Abstract
The pathogenic mechanism(s) contributing to loss of dopamine neurons in Parkinson’s disease (PD) remain obscure. Leucine-rich repeat kinase 2 (LRRK2) mutations are linked, as a causative gene, to PD. LRRK2 mutations are estimated to account for 10 % of familial and between 1 % and 3 % of sporadic PD. LRRK2 proximate single nucleotide polymorphisms have also been significantly associated with idiopathic/sporadic PD by genome-wide association studies. LRRK2 is a multidomain-containing protein and belongs to the protein kinase super-family. We constructed two inducible dopaminergic cell lines expressing either human-LRRK2-wild-type or human-LRRK2-mutant (G2019S). Phenotypes of these LRRK2 cell lines were examined with respect to cell viability, morphology, and protein function with or without induction of LRRK2 gene expression. The overexpression of G2019S gene promoted 1) low cellular metabolic activity without affecting cell viability, 2) blunted neurite extension, and 3) increased phosphorylation at S910 and S935. Our observations are consistent with reported general phenotypes in LRRK2 cell lines by other investigators. We used these cell lines to interrogate the biological function of LRRK2, to evaluate their potential as a drug-screening tool, and to investigate screening for small hairpin RNA-mediated LRRK2 G2019S gene knockdown as a potential therapeutic strategy. A proposed LRRK2 kinase inhibitor (i.e., IN-1) decreased LRRK2 S910 and S935 phosphorylation in our MN9DLRRK2 cell lines in a dose-dependent manner. Lentivirus-mediated transfer of LRRK2 G2019S allele-specific small hairpin RNA reversed the blunting of neurite extension caused by LRRK2 G2019S overexpression. Taken together, these inducible LRRK2 cell lines are suitable reagents for LRRK2 functional studies, and the screening of potential LRRK2 therapeutics.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-013-0208-3) contains supplementary material, which is available to authorized users.
Keywords: Parkinson’s disease (PD), Leucine-rich repeat kinase 2 (LRRK2), Dopaminergic cell lines, RNAi, Kinase assay, Cell viability
Introduction
The pathogenic mechanisms that cause the loss of dopamine neurons in Parkinson’s disease (PD) remain obscure. Leucine-rich repeat kinase 2 (LRRK2)/Dardarin mutations are linked, as a causative gene, to PD [1–4]. LRRK2 mutations are estimated to account for 10 % of familial and between 1 % and 3 % of sporadic PD [5–10]. LRRK2 proximate single nucleotide polymorphisms have also been significantly associated with idiopathic/sporadic PD by genome-wide association studies [3, 4, 11]. LRRK2 is a multi-domain containing protein and belongs to the protein kinase super-family [12, 13]. The 6 domains include: ankyrin repeats, leucine-rich repeats, a guanosine triphosphate-binding Ras of complex protein (ROC), a carboxy-terminal of ROC, a kinase domain, and a WD40 domain [14]. There are several variant forms of LRRK2 harboring mutations in different domains [1, 3], among which, the R1441C/G/H, Y1669C, I2020T, and G2019S mutations are known to be associated with PD [15]. These mutations are located within the ROC–carboxy-terminal of ROC-kinase domain of the LRRK2 protein, affecting the guanosine triphosphatase or the kinase activity; however, it is unclear how these changes influence the normal functions of wild-type (WT) LRRK2 [16]. However, the most frequent mutation is a single nucleotide mutation causing an amino acid substitution of glycine to serine (G2019S) [8, 11, 17]. This G2019S mutation leads to increased LRRK2 kinase activity [18–21]. Importantly, an inactivating mutation of the LRRK2 kinase domain, in concert with the G2019S mutation, has been shown to decrease neurotoxicity [13], thus implicating increased kinase activity as one of the mechanisms of LRRK2-associated PD pathogenesis.
Although the exact biological function(s) of LRRK2 and its role in biochemical pathways are under investigation, several potential substrates have been identified, including LRRK2, Akt1, ezrin/radxin/moesin (ERM) proteins, β-tubulin, eukaryotic initiation factor 4E-binding protein 1, and mitogen-activated kinase 3, 4, 6, and 7 [13, 22–34]. The functional implications of the majority of these potential substrates with respect to LRRK2 patho- and physiological actions remain uncertain. However, the work of Sheng et al. [34] has directly implicated LRRK2 Ser1292 in pathogenic effects in cultured cells.
As the LRRK2 G2019S mutation is causal and contributory to familial and sporadic/idiopathic PD respectively, together accounting for ~2 % of all PD in the North American and UK population [7, 8] and 20–40 % in certain populations [35–37], the development of models that may be predictive in the prosecution of new therapeutics is meritorious. Advancing the development of both small molecules and biologics for PD requires cellular models in which the varying and stable levels of a putative pathogenic gene product can be studied. Ideally, these studies should be undertaken in a dopaminergic background. In parallel, the examination of the WT form of the gene product in an identical context is required to ascribe the distinct pathogenic effects owing to the mutant form. Finally, the cellular models should prove useful for the demonstration that candidate therapeutics protect or reverse the pathogenic action due to putative mutant gene product.
In an effort to develop candidate therapeutics targeting LRRK G2019S we constructed two inducible dopaminergic MN9D cell lines expressing either human LRRK2-WT or human LRRK2-mutant (G2019S), each co-expressing green fluorescent protein (GFP). These LRRK2 cell lines were examined for cell viability, morphology, and LRRK2 functions with or without induction of gene expression [38]. In addition, we used these cell lines to investigate a previously described LRRK2 kinase inhibitor, IN-1 [38] and small hairpin RNA (shRNA)-mediated LRRK2 G2019S gene silencing. Our data, reported herein, indicate that these inducible LRRK2 cell lines are suitable for the study of LRRK2 function and for screening potential LRRK2 targeted therapeutics.
Materials and Methods
Reagents and Chemicals
Lipofectamine 2000, real-time polymerase chain reaction (PCR) universal human LRRK2 probe and Alexa-594 conjugated goat anti-rabbit IgG antibody (Ab) were from Life Technologies (Grand Island, NY, USA). Sodium bicarbonate, hygromycin, Dulbecco’s Modified Eagle Medium, doxycycline (DOX), sodium butyrate, 4’,6-diamidino-2-phenylindole, and trypan blue solutions were from Sigma-Aldrich (St. Louis, MO, USA). The MTS cell viability CellTiter 96 AQueous assay kit was from Promega (Madison, WI, USA). Rabbit monoclonal antihuman LRRK2 Ab, rabbit monoclonal antihuman phospho-LRRK2 S910 Ab, and rabbit monoclonal antihuman phospho-LRRK2 S935 Ab were from Epitomics (Burlingame, CA, USA). Rabbit polyclonal antibeta tubulin III Ab was from Covance (Chantilly, VA, USA). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG and HRP-conjugated goat anti-mouse IgG secondary Abs were from Jackson ImmunoResearch (West Grove, PA, USA). Mouse monoclonal anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) Ab was from Millipore (Billerica, MA, USA). Rabbit monoclonal pan-Akt (C67E7), rabbit polyclonal phospho-Akt (Ser473), and rabbit polyclonal ERM, rabbit monoclonal phospho-Ezrin(Thr567)/Radixin(Thr564)/Moesin(Thr558) (41A3) Abs were from Cell Signaling Technology (Danvers, MA, USA).
Chemical Synthesis
IN-1 was synthesized as described by Deng et al. [38].
DNA Plasmids
The full-length human LRRK2 WT or G2019S genes (7.6 kb) were subcloned in the vector plasmid with IRESeGFP, pBig2iFLAGSocs6IRES2eGFP plasmid, as described previously [39–41].
Stable Cell Lines
A mouse midbrain cell line, MN9D [42], was used for the LRRK2 stable cell line construction. MN9D cells were cultured in Dulbecco’s Modified Eagle Medium supplemented with 10 % fetal bovine serum, and 3.7 g/L sodium bicarbonate at 37 °C, 5 % carbon dioxide. MN9D cells were transfected with plasmid constructs that overexpress full-length human LRRK2 WT or mutant (G2019S) and GFP under the control of a tetracycline-inducible promoter. Low passage number MN9D cells (< 6 passage) were used for Lipofectamine 2000-mediated transfection of the LRRK2 WT or G2019S DNA according to the manufacturer’s specification. Transfected cells were selected with hygromycin (500 μg/mL). These constructs also express GFP (eGFP) using an internal ribosome entry sequence (IRES). LRRK2 WT, G2019S, and eGFP expression was induced with addition of DOX (2 μg/mL). Cell colonies with eGFP expression were selected, expanded, sorted (fluorescence-activated cell sorting), and confirmed for LRRK2 WT or G2019S expression.
Western Blotting
Total protein from MN9DLRRK2WT and MN9DLRRK2G2019S cells was extracted in RIPA buffer (50 mM Tris–hydrochloric acid, pH 7.4; 1 % NP-40; 0.25 % sodium deoxycholate; 150 mM sodium chloride) containing protease inhibitors (1 mM ethylenediaminetetraacetic acid; 1 mM phenylmethylsulphonyl fluoride) or using a RNA/DNA/Protein Purification Kit (Norgen, Thorold, ON, Canada). Equal amounts of total protein (20 μg) from each sample were subjected to denaturing polyacrylamide gel electrophoresis [4–20 % Bis-Tris gradient gel (BioRad, Hercules, CA, USA) or NuPAGE Novex 3–8 % Tris-Acetate Gel (Life Technologies)]. LRRK2 protein expression levels were detected using a rabbit polyclonal anti-human LRRK2 antibody (1:5000 dilution; Epitomics/MJFF#2 [c41-2]) followed by incubation with HRP-conjugated goat anti-rabbit IgG secondary antibody (1:2000) and chemiluminescent detection (Perkin Elmer, Waltham, MA, USA). Other Abs used were GAPDH (1:20,000), rabbit polyclonal anti-human phospho-LRRK2 S910 and S935 antibodies (1:5000), rabbit monoclonal pan-Akt (C67E7) (1:1000), rabbit polyclonal anti-phospho-Akt (Ser473)(1:1000), rabbit polyclonal ERM Ab (1:1000), rabbit monoclonal anti-phospho-Ezrin(Thr567)/Radixin(Thr564)/Moesin(Thr558) (41A3) Ab (1:1000), and rabbit anti-4E-binding protein 1 Ab (1:1000).
Immunocytochemistry and Neurite Extension Assay
MN9DLRRK2G2019S or MN9DLRRK2WT 5 × 104 cells/well were plated on polyethylenemine-coated coverslips and differentiated for 6 days with 2 mM sodium butyrate. On day 6, DOX was added to induce GFP and LRRK2 WT or G2019S expression. Forty-eight hours after the addition of DOX, cells were fixed in a 4 % paraformaldehyde, 4 % sucrose solution, permeabilized in 0.1 % Triton-X-100/PBS, and blocked in 10 % normal goat serum. Cells were probed with LRRK2 Ab (1:50) or rabbit polyclonal antibeta-tubulin III Ab (1:2000) followed by Alexa-594-conjugated goat anti-rabbit IgG Ab (1:200) and 4’,6-diamidino-2-phenylindole (300 nM) staining. The fluorescent labeled images were captured and analyzed with AxioVision software equipped AxioPlan2 Zeiss fluorescent microscope (Carl-Zeiss, Thornwood, NY, USA). For neurite measurements, MN9DLRRK2G2019S cells were plated on polyethylenemine-coated 12-mm coverslips in a 24-well plate and differentiated with 2 mM n-butyrate for 6 days. DOX (250 ng/mL) was added to induce LRRK2 G2019S expression for 48 h. Cells were treated with IN-1 or transduced with lenti-shRNA and subjected to immunocytochemical staining as described above. Pictures of 8 fields were taken for each sample and lengths of all beta-tubulin II-positive neurites were measured with Nikon NIS Elements software. About 40–100 neurites was measured for each condition.
RNA Interference With Lenti-shRNA
The shRNA p4 was designed according to the published LRRK2 G2019S allele-specific p4 sequence [43]. The shRNA p4 sequence 5’-GAGATTGCTGACTGCAGTACCTGACCCATGCTGTAGTCAGCAATCTCTT-3’ and the scrambled shRNA (shRNA) sequence 5’-GGAATACGTACGGCTTAGTCCTGACCCAACT AAGCCGTACGTATTCCTT-3’ were, respectively, cloned into the pENTR6/U6 vector (Life Technologies) and then subcloned into the pLenti6-/BLOCK-iT-DEST vector (Life Technologies) via Gateway cloning. The resulting plasmids were sequenced to confirmed accuracy. Lentivirus was packaged in 293 T cells by cotransfecting the cells with the above pLenti6-shRNA vectors and the ViraPower Packaging Mix (Life Technologies) according to the manufacturer’s specifications. MN9DLRRK2G2019S and MN9DLRRK2WT cells were plated in a 6-well plate and transduced with shRNAp4 or scrambled shRNA-expressing lentivirus at a multiplicity of infection of 50. MN9DLRRK2WT cells were induced with 250 ng/mL DOX before transduction owing to a very low level of LRRK2 WT expression. Cells were harvested 72 h after transduction using a RNA/DNA/Protein Purification Kit (Norgen Biotek). Quantitative real-time reverse transcription-PCR was performed to measure LRRK2 mRNA levels with universal human LRRK2 probe (Life Technologies,) and Applied Biosystems (Foster City, CA) 7900HT Fast Real-Time PCR System. For each sample, 1 μg of total RNA was used for complementary DNA synthesis. All data were normalized to mouse GAPDH expression as an internal control. Expression of shRNA was inferred from positive expression of the blasticidin gene within the same construct. The LRRK2 protein expression levels were examined by western blot assays as described above.
Results
Human LRRK2 Protein Expression in MN9DLRRK2 Stable Cell Lines with Doxycycline Induction
Full-length complementary DNA of LRRK2WT or LRRK2G2019S was amplified by high-fidelity PCR and cloned into the pBig2iFLAGSocs6IRES2eGFP, which is a tetracycline-responsive autoregulated bi-directional expression vector with an IRESeGFP cassette [39, 40]. Stably transfected human LRRK2 inducible cell lines were examined first for GFP expression and followed by fluorescence-activated cell sorting for GFP expression. Selected GFP-positive cells were expanded and confirmed for LRRK2 gene and protein expression. Undifferentiated MN9DLRRK2WT cells were induced with increasing concentrations of DOX (ranged from 0 to 4000 ng/mL) for 48 h. The transcription and translation of LRRK2 was turned on in response to DOX in a precise and dose-dependent manner (Fig. 1). The expression of LRRK2 mRNA in sodium butyrate-differentiated MN9DLRRK2WT cells followed a similar induction profile as the undifferentiated cells (Fig. 1A, top right). The expression level of LRRK2 mRNA in both undifferentiated (Fig. 1A, top left) and differentiated MN9DLRRK2WT cells (Fig. 1A, top right) increased approximately 10-fold in response to DOX over the range of 200–4000 ng/mL. The DOX-induced LRRK2 protein expression in MN9DLRRK2WT cells was also dose-dependent (Fig. 1B, left). Both undifferentiated and differentiated MN9DLRRK2G2019S cells displayed a similar dose-dependent induction of LRRK2 gene and protein expression (Fig. 1A, bottom panels, Fig. 1B, right) as MN9DLRRK2WT cells. However, the induction of LRRK2 mRNA expression in both undifferentiated (Fig. 1A, bottom left) and differentiated MN9DLRRK2G2019S cells (Fig. 1A, bottom right) was much greater than in the MN9DLRRK2WT cells, with approximately 120-fold mRNA induction in undifferentiated and 40-fold induction in differentiated cells in response to DOX. No human or mouse LRRK2 protein was detected in MN9D parental cells, which are of mouse origin, with or without DOX treatment (data not shown).
Fig. 1.
Doxycycline (DOX)-induced LRRK2 expression in MN9DLRRK2WT and MN9DLRRK2G2019S cells. DOX was added to the culture media 48 h before harvesting using an RNA/DNA/Protein Purification Kit (Norgen, Thorold, ON, Canada). For differentiated cells, cells were treated with 2 mM sodium butyrate for 6 days before the addition of DOX. a Quantitative real-time reverse transcription-polymerase chain reaction was performed to measure LRRK2 messenger RNA (mRNA) levels of DOX-induced undifferentiated and differentiated MN9DLRRK2WT (top panels) or MN9DLRRK2G2019S (bottom panels) cells. b Western blot analysis of LRRK2 expression upon DOX induction. For MN9DLRRK2WT cells, 20 μg of total protein was loaded for each sample; for MN9DLRRK2G2019S cells, 5 μg of total protein was loaded for each sample. β-Actin was used as a loading control. Quantitative determinations of intensities of LRRK2 signals normalized to β-actin were shown at the bottom of the Western blot
Cellular Expression Pattern of Human LRRK2 in MN9DLRRK2 Cell Lines
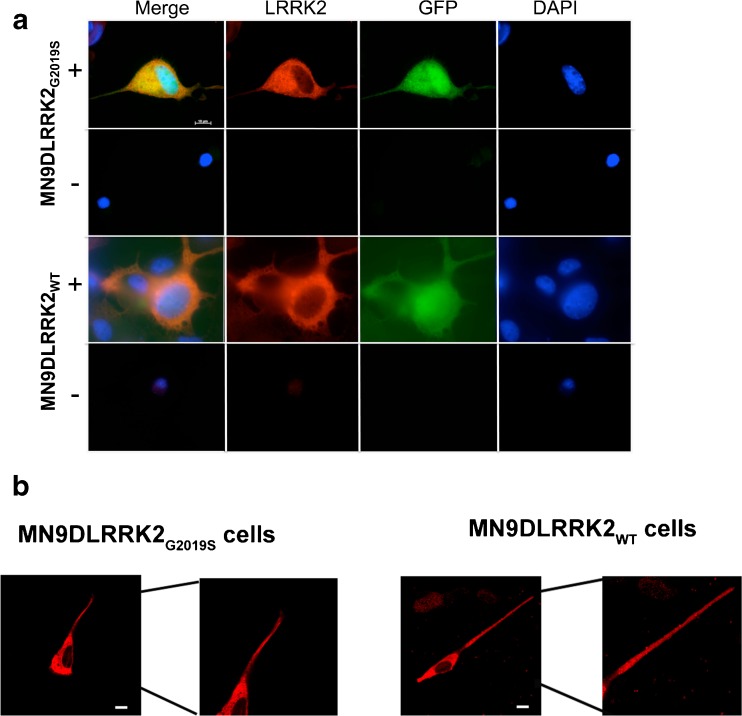
The stably-expressing cell lines were examined for the cellular pattern of human LRRK2 by immunocytochemistry. Stable cell lines grown on coverslips and differentiated to a neuronal phenotype for 6 days with sodium butyrate were either induced with DOX or remained uninduced (no DOX). Forty-eight hours later, cells were subjected to immunocytochemistry staining for human LRRK2. The human LRRK2 (Fig. 2A, red) was detected only in DOX-induced (+), GFP-positive (Fig. 2A, green) MN9DLRRK2WT and MN9DLRRK2G2019S cells (Fig. 2A). The LRRK2 expression appears cytosolic, with little or no nuclear staining. There was no significantly enriched subcellular expression of LRRK2 noted. LRRK2 was also detected in neurites following sodium butyrate differentiation (Fig. 2B). A marked blunting of neurite outgrowth was observed in MN9DLRRK2G2019S expressing cells, a phenotypic feature studied in subsequent experiments (see Fig. 4).
Fig. 2.
LRRK2 immunocytochemistry. a LRRK2 was detected (red) only in doxycycline (DOX) induced (+) green fluorescent protein (GFP) positive (green) MN9DLRRK2WT and MN9DLRRK2G2019S cells compared with cells grown in the absence of DOX (−). b LRRK2 was expressed in the neurites of sodium butyrate-differentiated DOX-induced cells. Parts of the image showing the neurites are enlarged to show LRRK2 expression. Image acquisition was with 40× magnification; scale bar is 10 μm. DAPI 4’,6-diamidino-2-phenylindole
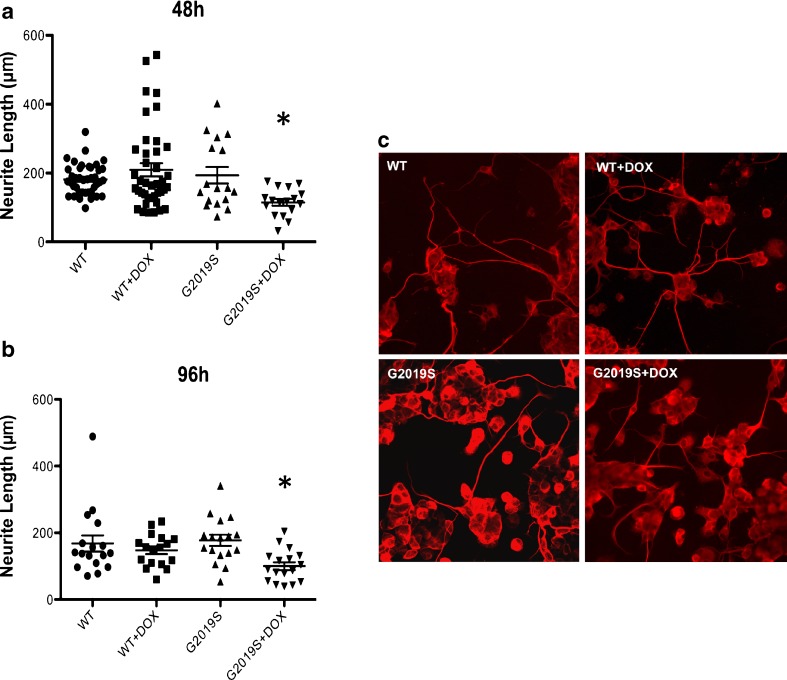
Fig. 4.
Neurite length is shortened in MN9DLRRK2G2019S cells with doxycycline (DOX) induction of LRRK2 G2019S. MN9DLRRK2WT or MN9DLRRK2G2019S cells were plated on polyethylenemine-coated 12-mm coverslips in a 24-well plate and differentiated with 2 mM sodium butyrate for 6 days. DOX (250 ng/mL) was added to the cells to induce LRRK2 expression for 48 h (a) or 96 h (b). Cells were fixed and immunocytochemically stained for β-tubulin. For each coverslip, pictures of 8 fields were taken and lengths of all neurites were measured with Nikon NIS Elements software. *p < 0.05 vs control; one-way analysis of variance. c Representative images of differentiated MN9DLRRK2WT or MN9DLRRK2G2019S cells with [wild-type (WT) + DOX, G2019S + DOX] or without (WT, G2019S) DOX induction are presented
LRRK2 Effects on Cell Viability
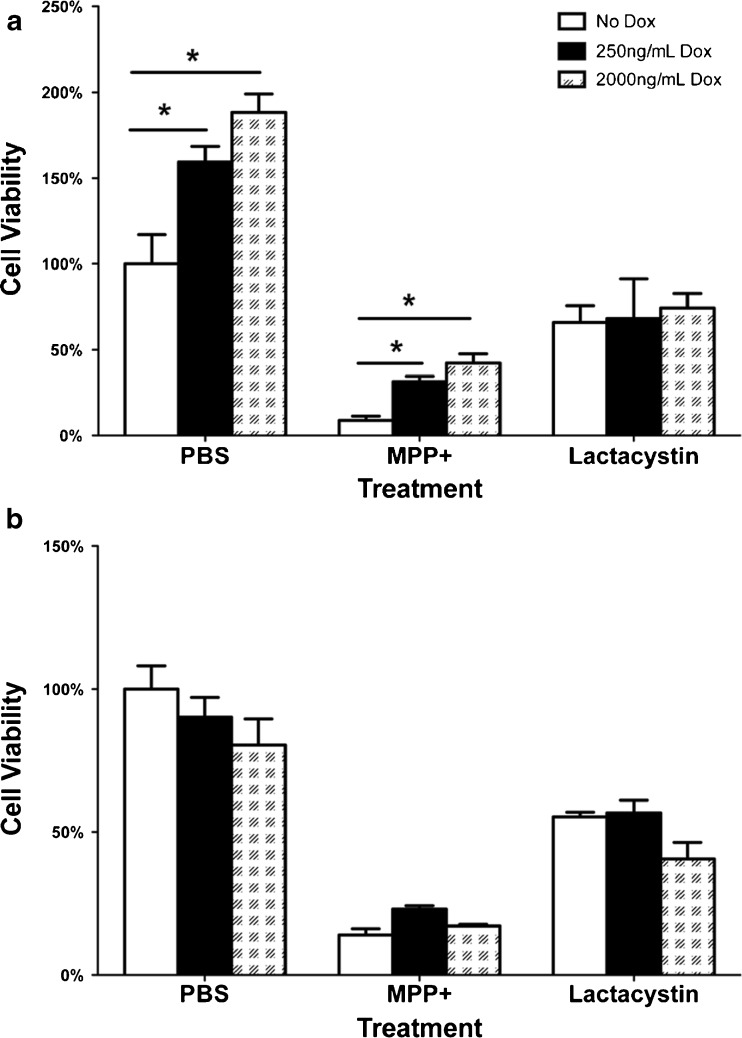
The cell viability of MN9DLRRK2WT and MN9DLRRK2G2019S cells following LRRK2 induction only or in combination of the addition of toxicants (MPP+ or lactacystin) were evaluated using an MTS assay. When LRRK2 was induced in the MN9DLRRK2WT cells, viability increased in a DOX dose-dependent manner relative to uninduced cells (Fig. 3A, PBS). When MN9DLRRK2WT were challenged with cytotoxic doses of MPP+ (Fig. 3A, MPP+), LRRK2 WT induction increased cellular viability in a DOX dose-dependent fashion. Cell viability with lactacystin treatment was not different between DOX treatment groups (Fig. 3A, lactacystin). In contrast, DOX induction of mutant LRRK2 expression (MN9DLRRK2G2019S) resulted in no changes in cellular viability at baseline or following toxicant challenge (Fig. 3B). Similar results were obtained with the lactate dehydrogenase cytotoxicity assay (i.e., DOX induction of LRRK2 WT, but not LRRK2 G2019S, increased cellular viability over time (data not shown).
Fig. 3.
Cell viability of MN9DLRRK2WT and MN9DLRRK2G2019S lines in response to LRRK2 induction only or in combination with 1-methyl-4-phenylpyridinium iodide (MPP) + or lactacystin. a MN9DLRRK2WT cells were plated in a 96-well configuration and induced with 250 ng/mL or 2000 ng/mL doxycycline (DOX) for 48 h, followed by incubation with either phosphate buffered saline (PBS), 500 μM MPP + or 5 μM lactacystin. Cell viability was determined by an 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay. b MN9DG2019S cells were plated in 96-well configuration and induced with 250 ng/mL or 2000 ng/mL DOX for 48 h, followed by incubation with either PBS, 500 μM MPP + or 5 μM lactacystin. Cell viability was determined by an MTS assay. *p < 0.05 vs No DOX; one-way analysis of variance
LRRK2 G2019S Expression Blunts Neurite Extension and is Reversed by Treatment With Kinase Inhibitor IN-1
Sodium butyrate-differentiated MN9DLRRK2 cells were induced with DOX and examined for neurite extension (Fig. 4). Quantitative neurite extension was undertaken at 48 (Fig. 4A) and 96 h (Fig. 4B) with (+DOX) or without DOX induction. MN9DLRRK2G2019S (G2019S), but not MN9DLRRK2WT (WT), cells showed a statistically significant blunting of neurite extension only when LRRK2 was induced by DOX. Photomicrographs of MN9D neurites from the different cell lines under the different conditions are shown in Fig. 4C.
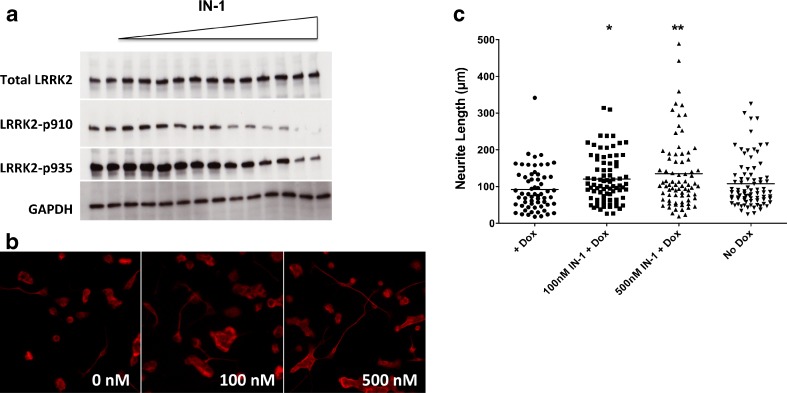
A specific and potent kinase inhibitor, IN-1 [38], was examined to determine whether inhibition of LRRK2 G2019S activity would alter the pathobiologic neurite phenotype. We first confirmed that IN-1 would reduce LRRK2 phosphorylation of endogenous serine residues at positions 910 and 935 (Fig. 5A). As anticipated, phosphorylation of both sites was reduced in an IN-1 concentration-dependent manner. Following immunocytochemistry for beta-tubulin III, we noted that blunted neurite extension mediated by LRRK2 G2019S overexpression was rescued by IN-1 treatment (Fig. 5B). Quantitative neurite extension studies confirmed the morphological changes and showed that the blunted neuritic phenotype was abrogated by IN-1 at both 100 and 500 nM. These data demonstrate that MN9DLRRK2G2019S cells have an (DOX) inducible neuritic phenotype (i.e., shorter length) that can be largely reversed by a potent and specific inhibitor of LRRK2, IN-1.
Fig. 5.
Evaluation of the LRRK2 inhibitor, IN-1, in MN9DLRRK2G2019S cells. a The LRRK2 inhibitor IN-1 decreases LRRK2 phosphorylation at amino acids 910 and 935. MN9DLRRK2G2019S cells were treated with kinase inhibitor IN-1 at various concentrations (30, 100, 300, and 3000 nM) for 90 mins followed by Western blot analysis of total LRRK2, phospho-LRRK2 at 910 and 935 expression. Equal amounts of total protein were loaded for each sample. b Neurite length shortening is reversed in MN9DLRRK2G2019S cells with the addition of the LRRK2 inhibitor IN-1. MN9DLRRK2G2019S cells were plated on polyethylenemine-coated 12-mm coverslips in a 24-well plate and differentiated with 2 mM sodium butyrate for 6 days. Doxycycline (DOX) (250 ng/mL) was added to the cells to induce LRRK2 G2019S expression for 48 h followed by the addition of IN-1 (100 nM or 500 nM). Twenty-four hours later, cells were fixed and immunocytochemically stained for β-tubulin. Representative images are presented for the IN-1 treatment at the concentration of 0 (+DOX), 100 nM and 500 nM. c Quantitative measurements of neurite lengths of MN9DLRRK2G2019S cells. For each coverslip, pictures of 8 fields were taken and lengths of all neurites were measured using Nikon NIS Elements software. MN9DLRRK2G2019S without DOX induction (no DOX) was used as a control. *p < 0.05 vs control; **p < 0.01 vs control; one-way analysis of variance. GAPDH glyceraldehyde 3-phosphate dehydrogenase
LRRK2 G2019S Mediated Blunting of Neurite Length is Reversed by Allele-Specific RNAi
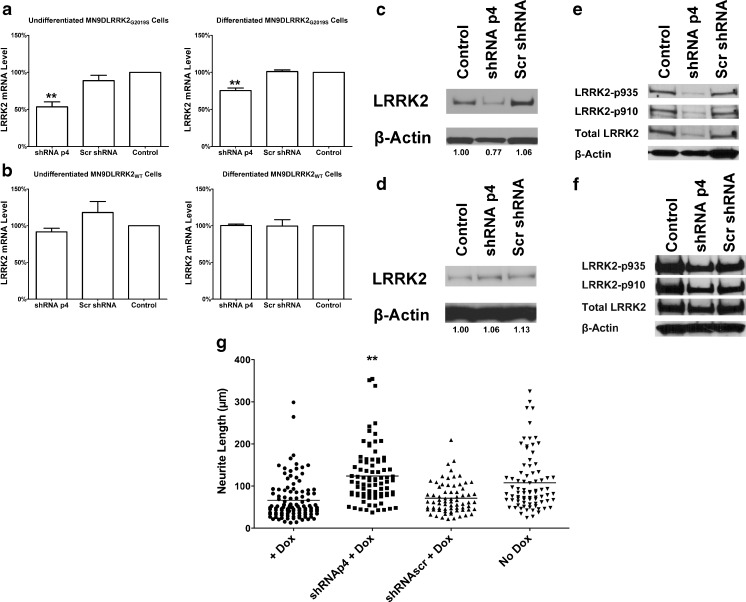
Undifferentiated and differentiated MN9DLRRK2 cells were transduced with lentiviral constructs (multiplicity of infection = 50) expressing a shRNA directed against the G2019S allele (p4) or a sequence scrambled control. PBS treatment was also included as a control for lentivirus transduction. In both undifferentiated and differentiated MN9DLRRK2G2019S cells lentiviral transduction of allele-specific p4, but not the control scrambled sequence, resulted in decreased expression of LRRK2 G2019S messenger RNA (mRNA) (Fig. 6A). By contrast, p4 transduction of MN9DLRRK2WT produced no significant decline in LRRK2 WT mRNA content in either undifferentiated or differentiated cells (Fig. 6B). Furthermore, the allele-specific knockdown of G2019S gene product led to a decrease in LRRK2 protein expression in MN9DLRRK2G2019S (Fig. 6C), but not in MN9DLRRK2WT cells (Fig. 6D). In addition, the LRRK2 phosphorylation at amino acids 910 and 935 was decreased in p4 transduced MN9DLRRK2G2019S (Fig. 6E), but not in the MN9DLRRK2WT cells (Fig. 6F). Last, we addressed whether lentiviral transduction could reverse the blunted neuritic phenotype engendered by LRRK2 G2019S induction. As shown in Fig. 6G, transduction with lentiviral p4 shRNA, but not the scrambled shRNA, reversed the neuritic shortening in DOX-induced differentiated MN9DLRRK2G2019S cells.
Fig. 6.
Evaluation of small hairpin RNA (shRNA)p4-mediated allele specific knockdown of LRRK2 G2019S in MN9DLRRK2G2019S cells. a The shRNAp4, but not scrambled shRNA (Scr shRNA), significantly decreased LRRK2 G2019S messenger RNA (mRNA) levels in both undifferentiated and differentiated MN9DLRRK2G2019S cells. Only the shRNAp4 exhibited a significant decrease in LRRK2 mRNA level [**p < 0.01, one-way analysis of variance (ANOVA)]. Cell differentiation was induced by treating MN9DLRRK2G2019S cells with 2 mM sodium butyrate for 6 days before transduction. Cells were then processed in the same way as undifferentiated cells. Similar to undifferentiated cells, only the shRNA p4 exhibited significant decrease in LRRK2 mRNA level (**p < 0.01 one-way ANOVA). b The shRNAp4 did not inhibit wild-type (WT) LRRK2 expression. No significant difference in LRRK2 mRNA level was observed in these experimental groups. Error bars indicate the standard error of the mean and represent 3 independent experiments. c Western blot showing a decrease of LRRK2 G2019S protein in Lenti-shRNAp4-transduced MN9DLRRK2G2019S cells. β-Actin is used as a loading control. Quantitative determinations of intensities of LRRK2 signals normalized to β-actin were shown at the bottom of the Western blot. d Western blot showing no change of WT LRRK2 protein in lenti-shRNAp4-transduced MN9DLRRK2WT cells. β-Actin is used as a loading control. Quantitative determinations of intensities of LRRK2 signals normalized to β-actin were shown at the bottom of the Western blot. e Western blot showing decreases of LRRK2 phosphorylation at amino acids 910 and 935 in lenti-shRNAp4-transduced MN9DLRRK2G2019S cells, but not in (f) MN9DLRRK2WT cells. g Neurite length shortening is reversed in MN9DG2019S cells following lenti-shRNAp4 transduction. MN9DLRRK2G2019S cells were plated on polyethylenemine-coated 12-mm coverslips in a 24-well plate and differentiated with 2 mM sodium butyrate for 6 days. Forty-eight hours before transduction, doxycycline (DOX) (250 ng/mL) was added to the cells to induce LRRK2 G2019S expression. Seventy-two hours after transduction, cells were fixed and immunocytochemically stained for β-tubulin. For each coverslip, pictures of 8 fields were taken and lengths of all neurites were measured using Nikon NIS Elements software. MN9DLRRK2G2019S without DOX induction (no DOX) was used as a control. **p < 0.01 vs control; one-way ANOVA
Discussion
The MN9DLRRK2 cell lines described herein can be used to study LRRK2 biology and serve as a cellular platform for LRRK2 therapeutics development. In the case of PD, there have been several LRRK2 animal models reported that include animals from invertebrates to nonhuman primates [44]; however, none demonstrate the progressive degenerative features of human PD. Such animal models may be useful for examination of specific questions of gene product function and/or disease pathogenesis, but appear limited in that they do not recapitulate the many features of PD. To initiate therapeutics development it is optimal to have a model where the cellular content of a putatively pathogenic mutant gene product can be compared across a range of steady-state levels. In addition, a comparator line harboring the nonmutant or WT version of the same gene product is ideal to ensure that the pathophysiology attributed to the mutant gene product can be distinguished, if possible, from that due to expression of the WT form.
Human LRRK2 PD-associated mutations are manifest as autosomal dominant [4], and account for a substantial proportion of familial PD and also are implicated in sporadic PD [5–10]. Additionally, genome-wide association studies studies have identified a single nucleotide polymorphism closely linked to the LRRK2 locus, suggesting a potential role in sporadic/idiopathic PD [45]. LRRK2-targeted therapeutics development appears to be a promising avenue for the potential treatment of symptomatic G2019S gene carriers. In some studies mutant LRRK2, particularly G2019S, is cytotoxic when over-expressed in cultured cells [18, 46], Drosophila [30, 47], Caenorhabditis elegans [12], and viral vector transduced mice [21]; however, no apparent neuronal loss was observed in transgenic mice carrying LRRK2 mutant genes alone [48–50]. This discordance raises the question as to what levels of LRRK2 gene product are most relevant for the study of PD pathogenesis and also for therapeutics development.
In our study, we generated inducible stable cell lines expressing LRRK2 G2019S, using a previously reported bicistronic and auto-regulated stable transfection strategy [39–41]. The parental MN9D cell line used to make inducible stable cell lines was selected because of its dopaminergic properties when differentiated [51] and because its murine origins allow for the detection, with appropriate Abs, of introduced genes expressing human LRRK2. We studied the following characteristics of the MN9DLRRK2 stably transfected cell lines: LRRK2 expression at the mRNA and protein levels, lactate dehydrogenase release, MTS reduction cell viability, cellular morphology, LRRK2 interaction with toxicants MPP + and lactacystin, effects of LRRK2 kinase inhibitor treatment, and effects of LRRK2 G2019S allele-specific shRNA gene knockdown.
We showed that increased expression of WT LRRK2 in MN9DLRRK2WT cells caused an apparent increase in cell viability. No significant change in cell viability was observed when LRRK2 was induced in MN9DLRRK2G2019 cells. When MN9DLRRK2WT cells were challenged with MPP + there was significant decline in cell viability in the absence of LRRK2 induction and, interestingly, cell viability significantly increased when LRRK2 was induced. MPP + treatment produced substantial decline in cell viability in MN9DLRRK2G2019S cells that was not ameliorated by the induction of LRRK2. It appeared that LRRK2 WT provided protection against a neurotoxin that caused mitochondrial dysfunction. These observations were in line with the recent C. elegans study, which showed that LRRK2 WT, but not LRRK2 mutant, protected dopaminergic neurons against rotenone or paraquat toxicity, agents which compromise mitochondrial function [52]. The underlying mechanisms by which LRRK2 WT protected mitochondrial dysfunction remained to be determined. The MN9DLRRK2 cell models enable further studies on the association of LRRK2 action and mitochondrial function. In both MN9DLRRK2WT and MN9DLRRK2G2019S cells treatment with lactacystin, a proteosomal inhibitor, caused a decline in cell viability that was unaffected by induction of either WT or G2019S mutant LRRK2, suggesting that in the MN9D cellular context LRRK2 does not affect proteosome function.
We also studied the effect of LRRK2 expression on MN9D neurite outgrowth after differentiation with sodium butyrate. As reported by other investigators in different cell types [53–59], we also observed that the overexpression of LRRK2 G2019S results in shortened neuritic extensions. We explored potential molecular contributors to this pathogenic neuritic phenotype. We sought to determine whether ERM phosphorylation may be involved given that in LRRK2 G2019S transgenic hippocampal neurons axonal length was reduced and required increased ERM phosphorylation [58]. However, in differentiated and DOX-induced MN9DLRRK2G2019S cells no change in overall ERM or ERM phosphorylation levels were observed (data not shown). Unlike primary hippocampal neurons that manifest marked axonal and dendritic features, MN9D neurites are less functionally specified [51]. Whether neurite blunting, which occurs in the absence of MN9DLRRK2G2019S cell death, is due to specific changes that alter the cytoskeleton and/or produce metabolic dysfunction is not known.
One of the goals for construction of the LRRK2 MN9D cell lines was to study candidate therapeutic strategies directed against the LRRK2 target. The G2019S mutant form of LRRK2 conveys an increase in kinase activity compared with the WT [13, 18, 23, 29, 30, 46, 60–63]. Not surprisingly, the causal role of dysregulated kinase activity of G2019S has spawned interest in the development of kinase inhibitors, which, if potent and specific, represent disease-modifying therapeutics for PD patients harboring the LRRK2 G2019S allele. One pharmacophore, IN-1 kinase inhibitor, was described by Deng et al. [38] as having an IC50 in the low nM range and high specificity for LRRK2 [38]. We evaluated IN-1 in both MN9DLRRK2WT and MN9DLRRK2G2019S cell lines. The data indicate that IN-1 does inhibit LRRK2 phosphorylation at both S910 and S935 without affecting the levels of total LRRK2 protein. In addition, we tested whether LRRK2 G2019S inhibition would reverse the pathogenic action of dysregulated kinase activity. Our observations indicate that this was, indeed, the case. Notably, IN-1 reversed the neurite blunting phenotype in a dose-dependent manner. The literature suggests that S910 and S935 are autophosphorylation sites on LRRK2 [64–66], although recent work reveals that S1292, at least in some cellular contexts, is also a pathogenically relevant LRRK2 autophophosphorylation site [34]. The IN-1 dose-dependent reduction of phosphoserine residues on LRRK2 in both cell lines support their use in the prosecution of small molecule kinase inhibitors. The ability to independently modulate LRRK2 levels during cell-based drug discovery efforts is another advantage of the MN9DLRRK2 cell lines in that LRRK2-mediated pathophysiological effects, and kinase inhibitor interdiction, may be more readily discovered at particular steady-state levels of LRRK2.
The MN9DLRRK2G2019S cell line was also used to evaluate LRRK2 transcript knockdown promoted by lentiviral transduction of shRNAs targeting the mutant allele. The results indicate that the p4 shRNA was effective in decreasing G2019S mRNA and protein levels in both undifferentiated and differentiated MN9DLRRK2G2019S cells. In addition, we showed that p4 was allele specific as it did not decrease WT LRRK2 expression. Most importantly, the neurite blunting phenotype engendered by LRRK2 G2019S was reversed by lentiviral p4 transduction. Prior work using RNA interference, introduced by transient transfection, has made evident that the G2019S target sequence, when embedded in a synthetic substrate, can be effectively targeted relative to WT sequence [43]. Our results with shRNA transduced by lentiviral vector are, to our knowledge, the first demonstration of allele selective knockdown of a native G2019S transcript and, importantly, the reduction in mutant gene product levels. This finding portends the rapid extrapolation of these constructs into an in vivo model.
Our key finding, blunted neurites upon G2019S induction without cytotoxicity, is in agreement with other reported results. Dächsel et al. [59] observed that expression of the G2019S mutant in primary neurons from transgenic mice resulted in diminished neurite outgrowth and branching [59]. Similarly, in transfected primary rat cortical neurons the forced expression of LRRK2 G2019S also resulted in neuritic shortening [56]. In MN9D cells, the induction of mutant G2019S gene does not cause cytotoxicity. While other investigators have reported toxicity when LRRK2 is overexpressed this may be function of the levels of gene product. In our stably transfected MN9DLRRK2G2019S cell line we observed a greater than 40-fold increase in transcript level and marked elevation of gene product. Unlike transiently transfected cells these MN9DLRRK2G2019S cells are selected for an integrated transgene and expanded in the absence of DOX. Whether this process of cell line construction or, alternatively, the nature of fusion cell line account for the absence of G2019S toxicity is unknown.
Our inducible stable MN9D cell lines appear useful for the study of LRRK2 biology in the context of a dopaminergic background and over a range of gene product levels. The absence of cytotoxicity and presence of a neuritic blunting phenotype in the MN9DLRRK2G2019S cells enable their use for the evaluation of candidate therapeutics, small molecule kinase inhibitors, and also RNA interference strategies.
Electronic supplementary material
(PDF 510 kb)
Acknowledgments
This work was supported RC2NS069450 (NIH/NINDS/ARRA) to HJF and Department of Defense grant USAMRMC11341009 to HJF. We thank Dr. M. Cookson and Ms. A. Kaganovich from NIA/NIH (Bethesda, MD, USA) for their valuable technical advice on neurite assay, and Amanda Edwards for her help with the evaluation of the cell lines.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Footnotes
Liang Huang and Mika Shimoji Joint first authors.
References
- 1.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Paisan-Ruiz C, Lang AE, Kawarai T, Sato C, Salehi-Rad S, Fisman GK, et al. LRRK2 gene in Parkinson disease: mutation analysis and case control association study. Neurology. 2005;65:696–700. doi: 10.1212/01.WNL.0000167552.79769.b3. [DOI] [PubMed] [Google Scholar]
- 3.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Funayama M, Hasegawa K, Kowa H, Saito M, Tsuji S, Obata F. A new locus for Parkinson's disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann Neurol. 2002;51:296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- 5.Zabetian CP, Samii A, Mosley AD, Roberts JW, Leis BC, Yearout D, et al. A clinic-based study of the LRRK2 gene in Parkinson disease yields new mutations. Neurology. 2005;65:741–744. doi: 10.1212/01.WNL.0000172630.22804.73. [DOI] [PubMed] [Google Scholar]
- 6.Tan EK, Skipper LM. Pathogenic mutations in Parkinson disease. Hum Mutat. 2007;28:641–653. doi: 10.1002/humu.20507. [DOI] [PubMed] [Google Scholar]
- 7.Nichols WC, Pankratz N, Hernandez D, Paisan-Ruiz C, Jain S, Halter CA, et al. Genetic screening for a single common LRRK2 mutation in familial Parkinson's disease. Lancet. 2005;365:410–412. doi: 10.1016/S0140-6736(05)17828-3. [DOI] [PubMed] [Google Scholar]
- 8.Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, et al. A common LRRK2 mutation in idiopathic Parkinson's disease. Lancet. 2005;365:415–416. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- 9.Farrer M, Stone J, Mata IF, Lincoln S, Kachergus J, Hulihan M, et al. LRRK2 mutations in Parkinson disease. Neurology. 2005;65:738–740. doi: 10.1212/01.WNL.0000169023.51764.b0. [DOI] [PubMed] [Google Scholar]
- 10.Di Fonzo A, Rohe CF, Ferreira J, Chien HF, Vacca L, Stocchi F, et al. A frequent LRRK2 gene mutation associated with autosomal dominant Parkinson's disease. Lancet. 2005;365:412–415. doi: 10.1016/S0140-6736(05)17829-5. [DOI] [PubMed] [Google Scholar]
- 11.Hardy J. Genetic analysis of pathways to Parkinson disease. Neuron. 2010;68:201–206. doi: 10.1016/j.neuron.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Tan YC, Poulose S, Olanow CW, Huang XY, Yue Z. Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson's disease R1441C/G mutants. J Neurochem. 2007;103:238–247. doi: 10.1111/j.1471-4159.2007.04940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, et al. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 14.Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson's disease: protein domains and functional insights. Trends Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Biskup S, West AB. Zeroing in on LRRK2-linked pathogenic mechanisms in Parkinson's disease. Biochim Biophys Acta. 2009;1792:625–633. doi: 10.1016/j.bbadis.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsika E, Moore DJ. Mechanisms of LRRK2-mediated neurodegeneration. Curr Neurol Neurosci Rep. 2012;12:251–260. doi: 10.1007/s11910-012-0265-8. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez D, Paisan Ruiz C, Crawley A, Malkani R, Werner J, Gwinn-Hardy K, et al. The dardarin G 2019 S mutation is a common cause of Parkinson's disease but not other neurodegenerative diseases. Neurosci Lett. 2005;389:137–139. doi: 10.1016/j.neulet.2005.07.044. [DOI] [PubMed] [Google Scholar]
- 18.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB, et al. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci U S A. 2005;102:18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson's disease. Nat Rev Neurosci. 2010;11:791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee BD, Shin JH, VanKampen J, Petrucelli L, West AB, Ko HS, et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nat Med. 2010;16:998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohta E, Kawakami F, Kubo M, Obata F. LRRK2 directly phosphorylates Akt1 as a possible physiological substrate: impairment of the kinase activity by Parkinson's disease-associated mutations. FEBS Lett. 2011;585:2165–2170. doi: 10.1016/j.febslet.2011.05.044. [DOI] [PubMed] [Google Scholar]
- 23.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, et al. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O'Neill E, Meitinger T, et al. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum Mol Genet. 2006;15:223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- 25.Hsu CH, Chan D, Greggio E, Saha S, Guillily MD, Ferree A, et al. MKK6 binds and regulates expression of Parkinson's disease-related protein LRRK2. J Neurochem. 2010;112:1593–1604. doi: 10.1111/j.1471-4159.2010.06568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito G, Okai T, Fujino G, Takeda K, Ichijo H, Katada T, et al. GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson's disease. Biochemistry. 2007;46:1380–1388. doi: 10.1021/bi061960m. [DOI] [PubMed] [Google Scholar]
- 27.Kamikawaji S, Ito G, Iwatsubo T. Identification of the autophosphorylation sites of LRRK2. Biochemistry. 2009;48:10963–10975. doi: 10.1021/bi9011379. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Moore DJ, Xiong Y, Dawson TM, Dawson VL. Reevaluation of phosphorylation sites in the Parkinson disease-associated leucine-rich repeat kinase 2. J Biol Chem. 2010;285:29569–29576. doi: 10.1074/jbc.M110.127639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, et al. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, et al. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gandhi PN, Wang X, Zhu X, Chen SG, Wilson-Delfosse AL. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J Neurosci Res. 2008;86:1711–1720. doi: 10.1002/jnr.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gloeckner CJ, Schumacher A, Boldt K, Ueffing M. The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. J Neurochem. 2009;109:959–968. doi: 10.1111/j.1471-4159.2009.06024.x. [DOI] [PubMed] [Google Scholar]
- 33.Kumar A, Greggio E, Beilina A, Kaganovich A, Chan D, Taymans JM, et al. The Parkinson's disease associated LRRK2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation. PLoS One. 2010;5:e8730. doi: 10.1371/journal.pone.0008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheng Z, Zhang S, Bustos D, Kleinheinz T, Le Pichon CE, Dominguez SL, et al. Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci Transl Med 2012;4:164ra1. [DOI] [PubMed]
- 35.Lesage S, Durr A, Tazir M, Lohmann E, Leutenegger AL, Janin S, et al. LRRK2 G2019S as a cause of Parkinson's disease in North African Arabs. N Engl J Med. 2006;354:422–423. doi: 10.1056/NEJMc055540. [DOI] [PubMed] [Google Scholar]
- 36.Saunders-Pullman R, Lipton RB, Senthil G, Katz M, Costan-Toth C, Derby C, et al. Increased frequency of the LRRK2 G2019S mutation in an elderly Ashkenazi Jewish population is not associated with dementia. Neurosci Lett. 2006;402:92–96. doi: 10.1016/j.neulet.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 37.Ozelius LJ, Senthil G, Saunders-Pullman R, Ohmann E, Deligtisch A, Tagliati M, et al. LRRK2 G2019S as a cause of Parkinson's disease in Ashkenazi Jews. N Engl J Med. 2006;354:424–425. doi: 10.1056/NEJMc055509. [DOI] [PubMed] [Google Scholar]
- 38.Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, et al. Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nat Chem Biol. 2011;7:203–205. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su X, Maguire-Zeiss KA, Giuliano R, Prifti L, Venkatesh K, Federoff HJ. Synuclein activates microglia in a model of Parkinson's disease. Neurobiol Aging. 2008;29:1690–1701. doi: 10.1016/j.neurobiolaging.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strathdee CA, McLeod MR, Hall JR. Efficient control of tetracycline-responsive gene expression from an autoregulated bi-directional expression vector. Gene. 1999;229:21–29. doi: 10.1016/S0378-1119(99)00045-1. [DOI] [PubMed] [Google Scholar]
- 41.Luo Y, Henricksen LA, Giuliano RE, Prifti L, Callahan LM, Federoff HJ. VIP is a transcriptional target of Nurr1 in dopaminergic cells. Exp Neurol. 2007;203:221–232. doi: 10.1016/j.expneurol.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Choi HK, Won LA, Kontur PJ, Hammond DN, Fox AP, Wainer BH, et al. Immortalization of embryonic mesencephalic dopaminergic neurons by somatic cell fusion. Brain Res. 1991;552:67–76. doi: 10.1016/0006-8993(91)90661-E. [DOI] [PubMed] [Google Scholar]
- 43.Sibley CR, Wood MJ. Identification of allele-specific RNAi effectors targeting genetic forms of Parkinson's disease. PLoS One. 2011;6:e26194. doi: 10.1371/journal.pone.0026194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dawson TM, Ko HS, Dawson VL. Genetic animal models of Parkinson's disease. Neuron. 2010;66:646–661. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Cheng R, Verbitsky M, Kisselev S, Browne A, Mejia-Sanatana H, et al. Genome-wide association study identifies candidate genes for Parkinson's disease in an Ashkenazi Jewish population. BMC Med Genet. 2011;12:104. doi: 10.1186/1471-2350-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 47.Liu Z, Wang X, Yu Y, Li X, Wang T, Jiang H, et al. A Drosophila model for LRRK2-linked parkinsonism. Proc Natl Acad Sci U S A. 2008;105:2693–2698. doi: 10.1073/pnas.0708452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Liu W, Oo TF, Wang L, Tang Y, Jackson-Lewis V, et al. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson's disease. Nat Neurosci. 2009;12:826–828. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong Y, Pisani A, Martella G, Karouani M, Yamaguchi H, Pothos EN, et al. R1441C mutation in LRRK2 impairs dopaminergic neurotransmission in mice. Proc Natl Acad Sci U S A. 2009;106:14622–14627. doi: 10.1073/pnas.0906334106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin X, Parisiadou L, Gu XL, Wang L, Shim H, Sun L, et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant alpha-synuclein. Neuron. 2009;64:807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi HK, Won L, Roback JD, Wainer BH, Heller A. Specific modulation of dopamine expression in neuronal hybrid cells by primary cells from different brain regions. Proc Natl Acad Sci U S A. 1992;89:8943–8947. doi: 10.1073/pnas.89.19.8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saha S, Guillily MD, Ferree A, Lanceta J, Chan D, Ghosh J, et al. LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans. J Neurosci. 2009;29:9210–9218. doi: 10.1523/JNEUROSCI.2281-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winner B, Melrose HL, Zhao C, Hinkle KM, Yue M, Kent C, et al. Adult neurogenesis and neurite outgrowth are impaired in LRRK2 G2019S mice. Neurobiol Dis. 2011;41:706–716. doi: 10.1016/j.nbd.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramonet D, Daher JP, Lin BM, Stafa K, Kim J, Banerjee R, et al. Dopaminergic neuronal loss, reduced neurite complexity and autophagic abnormalities in transgenic mice expressing G2019S mutant LRRK2. PLoS One. 2011;6:e18568. doi: 10.1371/journal.pone.0018568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plowey ED, Cherra SJ, 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 57.Heo HY, Kim KS, Seol W. Coordinate regulation of neurite outgrowth by LRRK2 and its interactor, Rab5. Exp Neurobiol. 2010;19:97–105. doi: 10.5607/en.2010.19.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parisiadou L, Xie C, Cho HJ, Lin X, Gu XL, Long CX, et al. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci. 2009;29:13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dachsel JC, Behrouz B, Yue M, Beevers JE, Melrose HL, Farrer MJ. A comparative study of Lrrk2 function in primary neuronal cultures. Parkinsonism Relat Disord. 2010;16:650–655. doi: 10.1016/j.parkreldis.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luzon-Toro B. Rubio de la Torre E, Delgado A, Perez-Tur J, Hilfiker S. Mechanistic insight into the dominant mode of the Parkinson's disease-associated G2019S LRRK2 mutation. Hum Mol Genet. 2007;16:2031–2039. doi: 10.1093/hmg/ddm151. [DOI] [PubMed] [Google Scholar]
- 61.Guo L, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG. The Parkinson's disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp Cell Res. 2007;313:3658–3670. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Covy JP, Giasson BI. Identification of compounds that inhibit the kinase activity of leucine-rich repeat kinase 2. Biochem Biophys Res Commun. 2009;378:473–477. doi: 10.1016/j.bbrc.2008.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anand VS, Reichling LJ, Lipinski K, Stochaj W, Duan W, Kelleher K, et al. Investigation of leucine-rich repeat kinase 2: enzymological properties and novel assays. FEBS J. 2009;276:466–478. doi: 10.1111/j.1742-4658.2008.06789.x. [DOI] [PubMed] [Google Scholar]
- 64.Nichols RJ, Dzamko N, Morrice NA, Campbell DG, Deak M, Ordureau A, et al. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization. Biochem J. 2010;430:393–404. doi: 10.1042/BJ20100483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, Wang QJ, Pan N, Lee S, Zhao Y, Chait BT, et al. Phosphorylation-dependent 14-3-3 binding to LRRK2 is impaired by common mutations of familial Parkinson's disease. PLoS One. 2011;6:e17153. doi: 10.1371/journal.pone.0017153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dzamko N, Deak M, Hentati F, Reith AD, Prescott AR, Alessi DR, et al. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14-3-3 binding and altered cytoplasmic localization. Biochem J. 2010;430:405–413. doi: 10.1042/BJ20100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 510 kb)