieling H, Bleich S, Otten J, Romer KD, Kornhuber J, de Zwaan M, Jacoby GE, Wilhelm J, Hillemacher T. Epigenetic downregulation of atrial natriuretic peptide but not vasopressin mRNA expression in females with eating disorders is related to impulsivity. Neuropsychopharmacology. 2008;33:2605–2609. doi: 10.1038/sj.npp.1301662. [DOI] [PubMed] [Google Scholar]
- Galusca B, Costes N, Zito NG, Peyron R, Bossu C, Lang F, Le Bars D, Estour B. Organic background of restrictive-type anorexia nervosa suggested by increased serotonin 1A receptor binding in right frontotemporal cortex of both lean and recovered patients: [18F]MPPF PET scan study. Biological Psychiatry. 2008;64:1009–1013. doi: 10.1016/j.biopsych.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Goldzak-Kunik G, Friedman R, Spitz M, Sandler L, Leshem M. Intact sensory function in anorexia nervosa. American Journal of Clinical Nutrition. 2012;95:272–282. doi: 10.3945/ajcn.111.020131. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Gelboin HV. Role of human cytochromes P450 in the metabolic activation of chemical carcinogens and toxins. Drug Metabolism Reviews. 1994;26:165–83. doi: 10.3109/03602539409029789. [DOI] [PubMed] [Google Scholar]
- Gribnau J, Grootegoed JA. Origin and evolution of × chromosome inactivation. Current Opinion in Cell Biology. 2012;24:397–404. doi: 10.1016/j.ceb.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Gut I, Danielova V, Holubova J, Soucek P, Kluckova H. Cytotoxicity of cyclophosphamide, paclitaxel, and docetaxel for tumor cell lines in vitro: effects of concentration, time and cytochrome P450-catalyzed metabolism. Archives of Toxicology. 2000;74:437–446. doi: 10.1007/s002040000141. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Huang Z, Roy P, Waxman DJ. Role of human liver microsomal CYP3A4 and CYP2B6 in catalyzing N-dechloroethylation of cyclophosphamide and ifosfamide. Biochemical Pharmacology. 2000;59:61–72. doi: 10.1016/s0006-2952(99)00410-4. [DOI] [PubMed] [Google Scholar]
- Janas-Kozik M, Stachowicz M, Mazurek U, Zajdel A, Wilczok A, Krupka-Matuszczyk I, Rybakowski JK. Preliminary study of the expression of genes connected with the orexigenic and anorexigenic system using microarray technique in anorexia nervosa. Neuropsychobiology. 2008;57:116–120. doi: 10.1159/000138913. [DOI] [PubMed] [Google Scholar]
- Kahl KG, Kruse N, Rieckmann P, Schmidt MH. Cytokine mRNA expression patterns in the disease course of female adolescents with anorexia nervosa. Psychoneuroendocrinology. 2004;29:13–20. doi: 10.1016/s0306-4530(02)00131-2. [DOI] [PubMed] [Google Scholar]
- Kaye WH, Gwirtsman H, George T, Ebert MH, Petersen R. Caloric consumption ativity levels after weight recovery in anorexia nervosa: a prolonged delay in normalization. International Journal of Eating Disorders. 1986;5:489–502. [Google Scholar]
- Kaye WH, Frank GK, McConaha C. Altered dopamine activity after recovery from restricting-type anorexia nervosa. Neuropsychopharmacology. 1999;21:503–506. doi: 10.1016/S0893-133X(99)00053-6. [DOI] [PubMed] [Google Scholar]
- Krauth C, Buser K, Vogel H. How high are the costs of eating disorders - anorexia nervosa and bulimia nervosa - for German society? The European Journal of Health Economics. 2002;3:244–250. doi: 10.1007/s10198-002-0137-2. [DOI] [PubMed] [Google Scholar]
- Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, Irizarry RA. Tackling the widespread and critical impact of batch effects in high-throughput data. Nature Reviews Genetics. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lob S, Pickel J, Bidlingmaier M, Schaaf L, Backmund H, Gerlinghoff M, Stalla GK. Serum leptin monitoring in anorectic patients during refeeding therapy. Experimental and Clinical Endocrinology and Diabetes. 2003;111:178–282. doi: 10.1055/s-2003-41286. [DOI] [PubMed] [Google Scholar]
- Marcos A, Nova E, Montero A. Changes in the immune system are conditioned by nutrition. European Journal of Clinical Nutrition. 2003;57:S66–69. doi: 10.1038/sj.ejcn.1601819. [DOI] [PubMed] [Google Scholar]
- Mika C, Holtkamp K, Heer M, Günther RW, Herpertz-Dahlmann B. A 2-year prospective study of bone metabolism and bone meneral density in adolescents with anorexia nervosa. Journal of Neural Transmission. 2007;114:1611–1618. doi: 10.1007/s00702-007-0787-4. [DOI] [PubMed] [Google Scholar]
- Millar HR, Wardell F, Vyvyan JP, Naji SA, Prescott GJ, Eagles JM. Anorexia nervosa mortality in Northeast Scotland, 1965-1999. American Journal of Psychiatry. 2005;162:753–757. doi: 10.1176/appi.ajp.162.4.753. [DOI] [PubMed] [Google Scholar]
- Mira M, Stewart PM, Vizzard J, Abraham S. Biochemical abnormalities in anorexia nervosa and bulimia. Annals of Clinical Biochemistry. 1987;24:29–35. doi: 10.1177/000456328702400104. [DOI] [PubMed] [Google Scholar]
- Mitchell AC, Leak RK, Zigmond MJ, Cameron JL, Mirnics K. Gene transcripts associated with BMI in the motor cortex and caudate nucleus of calorie restricted rhesus monkeys. Genomics. 2012;99:144–151. doi: 10.1016/j.ygeno.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mont L, Castro J. Anorexia nervosa: a disease with potentially lethal repercussions on the heart. Revista Española de Cardiología. 2003;56:652–653. doi: 10.1016/s0300-8932(03)76934-6. [DOI] [PubMed] [Google Scholar]
- Morgan JF, Bolton J, Sedgwick PM, Patel S, Lacey JH, Conway GS. The Journal of Clinical Endocrinology and Metabolism. 1999;84:2257. doi: 10.1210/jcem.84.6.5809-1. [DOI] [PubMed] [Google Scholar]
- Nakazato M, Hashimoto K, Shimizu E, Kumakiri C, Koizumi H, Okamura N, Mitsumori M, Komatsu N, Lyo M. Decreased levels of serum brain-derived neurotrophic factor in female patients with eating disorders. Biological Psychiatry. 2003;54:485–90. doi: 10.1016/s0006-3223(02)01746-8. [DOI] [PubMed] [Google Scholar]
- Narnaware YK, Peter RE. Effects of food deprivation and refeeding on neuropeptide Y (NPY) mRNA levels in goldfish. Comparative Biochemistry and Physiology. Part B, Biochemistry and Molecular Biology. 2001;129:633–637. doi: 10.1016/s1096-4959(01)00359-1. [DOI] [PubMed] [Google Scholar]
- Nova E, Gomez-Martinez S, Morande G, Marcos A. Cytokine production by blood mononuclear cells from in-patients with anorexia nervosa. British Journal of Nutrition. 2002;88:183–188. doi: 10.1079/BJNBJN2002608. [DOI] [PubMed] [Google Scholar]
- Ortega-Hernandez OD, Kivity S, Shoenfeld Y. Olfaction, psychiatric disorders and autoimmunity: is there a common genetic association? Autiomminuity. 2009;42:80–88. doi: 10.1080/08916930802366140. [DOI] [PubMed] [Google Scholar]
- Oshlack A, Robinson MD, Young MD. From RNA-seq reads to differential expression results. Genome Biology. 2010;11:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos FC, Ekbom A, Brandt L, Ekselius L. Excess mortality, causes of death and prognostic factors in anorexia nervosa. British Journal of Psychiatry. 2009;194:10–7. doi: 10.1192/bjp.bp.108.054742. [DOI] [PubMed] [Google Scholar]
- Plank M, Wuttke D, van Dam S, Clarke SA, de Magalhaes JP. A meta-analysis of caloric restriction gene expression profiles to infer common signatures and regulatory mechanisms. Molecular Biosystems. 2012;8:1339–1349. doi: 10.1039/c2mb05255e. [DOI] [PubMed] [Google Scholar]
- Rapps N, Giel KE, Söhngen E, Salini A, Enck P, Bischoff SC, Zipfel S. Olfactory deficits in patients with anorexia nervosa. European Eating Disorder Review. 2010;18:385–389. doi: 10.1002/erv.1010. [DOI] [PubMed] [Google Scholar]
- Raymond NC, Dysken M, Bettin K, Eckert ED, Crow SJ, Markus K, Pomeroy C. Cytokine production in patients with anorexia nervosa, bulimia nervosa, and obesity. International Journal of Eating Disorders. 2000;28:293–302. doi: 10.1002/1098-108x(200011)28:3<293::aid-eat6>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Scalfi L, Polito A, Bianchi L, Marra M, Caldara A, Nicolai E, Contaldo F. Body composition changes in patients with anorexia nervosa after complete weight recovery. European Journal of Clinical Nutrition. 2002;56:15–20. doi: 10.1038/sj.ejcn.1601290. [DOI] [PubMed] [Google Scholar]
- Schecklmann M, Pfannstiel C, Fallgatter AJ, Warnke A, Gerlach M, Ramonos M. Olfaction in child and adolescent anorexia nervosa. Journal of Neural Transmission. 2012;119:721–728. doi: 10.1007/s00702-011-0752-0. [DOI] [PubMed] [Google Scholar]
- Silber TJ, Chan M. Immunologic cytofluorometric studies in adolescents with anorexia nervosa. International Journal of Eating Disorders. 1996;19:415–418. doi: 10.1002/(SICI)1098-108X(199605)19:4<415::AID-EAT10>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Storch CH, Nikendei C, Schild S, Haefeli WE, Weiss J, Herzog W. Expression and activity of P-glycoprotein (MDR1/ABCB1) in peripheral blood mononuclear cells from patients with anorexia nervosa compared with healthy controls. International Journal of Eating Disorders. 2008;41:432–438. doi: 10.1002/eat.20519. [DOI] [PubMed] [Google Scholar]
- Støving RK, Andries A, Brixen K, Flyvbjerg A, Hørder K, Frystyk J. Leptin, ghrelin, and endocannabinoids: potential therapeutic targets in anorexia nervosa. Journal of Psychiatric Research. 2009;43:671–679. doi: 10.1016/j.jpsychires.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Sullivan PF. Mortality in anorexia nervosa. American Journal of Psychiatry. 1995;152:1073–1074. doi: 10.1176/ajp.152.7.1073. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics. 2006;141:261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- Tilg H, Moschen AR. Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nature Reviews. Immuniology. 2006;6:772–783. doi: 10.1038/nri1937. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1011. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckey RC. Progesterone synthesis by the human placenta. Placenta. 2005;26:273–281. doi: 10.1016/j.placenta.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Turecki G, Rouleau GA, Mari J, Joober R, Morgan K. Lack of association between bipolar disorder and tyrosine hydroxylase: a meta-analysis. American Journal of Medical Genetics. 1997;74:348–352. doi: 10.1002/(sici)1096-8628(19970725)74:4<348::aid-ajmg2>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Ulger Z, Gürses D, Ozyurek AR, Arikan C, Levent E, Aydo du S. Follow-up of cardiac abnormalities in female adolescents with anorexia nervosa after refeeding. Acta Cardiologica. 2006;61:43–49. doi: 10.2143/AC.61.1.2005139. [DOI] [PubMed] [Google Scholar]
- Umeki S. Biochemical abnormalities of the serum in anorexia nervosa. The Journal of Nervous and Mental Disease. 1988;176:503–506. doi: 10.1097/00005053-198808000-00009. [DOI] [PubMed] [Google Scholar]
- Usdan LS, Khaodhiar L, Apovian CM. The endocrinopathies of anorexia nervosa. Endocrine Practice. 2008;14:1055–1063. doi: 10.4158/EP.14.8.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D, Infante-Duarte C, Salbach-Andrae H, Burghardt R, Hamann I, Pfeiffer E, Lehmkuhl U, Ehrlich S. Preproenkephalin expression in peripheral blood mononuclear cells of acutely underweight and recovered patients with anorexia nervosa. Neuropsychobiology. 2010;62:151–157. doi: 10.1159/000318571. [DOI] [PubMed] [Google Scholar]
- Wu HS, Lin HT, Wang CK, Chiang YF, Chu HP, Hu MC. Human CYP11A1 promoter drives Cre recombinase expression in brain in addition to adrenals and gonads. Genesis. 2007;45:59–65. doi: 10.1002/dvg.20266. [DOI] [PubMed] [Google Scholar]
- Zipfel S, Lowe B, Reas DL, Deter HC, Herzog W. Long-term prognosis in anorexia nervosa: lessons from a 21-year follow-up study. Lancet. 2000;355:721–722. doi: 10.1016/S0140-6736(99)05363-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.