Abstract
Mounting evidence has recently underscored the importance of DNA methylation in normal brain functions. DNA methylation machineries are responsible for dynamic regulation of methylation patterns in discrete brain regions. In addition to methylation of cytosines in genomic DNA (5-methylcytosine; 5mC), other forms of modified cytosines, such as 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine, can potentially act as epigenetic marks that regulate gene expression. Importantly, epigenetic modifications require cognate binding proteins to read and translate information into gene expression regulation. Abnormal or incorrect interpretation of DNA methylation patterns can cause devastating consequences, including mental illnesses and neurological disorders. Although DNA methylation was generally considered to be a stable epigenetic mark in post-mitotic cells, recent studies have revealed dynamic DNA modifications in neurons. Such reversibility of 5mC sheds light on potential mechanisms underlying some neurological disorders and suggests a new route to correct aberrant methylation patterns associated with these disorders.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-013-0223-4) contains supplementary material, which is available to authorized users.
Key Words: DNA methylation, DNA demethylation, DNMT, TET, 5hmC, GADD45
Introduction
The methylation of cytosines at the 5-carbon position (5-methylcytosine; 5mC) in genomic DNA is a critical epigenetic mark in the metazoan genome. In multicellular organisms, cell type-specific landscapes of methylation patterns, the methylome, confer one of the bases for cell type-specific gene expression, resulting in vastly diverse cellular phenotypes and functions out of identical genetic materials. Moreover, DNA methylation plays important roles in genomic imprinting, X chromosome inactivation, and maintenance of genomic stability [1]. Appropriate generation, recognition, and erasure of 5mC are crucial for spatiotemporal regulation of gene expression and proper function of mammalian cells. Aberrant regulation or recognition of methylcytosine leads to various detrimental consequences, which have been observed in human diseases and animal models [2–5].
Historically, 5mC has been considered to be a stable epigenetic modification that becomes fixed throughout differentiation processes and can only be diluted through the DNA replication process during cell division. Several lines of evidence, however, indicated that active DNA demethylation could happen in post-mitotic cells [6, 7]. Efforts to search for the DNA demethylase resulted in the characterization of a potential DNA demethylation intermediate product, 5-hydroxymethylcytosine (5hmC), in Purkinje neurons and embryonic stem cells, along with methylcytosine hydroxylases of the ten–eleven translocation (TET) protein family [8, 9]. More recently, TET enzymes were shown to further oxidize 5hmC into 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), which can be readily repaired by DNA repair enzymes [10–12].
Aberrant 5mC patterns in certain genomic loci have been found to result in neuropsychiatric symptoms such as autistic behaviors or mental retardation [13–15] . Besides the functional requirement of 5mC, a number of studies reported that neurons have unique DNA modification features as opposed to the other cell types. Post-mitotic neurons have the highest level of 5hmC among mammalian cell types, suggesting that more dynamic turnover of 5mC exists in neurons, as well as the possibility that 5hmC may act as an independent epigenetic mark. Notably, neurons have a significant amount of 5mC in the nonCpG context in contrast to other somatic cell types where 5mC almost exclusively occurs at CG dinucleotides [16]. These exciting discoveries call for reconsideration of epigenetic neurological disorders in light of these new findings. In this review, we discuss our current understanding of DNA methylation in neurons and neurological disorders caused by dysregulation of DNA methylation. Moreover, we discuss genes that regulate DNA methylation and their potential as targets for pharmaceutical intervention.
Mechanism of DNA Methylation
The establishment and maintenance of the mammalian methylome, the genomic DNA methylation patterns, are orchestrated by 3 DNA methyltransferases (DNMTs): DNMT1, DNMT3a, and DNMT3b [17]. The 3 mammalian DNMTs share a C-terminal methyltransferase catalytic domain (Fig. 1), which transfers a methyl group from S-adenosyl methionine to the 5-carbon position of CpG cytosines in double-stranded DNA. Although their catalytic domains exhibit conserved sequence motifs, biochemical and genetic studies indicate that each DNMT has a specific function in DNA methylation processes. DNMT1 exhibits a substrate preference for hemi-methylated DNA over unmethylated DNA [18, 19]. During DNA replication, DNMT1 binds to replication forks and faithfully transmits the original DNA methylation patterns from the mother strand to the newly synthesized strand [3]. Because DNMT1 is responsible for maintaining methylation patterns during DNA replication, it has been referred to as the maintenance methyltransferase. In contrast, DNMT3a and DNMT3b have no preference toward hemi-methylated or unmethylated DNA, and, instead, have been implicated in de novo DNA methylation processes [4]. These 3 DNMTs cooperatively shape cell type-specific DNA methylation landscapes in the mammalian genome. Genetic ablation of any DNMT in mice results in an aberrant methylome and is lethal; DNMT1 or DNMT3b knockout mice die prenatally, and DNMT3a knockout mice die around 4 weeks after birth [4, 20]. In humans, deficiency in DNMT3b contributes to hypomethylation in pericentromeric DNA sequences and leads to immunodeficiency, centromere instability, and facial anomalies syndrome with varying degrees of mental retardation. Together, these phenotypic data highlight the importance of DNA methylation in the mammalian genome [15].
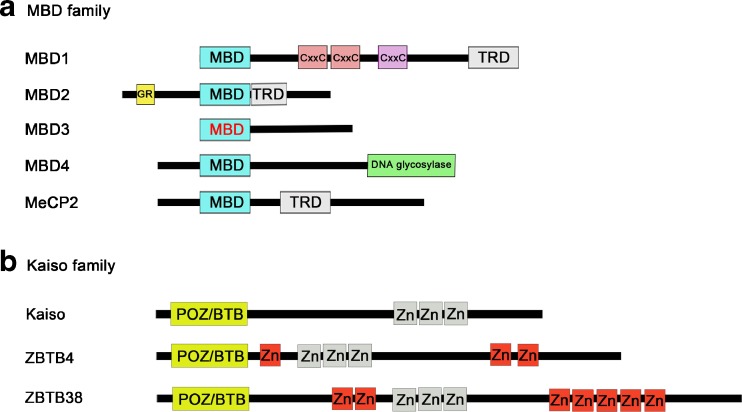
Fig. 1.
Mammalian methyl–CpG binding proteins. (a) MBD and Kaiso family proteins are capable of recognizing methylated DNA. Five classic MBD family members, namely MBD1, MBD2, MBD3, MBD4, and MeCP2 share a conserved MBD domain, with the exception of MBD3, which has a mutation in the MBD domain (red) that prevents it from binding methylated CpGs in mammals. MBDs can silence gene expression by recruiting various corepressor complexes through their transcription repression domains (TRD). MBD4 contains an additional C-terminal DNA glycosylase domain that is used for excision-based DNA repair and has been implicated in DNA repair and demethylation. (b)The Kaiso family is composed of Kaiso, ZBTB4, and ZBTB38, and shares homology in BTB/POZ (broad complex Tramtrack bric-a-brac/Pox virus and zinc finger) domain and 3 zinc finger motifs (depicted in gray). The 3 Kaiso zinc fingers are capable of binding both mCpG and specific nonmethylated DNA sequences
Epigenetic information encoded by DNA methylation patterns requires cognate binding proteins, termed “readers“, to translate this information into downstream biological processes. In mammals there are 3 families of methyl–CpG binding proteins that recognize methylated DNA: the Uhrf family, the methyl–CpG binding domain (MBD) family, and the Kaiso family. The primary function of the Uhrf family is to guide DNMT1 to hemi-methylated DNA and ensure faithful maintenance of DNA methylation patterns during cell division [21, 22]. In contrast, members of the MBD and Kaiso families mediate DNA methylation-dependent gene inactivation (Fig. 1) [23–26].
Within the MBD family, MBD1, MBD2, MBD4, and MeCP2 are able to associate with methylated DNA, whereas MBD3 lacks this capacity owing to its inactivated binding motif (Fig. 1) [23]. Both MBD1 and MeCP2 are constitutively expressed in the brain and play fundamental roles in neural development and synaptic plasticity [27, 28]. A primary feature of MBD family proteins is their ability to assemble other binding partners to form a repressor complex dedicated to transcriptional inactivation and heterochromatin formation. For example, MeCP2 can interact with SIN3a, histone deacetylase (HDAC)1, or HDAC2 via its transcriptional repression domain and maintain transcriptional silence in methylated genomic loci [29, 30]. Misinterpretation of DNA methylation patterns due to MeCP2 deficiency disrupts normal gene expression programs and, as a result, leads to Rett syndrome—a devastating neurodevelopmental disorder [27]. In addition to binding to methylated DNA, MBD4 has a DNA glycosylase domain that recognizes G:T/U mismatches [31, 32]. This particular characteristic suggests the possibility that MBD4 may act in DNA repair and possibly active DNA demethylation processes [33–35].
Another class of methylated DNA-binding proteins is the Kaiso family, which represents a subfamily of BTB/POZ transcription factors and is composed of Kaiso, ZBTB4, and ZBTB38 [36, 37]. Despite sharing the Kaiso-like zinc finger motifs, ZBTB4 and ZBTB38 are capable of recognizing a single methylated CpG dinucleotide, whereas Kaiso requires 2 consecutive methylated CpG dinucleotides (CGCGs) for binding. Importantly, unlike MBDs, the Kaiso family proteins also associate with unmethylated DNA in a sequence-specific manner; Kaiso and ZBTB4 can recognize the consensus Kaiso binding site, TCCTGCNA, while ZBTB38 binds to the CACCTG E-box motif. These notable differences in DNA-binding preference indicate that Kaiso family proteins may have diverse biological functions.
Recent studies have revealed Kaiso to be a key modulator of canonical Wnt signals as several Wnt gene targets, such as PPARδ, c-Myc, Cyclin D1, and Matrilysin, exhibited the consensus Kaiso binding sequences in the upstream region of the transcriptional start site [38]. Nonetheless, the Kaiso proteins seems to repress gene expression in a DNA methylation-dependent fashion by directing co-repressors and histone deacetylases to the Kaiso binding sites of target promoters. In situ hybridization analyses indicated that Kaiso family members exhibited distinct levels and patterns in the brain region; ZBTB4 was highly expressed in hippocampus, brainstem, olfactory bulb, piriform cortex, and cerebellum, whereas ZBTB38 was displayed in the brain and neuroendocrine organs [37]. Thus far, Kaiso has been shown to affect neural cell differentiation [39]; however, the neural function of Kaiso family proteins is largely unexplored. Further study of Kaiso family proteins in the central nervous system (CNS) and identification of their genomic targets will greatly enhance our understanding of how epigenetic machinery regulates brain functions.
Active DNA Demethylation
Accumulating data indicate that the mammalian methylome is subject to changes during development, aging, or even by a variety of environmental stimuli [40, 41]. While DNA methylation can be passively diluted during cell division, active DNA modifications can also occur in nondividing cells. For example, Guo et al. [7] demonstrated that methylation status in a large number of genomic loci in mature neurons in vivo is altered after synchronous neuronal activation. This re-patterning of DNA methylation likely resulted from combinatorial action of DNA methylation and demethylation processes. Recent studies have uncovered several potential pathways involved in DNA demethylation regulation and have begun to delineate mechanisms underlying active DNA demethylation (Fig. 2).
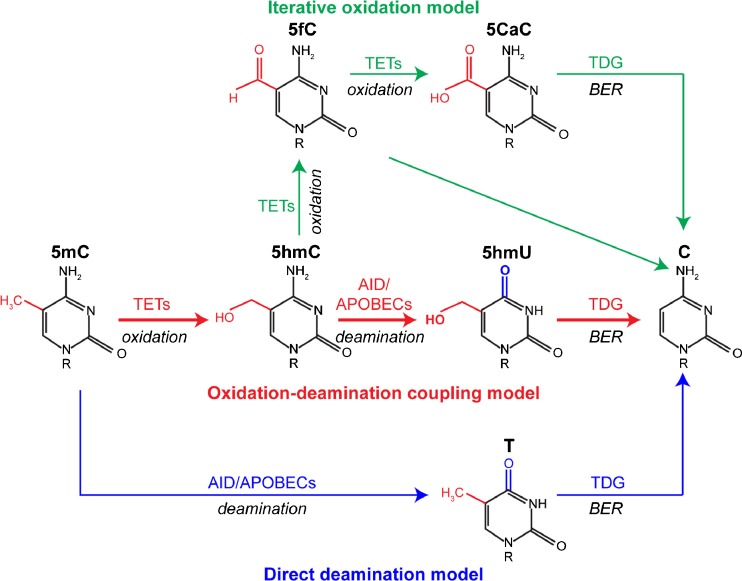
Fig. 2.
Mechanisms of active DNA demethylation. (a) Direct deamination model. 5-Methylcytosine (5mC) is deaminated to form thymine (T) by activation-induced deaminase (AID)/apolipoprotein B RNA-editing catalytic component (APOBEC) family proteins. The resultant T mismatch is then removed by methyl–CpG binding proteins 4 (MDB4) or thymine DNA glycosylases (TDG) via the base excision repair (BER) pathway. (b) Oxidation–deamination coupling model. 5mC is oxidized by the ten–eleven translocation (TET) family proteins to form 5-hydroxymethylcytosine (5hmC) and is then deaminized to 5-hydroxymethyluridine (5hmU) by the AID/APOBEC family. MDB4 or TDG can recognize 5hmU and activate the BER pathway to replace it with C. (c) Iterative oxidation model. 5mC can be hydroxylated by TET proteins to form 5hmC and further sequentially oxidized to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). The oxidation products 5fC and 5caC can be removed by TDG and the BER pathway, and filled with unmethylated cytosine
The first is the direct deamination model. Activation-induced deaminase (AID)/apolipoprotein B RNA-editing catalytic components (APOBECs) represent a family of cytosine deaminases and were initially identified as factors to deaminate cytosine (C) into uracil (U) in the immunoglobulin loci, leading to initiation of antibody diversification [42, 43]. Besides unmodified cytosine, recent studies also show that AID/APOBECs can convert 5mC into thymine (T) albeit with a reduced reactivity [44]. The resultant T:G mismatch can recruit 2 major DNA glycosylases—thymine DNA glycosylase (TDG) and MBD4—to excise thymine and yield an abasic site (AP; also known as apurinic/apyrimidinic site). This abasic site is then recognized by AP endonucleases to initiate the base excision repair (BER) pathway to reconstitute with unmethylated cytosine. In support of this model, AID deficiency perturbs the conversion of 5mC to T and, consequently, results in global genomic hypermethylation in the mouse germ line [45, 46].
The second is the oxidation–deamination coupling model. Recent studies have shown that 5mC can be hydroxylated into 5hmC by TET enzymes [8, 47]. Guo et al. [7] proposed a model that 5hmC is first deaminated into 5hmU, which can be recognized by TDG and MBD4 and followed by BER pathway processes.
The third is the iterative oxidation model. It was found that TET enzymes can further oxidize 5mC to sequentially form 5fC and 5caC [12]. Although it is possible that decarboxylation can convert 5caC directly to cytosine, the decarboxylase responsible for this conversion has not yet been identified in mammals. Instead, several lines of evidence suggest TDG recognizes and excises 5fC and 5caC [10, 11], and these DNA lesions go through the BER pathway, resulting in an unmethylated cytosine. Genome-wide analysis indicates that the genomic content of 5fC and 5caC is 1 or 2 orders of magnitude lower than the level of 5hmC [12]. This finding suggests a complex regulatory mechanism that controls the progression of oxidation and DNA demethylation. Importantly, the oxidation derivatives of 5mC (5hmC, 5fC, and 5caC) not only serve as intermediates of the active DNA demethylation process, but also potentially function as independent epigenetic marks to regulate gene expression [48, 49]. Neurons exhibit 5–10 times higher levels of 5hmC than other somatic tissues [9]. Recent studies have correlated substantial increases in brain 5hmC levels with aging [50, 51]. Together, these findings support the notion that 5hmC may play a role in neurodevelopment, as well as pathogenesis of neurodegenerative diseases.
The vertebrate TET family is composed of TET1, TET2, and TET3, and each member exhibits discrete expression patterns in tissues. Because of their potential involvement in DNA demethylation, TET family proteins must be tightly regulated in order to ensure accurate epigenetic function. Ectopic expression or knockdown of TET family members alters genomic methylation patterns and, consequently, causes developmental and physiological deficits. For example, knockdown of TET3 in Xenopus oocytes causes misregulation of developmental genes and abnormalities in neural development [52]. Also, dysregulation of cortical TET1 levels has been associated with psychosis [53]. Given the substantial amounts of 5hmC present in neurons through adulthood, several studies have begun to reveal the function of TET family proteins in the adult nervous system. Although loss of TET1 did not result in lethality or discernible developmental abnormalities, new evidence indicates TET1 is required for memory, cognitive function, and adult hippocampal neurogenesis. Using TET1 conventional and conditional knockout mice, Zhang et al. [54] demonstrated deficiency of TET1 restricted proliferation of neural progenitor cells (NPCs) in the hippocampal dentate gyrus that can diminish the pool of NPCs and ultimately reduce the number of newborn neurons. Furthermore, comparison of gene expression and DNA methylation profiles between WT and TET1 KO NPCs revealed that several neurogenesis-related genes in TET1 KO NPCs have hypermethylated promoters and decreased expression levels. These findings highlight the importance of TET1-mediated DNA demethylation in hippocampal adult neurogenesis, as well as the significance of this process in memory and cognition. Future studies will be necessary to functionally connect TET family proteins and pathogenesis of mental disorders.
The enzymatic activity of TET family proteins resides in the conserved C-terminal region and requires oxygen, 2-oxoglutarate (2-OG) and Fe2+ to oxidize methylated DNA. Accordingly, mutation of the iron-binding site of TET or treatment with competitive inhibitors of 2-OG abolishes TET function [8, 47, 55]. It is important to note that more than 60 2-OG dependent dioxygenases with differential 2-OG affinities exist in vertebrates. Of these proteins, Jumonji domain-containing histone demethylases are known to regulate gene expression via histone modifications [55]. Thereby, 2-OG competitors typically also have broad effects on multiple targets and affect both genome-wide histone methylation and DNA methylation patterns. Identification of highly selective inhibitors for individual TET family proteins will be useful in pharmacological studies and help elucidate the roles of TETs in human health and disease.
DNA Methylation and Gene Regulation
In the mammalian genome, CpG dinucleotides are predominant targets of DNA methylation. CpG dinucleotides are evolutionally selected against and are present throughout genome in lower frequency except in CpG islands where the density of CpG dinucleotides is unusually high. CpG islands are typically associated with promoters—approximately 70 % of annotated gene promoters have high CpG content [56]. While DNA methylation mainly occurs in CpG dinucleotides of repetitive elements and gene bodies, most CpG islands are devoid of DNA methylation. Even so, CpG islands become hypermethylated under certain circumstances and can repress gene activity. For example, CpG islands acquire DNA hypermethylation in order to silence gene expression during X chromosome inactivation and genomic imprinting [57, 58]. In the pathogenesis of many diseases, such as fragile X syndrome (FXS), inactivation or reduction of gene expression results from CpG island hypermethylation [59, 60]. Collectively, these findings, together with other evidence, indicate that methylation status of CpG islands is inversely correlated with transcriptional activity.
As opposed to the well established causal link between DNA methylation and transcriptional activity, the underlying molecular mechanisms remain elusive. It has been proposed that DNA methylation causes a failure of transcription by disrupting the ability of transcription factors to bind DNA [14]. Indeed, several factors including activating protein 2 (AP-2), cyclic AMP response element-binding (CREB) protein, c-Myc, and nuclear factor-kappa b (NF-κB), are known to be sensitive to methylated DNA and unable to activate transcription when target motifs are methylated [61]. Conversely, certain factors, such as MeCP2 and MBDs, exhibit strong preference toward methylated DNA. These proteins not only exert transcriptional repression activity by blocking transcription factor binding sites, but also form complexes with other regulators to restrict the accessibility of promoter sequences. For example, a MeCP2 complex can recruit HDACs that transform chromatin conformation into a repressive state [29]. Thus, it is generally accepted that DNA methylation works in concert with histone modifications to govern transcriptional environment and gene activity. Despite evidence that communication may exist between these 2 epigenetic modalities, there is no general rule defining whether DNA methylation precedes histone modifications or vice versa. In some situations, DNA methylation is not the dominant cause of gene inhibition, but maintains long-term transcriptional silencing.
Although DNA methylation has long been recognized as a repressive epigenetic mark at promoter regions, emerging evidence suggests that methylation in intragenic regions (gene bodies) is not necessarily associated with gene silencing. Instead, genome-wide analysis of proliferative tissues and cell lines demonstrated that methylation levels within gene bodies are positively correlated with transcriptional activity [62–64]. Such a relationship was not detected in human embryonic stem cells or brain tissue [65, 66], suggesting intragenic DNA methylation may have differential functions in distinct cell types. In support of this notion, Maunakea et al. [67] found that the use of alternative promoters within gene bodies, which partly contributes to expressing distinct messenger RNA isoforms in different brain regions, can be regulated by intragenic methylation. To date, the molecular mechanisms specifying functional differences between distinct regional DNA methylation (5’ proximal promoter versus intragenic region) have yet to be fully deciphered.
DNA Methylation and Brain Function
Considering the association between 5mC content and transcriptional activity, it is not surprising that DNA methylation has pivotal roles in regulating brain development and function. For example, changes in DNA methylation states are concomitant with fate choices of NPCs during embryonic cortical development [68]. In this scenario, glial fibrillary acidic protein (GFAP), a well-known astrocytic marker, is highly expressed during development of the astrocyte. Takizawa et al. [69] found that a CpG dinucleotide within a STAT3 binding element in the GFAP promoter is hypermethylated during neurogenesis at E11.5 and becomes hypomethylated within the period of astrocytogenesis at E14.5. The methylation status of this CpG seems to reflect binding accessibility at this site as it was clearly demonstrated that the methylated CpG dinucleotide is sufficient to disable STAT3 binding, and leads to transcriptional unresponsiveness of GFAP both in vitro and in vivo. Such spatiotemporal DNA methylation patterns not only create neural diversity in the brain, but also provide a rich repertoire of functional varieties.
In addition to lineage specification, studies have shown that DNA methylation can impose regulation on synaptic function and plasticity. While DNMTs have dynamic patterns of expression in the developing CNS, constitutive DNMT1 and DNMT3a expression is found in the adult brain [70]. Interestingly, although DNMT1 and DNMT3a presumably act on maintenance and de novo DNA methylation, respectively, a recent genetic study revealed DNMT1 and DNMT3a have functional redundancy in regulating syntactic plasticity and memory formation [71]. Furthermore, pharmacological blockade of DNMTs by 5-azacytidine (5-Aza) in hippocampal slices decreases genomic content of 5mC and reduces the frequency of miniature excitatory postsynaptic currents [72]. Another independent study found that these effects on synaptic plasticity are associated with hypomethylation of brain-derived neurotrophic factor (BDNF) and reelin (RELN) promoters [73]. These data not only highlight the role of DNMTs in regulating neuronal activity, but also suggest that DNA methylation on a subset of critical genes associated with plasticity is required for proper synaptic function.
The finding that DNA methylation plays a role in synaptic plasticity led to investigation of DNA methylation in learning, memory, and cognitive function. Contextual fear conditioning, a paradigm that can induce formation of hippocampal-dependent associative memories, was found to contemporaneously alter both the DNA methylome and the transcriptome [74]. Importantly, the changes in methylation status (both hyper- and hypomethylation) occurred on several memory-associated genes, including protein phosphatase 1 (PP1), reelin, and bdnf. The regulation of methylation status is owing, at least in part, to the de novo DNA methyltransferases DNMT3a and 3b, as they exhibit high expression in the hippocampus after fear conditioning [74]. Strikingly, although DNMT1 levels remain unchanged after fear conditioning, genetic studies reveal that it has functional overlap with DNMT3a in governing cognitive processes. Neither single knockout of DNMT1 or DNMT3a resulted in profound abnormalities in long-term plasticity in the hippocampal CA1 region, whereas double knockout mice exhibit impaired synaptic function and deficits in learning and memory [71]. Other studies also support a role for DNMT1 in behaviorally-induced epigenetic modification. For example, DNMT1 has increased expression when mice are placed in an enriched environment [75]. Inhibiting the activity of all DNMTs by 5-Aza-2-deoxycytidine (5-Daz) causes an aberrant methylome and disturb cognitive functions [74]. These cumulative findings not only emphasize the importance of DNMTs in brain function but also provide strong evidence for the role of DNA methylation in memory storage. Further studies are required to understand how active DNA methylation and demethylation processes work in concert to elicit changes in methylation patterns underlying formation of long-term memory.
DNA methylation serves as a molecular bridge between environmental stimuli and neural plasticity. A growing body of evidence supports the notion that environmental stress can confer long-lasting changes in physiology and behavior via DNA methylation in selected brain regions [76]. For example, recent work has shown that early life trauma or chronic adult stress can produce persisting methylation changes at glucocorticoid receptor (GR) and bdnf loci in neurons of the prefrontal cortex and hippocampus, with concomitant abnormalities in GR and BDNF expression [77]. Notably, dysregulation of GR and BDNF has been implicated in the pathogenesis of schizophrenia and mood disorders [78–80]. These results suggest that DNA methylation can influence neural processes and may be involved in the etiology of cognitive impairments and psychiatric illnesses [81]. In support of this idea, DNA hypomethylation was found in intron 7 of certain genetic FKBP5 variants in response to early life trauma or childhood abuse [82]. Given that FKBP5 is a key regulator of stress hormones, trauma victims with this epigenetic signature are more susceptible to development of post-traumatic stress disorder in adulthood owing to elevated FKBP5 expression.
5hmC and Brain Development
In addition to its role as an intermediate in active DNA demethylation, emerging data suggest that 5hmC acts as an independent epigenetic marker and may regulate neuronal development and function. For example, distinct cell identities in the CNS exhibit differential 5hmC levels [83]. In corticogenesis, maturing neurons in the cortical plate are characterized by elevated 5hmC levels compared with neural progenitor cells of the ventricular zone. High occupancy of CpGs with 5hmC is predominantly found in intragenic regions (gene bodies) of actively transcribed genes that are critical for neural differentiation [83]. To date, the relationship between 5hmC and gene transcription is not fully understood.
Additional data suggesting a functional link between 5hmC and neurodevelopment come from genome-wide maps of 5hmC in the mouse brain at different developmental stages [50, 51]. These studies reveal that mature neurons exhibit higher levels of 5hmC, and that global 5hmC content gradually increases with aging. The age-dependent acquisition of 5hmC does not occur randomly, but rather preferentially in developmentally activated genes related to neurodegenerative disorders, angiogenesis, and responses to hypoxia [50]. These findings suggest the possibility of active roles of 5hmC in both normal neuronal function and in the etiology of neurological disorders, such as Alzheimer’s disease and other dementias [84]. Further studies are required for identification of specific loci that acquire differential 5hmC status in different disease states and to define specific roles for 5hmC in disease pathogenesis. The emergence of distinct 5hmC distributions in given disease states might be used as a biomarker for risk assessment, and may facilitate the diagnosis, prognosis, and treatment of diseases.
To decipher the biological functions of 5hmC, studies have sought to identify 5hmC readers that bind 5hmC and alter downstream transcriptional activity. As such, it was recently discovered that MeCP2 directly interacts with 5hmC in vitro [48]. In contrast to its conventional role as a repressor of gene transcription, MeCP2 appears to facilitate gene activation in genomic loci with high 5hmC content. This finding supports the idea that MeCP2 has roles in both gene repression and activation [85]. However, the mechanistic explanation as to how MeCP2 leads to distinct transcriptional responsiveness via binding 5mC or 5hmC is still elusive. The interaction between MeCP2 and 5hmC seems to be crucial for neural development. The R133C mutation of MeCP2, which results in a milder form of Rett syndrome, was found to disrupt 5hmC binding by MeCP2. Nonetheless, other MeCP2 mutations associated with Rett syndrome did not disrupt 5hmC binding [48].
DNA Methylation and Mental Disorders
Many neurological diseases are associated with abnormal DNA methylation patterns (Table 1). Here, we briefly discuss the potential involvement of DNA methylation in cognitive impairment, psychiatric disorders, and FXS.
Table 1.
Neurological diseases with aberrant DNA methylome
| Neurological diseases | Symptoms | Methylation status | Impaired genes |
|---|---|---|---|
| Fragile X syndrome | Intellectual disability and autistic-like behaviors | Hypermethylation | FMR1 [86] |
| ICF1 syndrome | Mental retardation syndrome | Hypomorphic mutation of DNMT3B | DNMT3B [15, 87] |
| ICF2 syndrome | Mental retardation syndrome | Hypomethylation | ZBTB24 [88] |
| Rett syndrome | Repetitive stereotyped hand movements, seizures, loss of speech, autistic-like behaviors, cerebral palsy | Loss of activity of MeCP2 | MeCP2 [27] |
| HSAN1 | Various neuropathies and onset of dementia in the third or fourth decade of life | Hypomethylation | DNMT1 [89] |
| ADCA-DN | Autosomal dominant cerebellar ataxia, deafness and narcolepsy, early-onset dementia | Hypomethylation | DNMT1 [90] |
| Alzheimer’s disease | Progressive dementia | Hypermethylation | Neprilysin (NEP) [91] |
| Parkinson’s disease | Shaking, rigidity, slowness of movement and difficulty with walking and gait | Hypomethylation | Loss of DNMT1 [92] SNCA [93] |
| Amyotrophic lateral sclerosis | Muscle weakness and atrophy | Hypermethylation | Increased DNMT1
DNMT3a [94] |
| Stroke | Sudden onset focal dysfunction | Hypermethylation Hypomethylation Hypermethylation Hypomethylation |
Global methylation [95] NKCC1 [96] TSP-1 [97] ER [98] |
| Epilepsy | Seizure | Hypermethylation Hypomethylation |
Reelin [99] Phc2 [100] |
| Multiple sclerosis | Multiple neurological symptoms: visual alterations, muscular weakness, muscular rigidity, tremors, and urinary, intestinal, and cognitive abnormalities, sexual dysfunction | Hypomethylation | PADl2 [101] |
ICF1 immunodeficiency, centromeric instability, and facial anomalies syndrome type 1, ICF2 immunodeficiency, centromeric instability, and facial anomalies syndrome type 2, HSAN1 hereditary sensory and autonomic neuropathy type 1, ADCA-DN autosomal dominant cerebellar ataxia, deafness and narcolepsy
Emerging evidence suggests that certain mental illnesses may be caused by dysregulation of the epigenetic landscape in the brain. For example, age-dependent decline of the splice variant DNMT3a2 in the hippocampus was recently linked to cognitive impairments [102]. Additionally, knockdown of DNMT3a2 expression in younger animals leads to cognitive deficits in fear and object–place recognition memory, demonstrating the necessity of DNMT3a2 in memory formation. While the cause of decreased DNMT3a2 expression during normal aging remains obscure, the consequent cognitive impairments can be alleviated by reintroduction of DNMT3a2. These results suggest that cognitive decline resulting from reduced levels of DNMT3a2 may be reversible, raising the possibility for pharmacological interventions.
Recent studies also implicate aberrant DNA methylation in various psychiatric disorders [81]. Postmortem methylome profiling of brains from patients with schizophrenia or bipolar disorder has revealed profound changes in the DNA methylation landscape, including genes that are functionally linked to disease pathogenesis [103]. Particularly, hypermethylation of glutamic acid decarboxylase 67 (GAD67) and RELN promoter regions was linked to decreased expression of these genes in schizophrenia patients [104]. This epigenetic feature is likely the cause of decreased expression of GAD67 and RELN as in vitro methylation of these promoters restricts their expression. Furthermore, administration of antipsychotic drugs, such as valproate, clozapine or sulpiride, can reduce L-methionine-induced hypermethylation of RELN and GAD67 promoters in the mouse frontal cortex and striatum [105]. Collectively, these results highlight the importance of tightly regulated DNA methylation processes in regulating site-specific methylation patterns. Disruption of this regulation confers an aberrant methylome, triggering susceptibility to psychiatric disorders.
Changes in DNA methylation patterns caused by up-regulation of DNMTs or environmental stress have been proposed as a potential etiologic factor in schizophrenia. While DNMTs exhibit persistent and dynamic expression in both the developing and mature CNS, in situ hybridization studies have revealed DNMT1 and DNMT3a transcripts are preferentially increased in gamma aminobutyric acid (GABA)ergic interneurons in cortical layers I and II of schizophrenia and bipolar brains [106, 107]. Notably, deficits in GABAergic signaling cause increased excitatory activity of glutamatergic pyramidal neurons which can, in turn, alter the balance of excitatory and inhibitory neurotransmission—a phenomenon that has been described in both schizophrenia and bipolar disorder. Elevated levels of DNMT1 and DNMT3a in GABAergic interneurons is an attractive pathological mechanism as it explains observations of DNA hypermethylation followed by downregulation of RELN, GAD67 and other genes that are crucial for proper function of the GABAergic system. To date, the cause of increased DNMT expression in schizophrenia brains remains unknown and whether its increased expression is pathogenic for schizophrenia requires further investigation.
FXS is marked clinically by inherited intellectual disability and autistic-like behaviors. It is believed that the cause of this disorder is due to an expansion of CGG trinucleotide repeats in the 5’ untranslated region of fragile X mental retardation 1 (FMR1) gene. FXS patients have more than 200 copies (normal individuals typically have 5–59 repetitions [59, 60, 108]). These CGG repeats constitute the CpG island of FMR1 and become hypermethylated when the number of repeats is expanded. As a result, DNA methylation-mediated epigenetic silencing leads to inactivation of the FMR1 gene. The molecular explanation as to how expansion of CGG repeats causes hypermethylation of the FMR1 CpG island is unclear. Nonetheless, this process is developmentally regulated. Examination of chorionic villi and embryonic cells from affected fetuses with FXS indicate active transcription of FMR1 until early embryogenesis [109]. Additionally, studies of FXS-derived human embryonic stem cells reveal that FMR1 is active and unmethylated in undifferentiated cells, but becomes silent upon gain of hypermethylation in differentiated cells [110]. These findings imply that the aberrant epigenetic status of the mutant FMR1 allele can be reset, likely through DNA demethylation processes, during germline transmission. Unraveling the corresponding mechanisms may provide insight into ways of reversing methylation of FMR1 and reestablishing FMR1 expression in later developmental stages.
Pharmacology of Epigenetics
The observation over recent years that DNA methylation is a reversible covalent modification suggests possible methods of correcting aberrant methylation patterns associated with disease. To date, a number of pharmacological agents that modify the epigenome—including inhibitors of DNA methylation and various histone modifications—have been developed and used both for research and the clinic (Table 2) [120]. Among them, 5-Aza, 5-Daz (Decitabine), and RG108 are pan-DNMT inhibitors commonly used to delineate roles of DNMTs in neuronal function. Both 5-Aza and 5-Daz are nucleotide analogs of cytidine. Mechanistically, it is believed that 5-Aza and 5-Daz must be integrated into genome before irreversibly binding to the catalytic sites of DNMTs. Although several studies report that treatment of neurons with 5-Aza or 5-Daz results in reduced methylation at certain gene promoters, the demethylation effect of 5-Aza and 5-Daz in post-mitotic neurons is still under debate. A major unresolved question is how these cytidine analogs could exert inhibitory function in post-mitotic neurons. One possibility is via DNA repair during the DNA demethylation process [13]. However, RG108 is a non-nucleoside DNMT inhibitor, which blocks the catalytic sites of DNMTs [120].
Table 2.
DNA methylation and demethylation inhibitors
| Pathway | Drug | Types | Targets | Reference |
|---|---|---|---|---|
| DNA methylation | ||||
| Cytosine methylation | 5-Aza-cytidine | Small molecule | DNMTs | [111] |
| Decitabine | Small molecule | DNMTs | [111] | |
| RG108 | Small molecule | DNMTs | [94] | |
| Zebularine | Small molecule | DNMTs | [112] | |
| MG98 | Oligonucleotide | DNMT1 | [113] | |
| miR-29 | miR | DNMT 3a, 3b | [114] | |
| miR-126 | miR | DNMT1 | [115] | |
| miR-148 | miR | DNMT1 and 3b | [116, 117] | |
| miR-152 | miR | DNMT1 | [118] | |
| DNA demethylation | ||||
| 5-methylcytosine oxidation | 2HG | Small molecule | TET family a-KG-dependent dioxygenases | [55, 119] |
| DMOG | Small molecule | TET family a-KG-dependent dioxygenases | [55] | |
| Base excision repair | ABT-888 | Small molecule | poly(ADP-ribose) polymerase | [7] |
| CRT0044876 | Small molecule | APE1 | [7] | |
miR microRNA, DMOG dimethyloxaloylglycine, DNMT DNA methyltransferase, TET ten–eleven translocation, a-KG alpha-ketoglutarate, APE1 apurinic/apurimidinic endonuclease 1
Antisense inhibitors compose another tool box to selectively modulate individual DNMTs. For example, MG98 is an antisense oligonucleotide that hybridizes to the 3′ untranslated region of DNMT1 messenger RNA [113]. Upon hybridization, the synthetic oligonucleotide can specifically decrease expression of DNMT1 without affecting transcription/translation of other DNMTs. The well-designed synthetic oligonucleotides against DNMTs may provide target specificity and thereby alter individual DNMT-dependent methylation patterns. Similarly, noncoding RNAs, such as short interference RNAs and microRNAs (miRs), can also be used to inhibit DNMT activity. For example, recent studies have shown that miR-29b targets DNMT3a and DNMT3b and governs the methylation state of genomic DNA [114, 121]. The level of miR-29b is inversely associated with DNA methylation; overexpression of miR-29b leads to hypomethylation, whereas inhibition of miR-29a by antisense oligonucleotides results in hypermethylated DNA [121]. miR-29b may therefore act as an additional mechanism by which cells modulate levels of genomic DNA methylation. Importantly, miR-29b also targets additional genes and this lack of specificity may limit the use of miR-29 clinically.
Genomic methylation levels are tightly controlled by DNA methylation and demethylation processes. Recent studies have begun to delineate the molecular mechanisms underlying DNA demethylation and have uncovered several key players. These DNA demethylases exhibit global DNA demethylation properties during early stages of embryogenesis, but confer locus-specific demethylation in neurons. For example, Gadd45b is transiently induced in hippocampal neurons in response to environmental stimuli and, in turn, demethylates specific promoter regions, including BDNF and fibroblast growth factor 1 (FGF1) [122]. Similarly, exogenous expression of TET1 or AID in the dentate gyrus promotes region-specific DNA demethylation [7]. Guiding DNA demethylases to particular genomic regions most likely requires a complex regulatory system. The CXXC domain of TET family proteins has a high binding affinity for clustered unmethylated CpGs. Sequence-specific binding proteins can also recruit DNA demethylase machinery to specific genomic loci. Further understanding of substrate preferences for different DNA demethylases will generate additional insight into mechanisms of reversing DNA methylation at specific genomic loci.
Conclusion and Perspective
Discrete brain regions have dynamic genomic methylation patterns, which are defined by both DNA methylation and demethylation machineries. Such variation in methylation patterns allows for a rich diversity of neuronal subtypes, and differential function and activity amongst neurons of the same subtype. Thus, it is critical that the methylome is tightly regulated from development into adulthood. Aberrant DNA methylation patterns dramatically change physiological characteristics of neurons and have been found to be associated with several mental and neurological disorders. The origins of these conditions might be caused by dysregulation of DNA methylation/demethylation processes or by misinterpretation of methylated DNA sequences.
Identification of active DNA demethylation mechanisms indicates that DNA methylation in post-mitotic neurons is reversible. Importantly, this process has the potential to correct disease-associated methylation patterns, and sheds light on prevention and therapy of neurological disorders. To date, several regulators capable of promoting DNA demethylation have been identified, including Gadd45 and TET families. Ectopic expression of these families of proteins has been shown to induce local DNA demethylation. These data suggest that members of Gadd45 and TET families may become important tools and new therapeutic targets for reverting methylation at defined genomic regions in the future.
Electronic supplementary material
(PDF 1224 kb)
Acknowledgments
This work was supported by Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, and the Maryland Stem Cell Research Fund (MSCRF) to G.L.M., SAFRI to H.S., by postdoctoral fellowship from MSCRF to Y.W and by Samsung Scholarship to J.S. We thank Jared Cregg and Kimberley Christian for critical comments on the manuscript.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 3.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 4.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 5.Xu GL, Bestor TH, Bourc'his D, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 6.Martinowich K, Hattori D, Wu H, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 7.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dittrich B, Robinson WP, Knoblauch H, et al. Molecular diagnosis of the Prader-Willi and Angelman syndromes by detection of parent-of-origin specific DNA methylation in 15q11-13. Hum Genet. 1992;90:313–315. doi: 10.1007/BF00220089. [DOI] [PubMed] [Google Scholar]
- 14.Driscoll DJ, Waters MF, Williams CA, et al. A DNA methylation imprint, determined by the sex of the parent, distinguishes the Angelman and Prader-Willi syndromes. Genomics. 1992;13:917–924. doi: 10.1016/0888-7543(92)90001-9. [DOI] [PubMed] [Google Scholar]
- 15.Jin B, Tao Q, Peng J, et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet. 2008;17:690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
- 16.Lister R, Mukamel EA, Nery JR, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet 2000;9:2395-2402. [DOI] [PubMed]
- 18.Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- 19.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 20.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 21.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 22.Sharif J, Muto M, Takebayashi S, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 23.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 25.Lewis JD, Meehan RR, Henzel WJ, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 26.Clouaire T, Stancheva I. Methyl-CpG binding proteins: specialized transcriptional repressors or structural components of chromatin? Cell Mol Life Sci. 2008;65:1509–1522. doi: 10.1007/s00018-008-7324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 28.Zhao X, Ueba T, Christie BR, et al. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci U S A. 2003;100:6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 30.Van den Veyver IB, Zoghbi HY. Methyl-CpG-binding protein 2 mutations in Rett syndrome. Curr Opin Genet Develop. 2000;10:275–279. doi: 10.1016/s0959-437x(00)00083-6. [DOI] [PubMed] [Google Scholar]
- 31.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 32.Bellacosa A, Cicchillitti L, Schepis F, et al. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc Natl Acad Sci U S A. 1999;96:3969–3974. doi: 10.1073/pnas.96.7.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet 2009;43:143-166. [DOI] [PMC free article] [PubMed]
- 35.Bellacosa A. Role of MED1 (MBD4) Gene in DNA repair and human cancer. J Cell Physiol. 2001;187:137–144. doi: 10.1002/jcp.1064. [DOI] [PubMed] [Google Scholar]
- 36.Prokhortchouk A, Hendrich B, Jorgensen H, et al. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26:169–181. doi: 10.1128/MCB.26.1.169-181.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JI, Kim SW, Lyons JP, et al. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Develop Cell. 2005;8:843–854. doi: 10.1016/j.devcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 39.Martin Caballero I, Hansen J, Leaford D, Pollard S, Hendrich BD. The methyl-CpG binding proteins Mecp2, Mbd2 and Kaiso are dispensable for mouse embryogenesis, but play a redundant function in neural differentiation. PLoS One. 2009;4:e4315. doi: 10.1371/journal.pone.0004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heyn H, Li N, Ferreira HJ, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christensen BC, Houseman EA, Marsit CJ, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 43.Wang JH. The role of activation-induced deaminase in antibody diversification and genomic instability. Immunol Res. 2013;55:287–297. doi: 10.1007/s12026-012-8369-4. [DOI] [PubMed] [Google Scholar]
- 44.Nabel CS, Jia H, Ye Y, et al. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popp C, Dean W, Feng S, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yildirim O, Li R, Hung JH, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song CX, Szulwach KE, Fu Y, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szulwach KE, Li X, Li Y, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Y, Xu C, Kato A, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell 2012;151:1200-1213. [DOI] [PMC free article] [PubMed]
- 53.Dong E, Gavin DP, Chen Y, Davis J. Upregulation of TET1 and downregulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Transl Psychiatry. 2012;2:e159. doi: 10.1038/tp.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang RR, Cui QY, Murai K, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13:237–245. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 58.Caspary T, Cleary MA, Baker CC, Guan XJ, Tilghman SM. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol Cell Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 60.Verkerk AJ, Pieretti M, Sutcliffe JS, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 61.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Develop. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 62.Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet. 2011;20:670–680. doi: 10.1093/hmg/ddq513. [DOI] [PubMed] [Google Scholar]
- 63.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 64.Ball MP, Li JB, Gao Y, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo JU, Ma DK, Mo H, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xie W, Barr CL, Kim A, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teter B, Osterburg HH, Anderson CP, Finch CE. Methylation of the rat glial fibrillary acidic protein gene shows tissue-specific domains. J Neurosci Res. 1994;39:680–693. doi: 10.1002/jnr.490390609. [DOI] [PubMed] [Google Scholar]
- 69.Takizawa T, Nakashima K, Namihira M, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Develop Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 70.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 71.Feng J, Zhou Y, Campbell SL, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2001;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Levenson JM, Roth TL, Lubin FD, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2005;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 74.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 75.Rampon C, Tang YP, Goodhouse J, Shimizu E, Kyin M, Tsien JZ. Enrichment induces structural changes and recovery from nonspatial memory deficits in CA1 NMDAR1-knockout mice. Nat Neurosci. 2000;3:238–244. doi: 10.1038/72945. [DOI] [PubMed] [Google Scholar]
- 76.Murgatroyd C, Patchev AV, Wu Y, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 77.Roth TL, Zoladz PR, Sweatt JD, Diamond DM. Epigenetic modification of hippocampal Bdnf DNA in adult rats in an animal model of post-traumatic stress disorder. J Psychiatric Res. 2011;45:919–926. doi: 10.1016/j.jpsychires.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aloe L, Iannitelli A, Angelucci F, Bersani G, Fiore M. Studies in animal models and humans suggesting a role of nerve growth factor in schizophrenia-like disorders. Behav Pharmacol. 2000;11:235–242. doi: 10.1097/00008877-200006000-00007. [DOI] [PubMed] [Google Scholar]
- 79.Angelucci F, Brene S, Mathe AA. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry. 2005;10:345–352. doi: 10.1038/sj.mp.4001637. [DOI] [PubMed] [Google Scholar]
- 80.Sinclair D, Tsai SY, Woon HG, Weickert CS. Abnormal glucocorticoid receptor mRNA and protein isoform expression in the prefrontal cortex in psychiatric illness. Neuropsychopharmacology. 2011;36:2698–2709. doi: 10.1038/npp.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology. 2013;38:138–166. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klengel T, Mehta D, Anacker C, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hahn MA, Qiu R, Wu X, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van den Hove DL, Chouliaras L, Rutten BP. The role of 5-hydroxymethylcytosine in aging and Alzheimer's disease: current status and prospects for future studies. Curr Alzheimer Res. 2012;9:545–549. doi: 10.2174/156720512800618008. [DOI] [PubMed] [Google Scholar]
- 85.Chahrour M, Jung SY, Shaw C, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oberle I, Rousseau F, Heitz D, et al. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 87.Hansen RS, Wijmenga C, Luo P, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de Greef JC, Wang J, Balog J, et al. Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am J Hum Genet. 2011;88:796–804. doi: 10.1016/j.ajhg.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klein CJ, Botuyan MV, Wu Y, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Winkelmann J, Lin L, Schormair B, et al. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet. 2012;21:2205–2210. doi: 10.1093/hmg/dds035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen KL, Wang SS, Yang YY, Yuan RY, Chen RM, Hu CJ. The epigenetic effects of amyloid-beta(1-40) on global DNA and neprilysin genes in murine cerebral endothelial cells. Biochem Biophys Res Commun. 2009;378:57–61. doi: 10.1016/j.bbrc.2008.10.173. [DOI] [PubMed] [Google Scholar]
- 92.Desplats P, Spencer B, Coffee E, et al. Alpha-synuclein sequesters Dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. The J Biol Chem. 2011;286:9031–9037. doi: 10.1074/jbc.C110.212589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jowaed A, Schmitt I, Kaut O, Wullner U. Methylation regulates alpha-synuclein expression and is decreased in Parkinson's disease patients' brains. J Neurosci. 2010;30:6355–6359. doi: 10.1523/JNEUROSCI.6119-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M, Martin LJ. Epigenetic regulation of motor neuron cell death through DNA methylation. J Neurosci. 2011;31:16619–16636. doi: 10.1523/JNEUROSCI.1639-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Endres M, Meisel A, Biniszkiewicz D, et al. DNA methyltransferase contributes to delayed ischemic brain injury. J Neurosci. 2000;20:3175–3181. doi: 10.1523/JNEUROSCI.20-09-03175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee HA, Hong SH, Kim JW, Jang IS. Possible involvement of DNA methylation in NKCC1 gene expression during postnatal development and in response to ischemia. J Neurochem. 2010;114:520–529. doi: 10.1111/j.1471-4159.2010.06772.x. [DOI] [PubMed] [Google Scholar]
- 97.Hu CJ, Chen SD, Yang DI, et al. Promoter region methylation and reduced expression of thrombospondin-1 after oxygen-glucose deprivation in murine cerebral endothelial cells. J Cereb Blood Flow Metab. 2006;26:1519–1526. doi: 10.1038/sj.jcbfm.9600304. [DOI] [PubMed] [Google Scholar]
- 98.Wilson ME, Westberry JM. Regulation of oestrogen receptor gene expression: new insights and novel mechanisms. J Neuroendocrinol. 2009;21:238–242. doi: 10.1111/j.1365-2826.2009.01830.x. [DOI] [PubMed] [Google Scholar]
- 99.Kobow K, Jeske I, Hildebrandt M, et al. Increased reelin promoter methylation is associated with granule cell dispersion in human temporal lobe epilepsy. J Neuropathol Exp Neurol. 2009;68:356–364. doi: 10.1097/NEN.0b013e31819ba737. [DOI] [PubMed] [Google Scholar]
- 100.Miller-Delaney SF, Das S, Sano T, et al. Differential DNA methylation patterns define status epilepticus and epileptic tolerance. J Neurosci. 2012;32:1577–1588. doi: 10.1523/JNEUROSCI.5180-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mastronardi FG, Noor A, Wood DD, Paton T, Moscarello MA. Peptidyl argininedeiminase 2 CpG island in multiple sclerosis white matter is hypomethylated. J Neurosci Res. 2007;85:2006–2016. doi: 10.1002/jnr.21329. [DOI] [PubMed] [Google Scholar]
- 102.Oliveira AM, Hemstedt TJ, Bading H. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat Neurosci. 2012;15:1111–1113. doi: 10.1038/nn.3151. [DOI] [PubMed] [Google Scholar]
- 103.Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Human Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guidotti A, Auta J, Davis JM, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- 105.Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci U S A. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Veldic M, Caruncho HJ, Liu WS, et al. DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc Natl Acad Sci U S A. 2004;101:348–353. doi: 10.1073/pnas.2637013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhubi A, Veldic M, Puri NV, et al. An upregulation of DNA-methyltransferase 1 and 3a expressed in telencephalic GABAergic neurons of schizophrenia patients is also detected in peripheral blood lymphocytes. Schizophr Res. 2009;111:115–122. doi: 10.1016/j.schres.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- 109.Sutcliffe JS, Nelson DL, Zhang F, et al. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 110.Eiges R, Urbach A, Malcov M, et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 111.Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 112.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Amato RJ. Inhibition of DNA methylation by antisense oligonucleotide MG98 as cancer therapy. Clin Genitourin Cancer. 2007;5:422–426. doi: 10.3816/CGC.2007.n.029. [DOI] [PubMed] [Google Scholar]
- 114.Amodio N, Leotta M, Bellizzi D, et al. DNA-demethylating and anti-tumor activity of synthetic miR-29b mimics in multiple myeloma. Oncotarget. 2012;3:1246–1258. doi: 10.18632/oncotarget.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao S, Wang Y, Liang Y, et al. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011;63:1376–1386. doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]
- 116.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zuo J, Xia J, Ju F, et al. MicroRNA-148a can regulate runt-related transcription factor 3 gene expression via modulation of DNA methyltransferase 1 in gastric cancer. Mol Cell. 2013;35:313–319. doi: 10.1007/s10059-013-2314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- 119.Koivunen P, Lee S, Duncan CG, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;97:1498–1506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 121.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ma DK, Jang MH, Guo JU, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1224 kb)