Abstract
Amyotrophic lateral sclerosis (ALS) is the third most common adult-onset neurodegenerative disease. A diagnosis is fatal owing to degeneration of motor neurons in brain and spinal cord that control swallowing, breathing, and movement. ALS can be inherited, but most cases are not associated with a family history of the disease. The mechanisms causing motor neuron death in ALS are still unknown. Given the suspected complex interplay between multiple genes, the environment, metabolism, and lifestyle in the pathogenesis of ALS, we have hypothesized that the mechanisms of disease in ALS involve epigenetic contributions that can drive motor neuron degeneration. DNA methylation is an epigenetic mechanism for gene regulation engaged by DNA methyltransferase (Dnmt)-catalyzed methyl group transfer to carbon-5 in cytosine residues in gene regulatory promoter and nonpromoter regions. Recent genome-wide analyses have found differential gene methylation in human ALS. Neuropathologic assessments have revealed that motor neurons in human ALS show significant abnormalities in Dnmt1, Dnmt3a, and 5-methylcytosine. Similar changes are seen in mice with motor neuron degeneration, and Dnmt3a was found abundantly at synapses and in mitochondria. During apoptosis of cultured motor neuron-like cells, Dnmt1 and Dnmt3a protein levels increase, and 5-methylcytosine accumulates. Enforced expression of Dnmt3a, but not Dnmt1, induces degeneration of cultured neurons. Truncation mutation of the Dnmt3a catalytic domain and Dnmt3a RNAi blocks apoptosis of cultured neurons. Inhibition of Dnmt catalytic activity with small molecules RG108 and procainamide protects motor neurons from excessive DNA methylation and apoptosis in cell culture and in a mouse model of ALS. Thus, motor neurons can engage epigenetic mechanisms to cause their degeneration, involving Dnmts and increased DNA methylation. Aberrant DNA methylation in vulnerable cells is a new direction for discovering mechanisms of ALS pathogenesis that could be relevant to new disease target identification and therapies for ALS.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-013-0205-6) contains supplementary material, which is available to authorized users.
Keywords: Chromatin modification, DNA methyltransferase, 5-methylcytosine, mitochondria, motor neuron, RG108.
Introduction
Amyotrophic lateral sclerosis (ALS) is a progressive and severely disabling fatal adult-onset neurologic disease characterized by initial muscle spasticity, cramps, and fasciculations, and then muscle weakness, atrophy, and eventual paralysis and death typically 3–5 years after symptoms begin [1, 2]. More than 5,000 people in the USA are diagnosed with ALS each year (http://www.alsa.org), and the global incidence of ALS is about 2 per 100,000 persons [3]. The average age of disease onset is 55–65 years of age, and it occurs more frequently in men than in women (http://www.alsa.org). The cause of the spasticity, paralysis, and death is believed to be progressive degeneration and elimination of upper motor neurons in cerebral cortex, and lower motor neurons in brainstem and spinal cord [1, 4, 5]. Other than life support management, no effective treatments exist for ALS [2, 3, 6]. Most individuals (~90 %) who develop classical ALS are identified as sporadic, with few known genetic contributions [1]. Familial forms of ALS are autosomal dominant or autosomal recessive, and make up ~10 % or less of all ALS cases. ALS-linked gene mutations occur in superoxide dismutase-1 (SOD1), ALS2 (Alsin), p150glueddynactin, senataxin, fused in sarcoma, tar-DNA binding protein-43, chromosome 9 open reading frame 72 (C9orf72), and in other genes that may be susceptibility loci [5, 7, 8]. The identification of C9orf72 mutations in patients without a family history of ALS [9] challenges the traditional binary classification of the disease into familial and sporadic forms. Moreover, sporadic disease is clinically and neuropathologically indistinguishable from familial forms, thus raising the possibility that the disease is caused by interplay of multiple genes or epimutations [10]. Disease discordance in monozygotic twins suggests a strong contribution of epigenetic factors in ALS [11]. The greatest known risk factor for ALS is aging [5, 8]. It is intriguing that major epigenetic differences arise in skeletal muscle, fat, and lymphocytes during the lifetime of monozygotic twins [12].
The molecular pathogenesis of ALS is not understood [5, 13–15], contributing to the lack of appropriate and legitimate disease target identification, and effective mechanism-based therapies to treat this fatal disease. Many theories have implicated perturbations in neurotrophin availability, axonal transport, glutamate receptors and transporters, protein quality control, mitochondria, bioenergetics, antioxidant status, apoptosis, inflammation, and autoimmunity in the mechanisms of ALS pathogenesis [5, 6, 16]. Many of these theories have been springboards for drug and non-drug trials, but all interventions tested, except one, have failed to meet criteria for efficacy. Riluzole, the only Food and Drug Administration (FDA)-approved drug for treating ALS [2], has actions likely through its sodium channel antagonist properties. However, riluzole has only marginal efficacy in some patients [2]. New disease mechanism-based molecular targets are needed for ALS.
DNA Methylation as an Epigenetic Mechanism
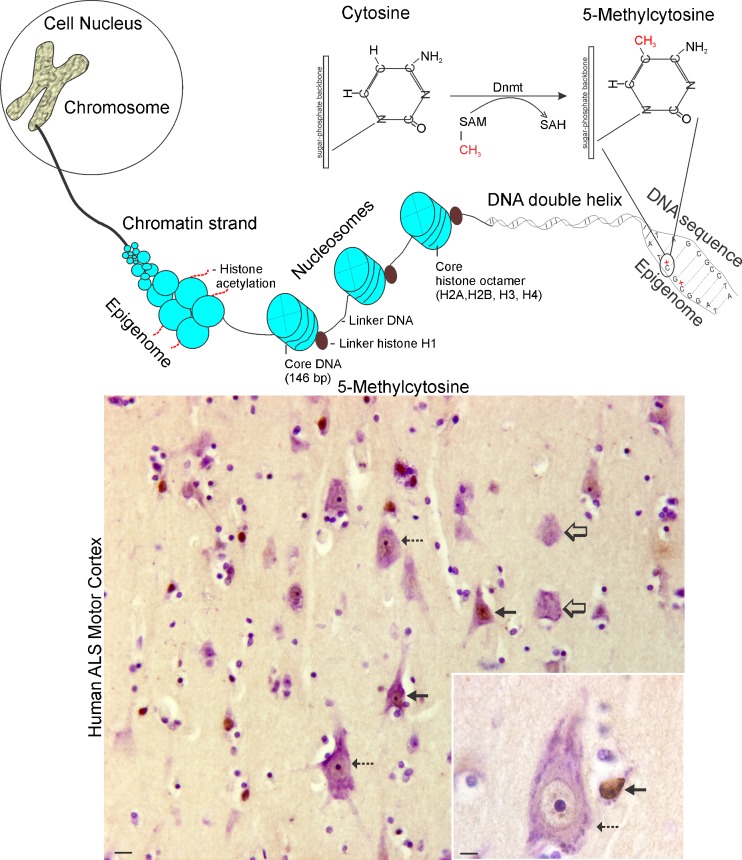
A major hurdle for the identification of genetic causes of sporadic ALS is the inability to find presumed germline genetic variants in patients and their kindred [17]. However, the identification of C9orf72 repeat expansions in apparently sporadic cases of ALS has provided an explanation for some cases [9]. Another possibility is a role of epigenetics. Epigenetic mechanisms modify chromatin structure (Fig. 1) and can mimic genetic change where Mendelian inheritance of DNA sequence is lacking. Epigenetic modifications are reversible, heritable, and nonheritable changes in DNA or chromatin structure, but not in DNA sequence, that can modulate gene activity in a context-dependent manner (Fig. 1) [18–20]. Chromosomes are packaged chromatin, and the nucleosome is the basic unit of chromatin structure, comprised of a 146-base pair stretch of DNA wrapped 1.7 times around an octameric core of histone protein pairs (H2A, H2B, H3, and H4) [21]. There are different types of epigenetic modifications, such as DNA methylation and histone acetylation (Fig. 1). These covalent modifications of DNA and histone proteins regulate the degree of compaction or relaxation of chromatin, and permit or restrict access of regulatory molecules to DNA. Biological processes related to epigenetics include gene transcriptional regulation, imprinting, X-chromosome inactivation, pluripotency, differentiation, noncoding RNA function, and synaptic plasticity [20, 22]. DNA methylation, the topic of this review, is the major direct modification of eukaryotic DNA and is known to have profound effects on the regulation of gene expression [20, 23–25]. There are two related, but functionally different, forms of DNA methylation that occur at cytosine moieties identified as 5-methylcytosine methylation (5mC) and 5-hydroxymethylcytosine methylation (5hmC) [20]. DNA methylation can act synergistically with chromatin modification (Fig. 1) by histone deacetylation to cause gene transcriptional repression by directly interfering with binding of transcription factors to DNA [23, 24].
Fig. 1.
Chromatin organization, the epigenome, and aberrant neuronal DNA methylation human ALS. (Top) Eukaryotic chromosomes are packages of DNA and protein located in the cell nucleus. The DNA, comprised of a double helix and specific types of proteins called histones are organized into chromatin strands. The substructure of the chromatin strand is the nucleosome. The histone proteins of the chromatin strand are subject to epigenetic modification through acetylation. The DNA double helix can be epigenetically modified by methylation of cytosine residues in the primary sequence. The enzymes responsible for cytosine methylation are DNA methyltranferases (Dnmts) that transfer a methyl group from the donor S-adenosylmethionine (SAM), generating S-adenosylhomocysteine (SAH), to carbon-5 in the cytosine moiety to form 5-methycytosine (5mC). (Bottom) 5mC can be detected in the human central nervous system with highly specific monoclonal antibodies. In these images of human cerebral cortex, 5mC was detected using an immunoperoxidase method (5mC immunoreactivity is brown) and then the sections were counterstained with cresyl violet to show histologic details. Individual pyramidal neurons in amyotrophic lateral sclerosis (ALS) motor cortex have varying intensities of nuclear staining for 5mC, including dark (solid black arrows), moderate (hatched arrows), or low/undetectable (open arrows). The inset shows a large upper motor neuron Betz cell (hatched arrow) with moderate chromatin 5mC and a nearby small neuron (solid black arrow), likely to be an interneuron, with a high chromatin 5mC. Several nearby glial cells (round nuclei) have low or undetectable 5mC. The details of this study showing changes in human ALS motor cortex compared with age-matched controls have been reported [70]. Scale bars: panorama 17 μm; inset 8 μm
DNA methylation is a covalent modification whereby DNA methyltransferases (Dnmts), the enzymes responsible for the formation of 5mC, catalyze methyl group transfer from the endogenous donor S-adenosyl-methionine to carbon-5 of the substrate cytosine residue (Fig. 1) [26–29]. There are ~3 × 107 residues of 5mC in the mammalian genome [28], but 5mC content in the genome varies more than 100-fold across species [30]. Generally, when DNA methylation is involved in gene silencing, the 5mCs are located in cytosine–phosphate–guanine (CpG) dinucleotides (the phosphate is the phosphodiester bond between the cytosine and guanine nucleotides) within the 5’ regulatory regions of genes upstream from the transcriptional start sites [20, 31, 32]. These CpG-rich areas are referred to as CpG islands [31, 32], and are unique in their ability to carry genetic and epigenetic information. A CpG island in DNA has been defined as a region of 200 base pairs where CGs make up more than 50 % of the region and CpGs are greater than 60 % of the observed/expected ratio within that region [33]. About 60 % of genes in the human genome contain CpG islands in their promoter regions [20, 28, 34]. The CpG-rich areas in the promoter region of genes are one of the initiation sites where transcription factors bind to the DNA, allowing transcription of the corresponding gene. DNA methylation changes the protein–DNA interactions that lead to alterations in chromatin structure and rate of gene transcription [21, 35]. When promoter regions contain 5mC in their CpG islands, transcription factors are unable to bind DNA, thus causing gene silencing by inhibition of gene transcription [31, 32, 36]. The transcriptional repression of genes involves the 5mC operating in a stable multiprotein complex of corepressors to modify chromatin structure and inhibit transcription.
NonCpG cytosine methylation has also been observed in human somatic tissues and is particularly prevalent in human brain and skeletal muscle [37, 38], but cultured differentiated human cells may have different patterns of non-CpG methylation compared with tissues [39]. Non-CpG 5mC can occur in a CAC sequence context in human brain, which is distinct from the CAG trinucleotide context found in embryonic stem cells [37]. 5mCs can also be located within non-promoter intra- and intergenic sequences, and DNA repeat sequences [40]. In contrast to promoter 5mC, the presence of 5mC in intragenic regions (the gene body) can be correlated positively with gene expression [41].
Another epigenetically modified DNA base is 5hmC. Substantial amounts of 5hmC, sometimes called the fifth base, occur in certain tissues, such as brain [42, 43], but its formation and physiological significance is controversial. 5hmC is confined mostly to the 5’ regions of genes [44] where it is thought to oppose the role of 5mC-mediated transcriptional repression by inhibiting the binding of methyl-CpG binding protein-2 and other DNA binding proteins; thus, 5hmC functions as a facilitator of gene-expression activity [44]. 5hmC might also be an intermediate of an active oxidative demethylation pathway [45], although metabolite evidence for this pathway is lacking [46]. Ten-eleven translocation methylcytosine dioxygenases (Ten-eleven translocation 1–3) are thought to be the oxidases that form 5hmC from 5mC [43]. Other active DNA demethylation pathways appear to involve excision of 5mC by DNA base excision repair with thymine DNA glycosylase, modification of the 5mC base by activation-induced cytidine deaminase, and apolipoprotein B mRNA editing enzyme followed by base excision repair, and nucleotide excision repair involving GADD45A [45].
5mC has other actions in the genome in addition the regulation of gene expression. The methylation of cytosines causes spontaneous mutations in the genome [30]. CG methylation sites are hotspots for most C>T and G>A transition mutations in the human genome. Age-related accumulation of 5mC-triggered mutations in tumor suppressor genes, cell cycle genes, and oncogenes is a major pathway for developing cancer, accounting for about 60 % of all spontaneous somatic mutations that inactivate the human p53 gene [30]. Thus, DNA methylation has been considered a basic mechanism for aging because it destabilizes the genome and organismal function over time [30].
Mammalian Dnmts in the Central Nervous System
Dnmts catalyze DNA methylation (Fig. 1) and are thus critical for the modulation of gene transcription [26–29, 31]. The Dnmt family has five members [28, 29, 31]. Dnmt1 is an abundant isoform in proliferating cells and displays a preference for hemi-methylated substrates, and is targeted to replication forks, acting to maintain methylation patterns during cell replication [47] and to DNA repair sites [48]. Dnmt1 might also have nonmethyltransferase catalytic functions through protein–protein and protein–DNA interactions that operate through its large N-terminal domain [49, 50]. Dnmt1 binds histone deacetylase-2 and Dnmt1-associated protein-1 to form a complex that directly mediates transcriptional repression of chromatin [49]. Dnmt2 (also called transfer RNA methyltransferase-1) transfers methyl groups to RNA not DNA, despite its sequence similarities to authentic Dnmts [51]. Dnmt3a and Dnmt3b function as de novo methyltransferases because they methylate hemi-methylated DNA and also completely unmethylated DNA [52, 53]. Dnmt3a and Dnmt3b are known to control non-CpG methylation [38, 39]. Dnmt3a can also function as a DNA demethylase [54]. Dnmt3L is a catalytically inactive homologue of Dnmt3a and Dnmt3b that functions as an essential regulatory cofactor for tetrameric complex assembly [55].
Mutations in the different Dnmt genes cause distinct human diseases. Mutations in the Dnmt3b gene cause immunodeficiency centromeric region instability and facial anomalies syndrome [56]. Some mutations in the Dnmt1 gene are linked to adult-onset neurodegenerative disorders. Dnmt1 mutations have been identified as a cause of hereditary sensory and autonomic neuropathy type 1 (HSAN1) [57]. Mutations associated with HSAN1 are in the coding sequence of dnmt1, corresponding to the nuclear localization targeting sequence and sites critical for catalytic activity and stability [57]. Peripheral blood cells from HSAN1 cases show hypomethylation at numerous gene promoters and DNA repeat sequences [57]. Substitution mutations in Dnmt1 also cause a polymorphic disease identified as autosomal dominant cerebellar ataxia, deafness, and narcolepsy [58]. These mutations are located in exon 21, corresponding to the replication foci targeting sequence [58]. It is unclear how these loss-of-function mutations in Dnmt1 affect the nervous system. With traditional thinking, cycling cells would be most affected given the function of Dnmt1 in methylating newly synthesized DNA strands [47]. However, Dnmt1 has been shown to undergo replication-independent chromatin loading [59] and is found at DNA repair sites [48], which are very relevant to postmitotic neurons.
Dnmts have differential roles in nervous system development and function [60], and DNA hyper- and hypomethylation can both cause neurodegeneration. Complete unconditional ablation of Dnmt1 and Dnmt3b causes embryonic lethality [52, 61], while ablation of Dnmt3a results in perinatal lethality around 3–4 weeks of age [52]. In heterozygous Dnmt1-null mice, brain injury after stroke is less compared with wild-type mice [62], supporting a role for DNA methylation in neurodegeneration. Conditional knockout of Dnmt1 in neuroblasts at E12 results in mosaic mice with 95 % hypomethylation in brain cells and perinatal death caused by respiratory failure, or mice with 30 % hypomethylation in brain cells and viability [63]. Conditional knockout of Dnmt1 in forebrain neuroblasts at E13.5 results in mice with ~90 % hypomethylation in forebrain tissue and normal viability and lifespan, but severe cerebral cortical neurodegeneration and atrophy [64]. Conditional knockout of Dnmt1 in postmitotic neurons does not affect global DNA methylation in brain or mouse viability [63]. In contrast, mice with conditional knockout of Dnmt3a in neuronal precursors results in about 50 % global hypomethylation in cerebral cortex and mouse death by 10 months of age [65]. Dnmt3a-null mice show impaired postnatal neurogenesis related to faulty transcription of neurogenic genes, rather than abnormal proliferation and survival of postnatal neural stem cells [41].
Several studies have found high levels of Dnmt enzyme activity, mRNA, and protein in adult mammalian central nervous system (CNS) [66–70]. Mature neurons are enriched in Dnmts [66–70]. We have shown that Dnmt1 and Dnmt3a are expressed in adult mouse brain and spinal cord, and in adult human cerebral cortex and spinal cord, and that mouse and human spinal motor neurons are enriched in these proteins [70]. The functions of Dnmts in terminally-differentiated postmitotic neurons are not understood. A possibility is that Dnmts in the adult mammalian brain re-methylate newly incorporated cytosines from G–T mismatch DNA repair after 5mC deamination [67]. We have found that Dnmt1 and Dnmt3a have distinct subcellular localizations in neurons [70], suggesting non-redundant functions. Our discovery that Dnmt3a, but not Dnmt1, is located in presynaptic axon terminals in the adult CNS is exciting because it identifies a local presynaptic possibility for Dnmt3a in regulating synaptic function. Studies have shown that Dnmts function in synaptic plasticity [22, 71] and behavioral plasticity [72]. Mice with conditional knockout of Dnmt1 and Dnmt3a, and with hypomethylation in forebrain show deficits in learning and memory distinct from single knockout mice, which show similar changes on behavioral tasks, suggesting that Dnmt1 and Dnmt3a have some overlapping functions in neurons [73].
Mitochondrial homeostasis is another possible function of Dnmts in neurons [70]. Dnmts are present in mitochondria, but it seems controversial as to which isoform is present in mitochondria. Dnmt1, but not Dnmt3a, was reported to be found in mitochondria from cultured mouse embryonic fibroblast and human colon carcinoma cell lines [74]. We have found Dnmt3a enriched in mitochondria from human and mouse CNS and, specifically, in neuronal mitochondria in vivo [70]. Dnmt1 is much less abundant than Dnmt3a in CNS mitochondria [70]. We suspect there is tissue- and cell-specific distribution of Dnmt isoforms in mitochondria. 5mC is also found in neuronal mitochondria in vivo as seen by immunohistochemistry [70], indicating in vivo methylation of mitochondrial DNA. Methylated cytosine residues have been detected in mouse and human mitochondrial DNA, as determined by methylation-sensitive restriction enzyme digestion [75, 76], but it has not been known how mitochondrial DNA methylation is achieved. The presence of Dnmt1 and Dnmt3a in mitochondria [70, 74] is a likely mechanism for mitochondrial DNA methylation. The existence of Dnmt3a and 5mC in mitochondria enhances the idea of DNA methylation involvement in synaptic mechanisms by implicating epigenetic modification of the neuronal mitochondrial DNA as a new possible function of Dnmt3a in adult neurons.
Methylomics in ALS
We hypothesize that the neurodegeneration in ALS may have an epigenetic contribution [70]. Other laboratories have proposed that epigenetic mechanisms might contribute to the pathophysiology of other age-related neurodegenerative disorders, such as Alzheimer’s disease [17]. Epigenetic mechanisms involving DNA methylation might explain how variation in gene activity arising during development or from environmental and lifestyle factors is not genetically transmissible in sporadic forms of ALS. Few articles exist on DNA methylation in human ALS. Methylation in sporadic ALS has been examined previously only in two candidate genes—SOD1 and VEGF [77]—in members of the metallothionein gene family [78], and in the astroglial glutamate transporter EAAT2 gene promoter [79]. None of these studies found differences in methylation patterns in sporadic ALS cases compared with age-matched control cases.
A study reported on a genome-wide analysis of brain DNA methylation in sporadic ALS accomplished by chromatin immunoprecipitation followed by microarray hybridization using GeneChip Human Tiling 2.0R arrays [80]. The brain tissue analyzed was dorsolateral prefrontal cortex [80]. Unlike Brodmann area 4 (motor cortex), this region of cerebral cortex (Brodmann area 46) is not usually affected in classical ALS [1] unless there is dementia [9]. Because DNA methylation signatures are likely to vary in different parts of the human CNS [81], tissue samples from affected and unaffected regions should be studied from the same individual. Nevertheless, this provocative study identified differentially methylated genes in ALS cases compared with control cases [80]. Calcium channel genes (CACNA1B and CACNA1C) were hypermethylated, while the genes for neurexin-1 (NRXN1), glial cell-derived neurotrophic factor (GDNF) receptors (GFRA1 and GFRA2), and phospholipid metabolism (PLA2G4C) were hypomethylated [80]. CACNA1C has at least 55 exons and encodes an α1 subunit of the L-type voltage-gated calcium channel, also referred to as Cav1.2, and is known to undergo neuronal cell-type specific DNA methylation patterns [82]. Changes in the regulation of these genes suggest anomalies in ion channel and plasma membrane function in ALS. Consistent with earlier work, promoter regions of SOD1, VEGF, EAAT2, and metallothioneins I and II were not differentially methylated in ALS. Of further interest was the finding that only about half of the brain DNA methylation was found in gene domain regions, with about 30 % of the DNA methylation found within the gene body and 8 % in promoter regions (distal and proximal) [80]. Surprisingly, this study found that inter-individual variation in human brain DNA methylation is large and no altered DNA methylation pattern was common to all sporadic ALS cases [80].
In a more recent study, CpG methylation microarrays were used to profile postmortem spinal cord (ventral horn gray and white matter) DNA extracts from sporadic ALS cases [83]. Bisulfite-converted DNA was amplified and hybridized to Infinium Human Methylation27 DNA BeadChip arrays. These arrays determine the methylation of 27,578 CpG sites spanning 14,495 human genes. Hypo- or hypermethylation was found in 112 genes in ALS cases [83]. Many of the differentially methylated genes found in sporadic ALS spinal cord were associated with immune and inflammatory responses, and suggested infiltration of vascular-derived cells and activation of resident microglia [83]. Some of the changes identified by DNA methylation microarray were confirmed at the RNA level by real-time polymerase chain reaction.
A perennial difficulty of cortical and spinal cord tissue homogenate-based assays is the lack of cellular resolution, and the CNS has extraordinary cellular heterogeneity. Cellular resolution is necessary for more precise interpretation of changes in tissue DNA methylation in ALS. Future studies should employ laser capture microdissection of specific cell populations in human ALS tissues to maximize information gleaned from assessments of the DNA methylome [84]. Motor neuron-specific DNA methylation profiling in human ALS can provide direct information on the genetic changes leading to neurodgeneration and neuronal cell death, and on the whole be directly relevant to the formulation of new therapeutic interventions.
The most common known genetic cause of ALS is the recently discovered noncoding hexanucleotide repeat expansion in the C9orf72 gene [9, 85]. This mutation accounts for 24–37 % of familial ALS cases and ~6 % of sporadic ALS cases in Caucasians. The functional consequences of the mutated repeat expansion in C9orf72 are hypothetical and implicate toxic gain-of-function properties related to sequestering of RNA binding proteins by RNA species containing the repeat expansion or to anomalous nonATG-initiated translation from the repeat expansion leading to aggregation of dipeptide-repeat proteins [85]. Another possibility is a loss-of-function mechanism caused by the expansion resulting in reduction of mRNA [85]. Recently, an exciting discovery was made showing that CpG islands in the 5’ regions of the C9orf72 repeat expansion of individuals with ALS were hypermerthylated, and the corresponding mRNA was down-regulated [86]. Furthermore, greater levels of CpG island methylation were associated with more aggressive disease. This report is exciting because it supports the hypothesis that C9orf72 repeat expansion causes a loss-of-function, and it demonstrates that mutant genes causing ALS can be silenced epigenetically.
Loss-of function deletions and mutations in the coding sequence of ALS2 can cause autosomal recessive juvenile onset ALS [87, 88]. The ALS2 gene encodes for Alsin, which is a guanine nucleotide-exchange factor for the small GTPase Rab5. A recent genome-wide study of promoter methylation of individuals who experienced severe psychosocial trauma in early life and age-matched controls identified differentially methylated promoters in brain tissue from case samples [89]. The ALS2 gene promoter showed the most significant hypermethylation of 248 promoters [89]. This study is the first to provide evidence that rearing history can influence the activity of genes that are involved in the mechanisms of ALS.
Cellular Localizations of Aberrant DNA Methylation and Dnmts in Human ALS
To put into specific cellular context potential DNA methylation changes in the human ALS nervous system, we have used immunohistochemistry to localize 5mC and Dnmts [70]. 5mC accumulated in vulnerable motor neurons in spinal cord and motor cortex, and dissipated in astrocytes. Dnmt1 and Dnmt3a also accumulated in cortical and spinal motor neurons [70]. Western blotting confirmed these general changes in human ALS tissue. A limitation of this work is that the human ALS tissue is studied at end-stage disease, so causal relationships between deregulated DNA methylation and Dnmt changes are difficult to identify. Cause–effect relationships need to be studied in experimental model systems. Using a combination of cell culture and animal experimental designs we have shown that Dnmt activity can drive neuronal degeneration [70]. Increased activity of Dnmt3a appears to have a particularly prominent pro-apoptotic role in model systems of neuron degeneration that involve p53 and caspase-3 activation [70].
Endogenous Dnmts Mediate Apoptosis of Adult Mouse Motor Neurons In Vivo
To examine the potential pro-apoptotic role of Dnmts in vivo, we used a model of unilateral sciatic nerve avulsion that induces robust p53-regulated apoptosis of adult spinal motor neurons [90]. Although this animal model of acquired motor neuron injury is not a genetic model of familial ALS harboring mutations in human SOD1, p150glued dynactin, or tar-DNA binding protein-43 genes, it is very relevant to human ALS because, unlike some of the transgenic mouse models of ALS [91], the motor neurons degenerate through an apparent programmed cell death process involving p53 and caspase-3 activation [90] as in human ALS [92, 93]. Dnmt1 was up-regulated in both lesioned and nonlesioned sides of spinal cord, while Dnmt3a was up-regulated in the lesioned side only [70]. As a read-out for DNA methylation directly in motor neurons during apoptosis, 5mC was detected using an antibody. Obvious changes in the chromatin patterns of 5mC were seen as early as 2 days after motor neuron injury and significant increases in the levels of immunoreactivity were detected as soon as 3 days after injury [70]. Treatment of mice with RG108 blocked the injury-induced enhancement of DNA methylation in motor neurons and afforded sustained, near-complete protection of motor neurons against apoptosis [70].
Enforced DNMT3a Drives Apoptosis of Cultured Motor Neuron-like Cells
In the course of cell transfection experiments for Dnmt antibody validation, we observed that enforced expression of Dnmt3a induced apoptosis in cultured NSC34 cells (a mouse spinal cord neuron-hybridoma cell line capable of differentiating into cells with some motor neuron-like features) [70]. We then engineered mammalian expression constructs for Dnmt3a-green fluorescent protein (GFP) fusions. NSC34 cells were transfected with the Dnmt–GFP constructs or a GFP expression plasmid as a control. NSC34 cells over-expressing Dnmt3a for 24 h showed significant increases in p53 activation and caspase-3 enzyme activity. These events are typically associated with apoptosis; moreover, the nuclei of these cells showed chromatin condensation with characteristics of apoptosis. To determine if the neuronal degeneration was related specifically to the methyltransferase activity of Dnmt3a, we engineered an expression construct (Dnmt3aΔ–GFP) containing the Dnmt3a gene with a deletion in the DNA sequence encompassing >93 % of the C-terminal catalytic domain of the encoded protein. NSC34 cells transfected with the deletion plasmid Dnmt3aΔ-GFP survived, differentiated into neuron-like cells, and remained viable [70].
Endogenous Dnmts Mediate Apoptosis in Cultured Motor Neuron-like Cells
We performed experiments to determine if endogenous Dnmts function in neuronal apoptosis. During apoptosis induced by the topoisomerase-I inhibitor, DNA strand-break inducer camptothecin (CPT), Dnmt1, and Dnmt3a protein levels and enzyme activity were significantly up-regulated in differentiated NSC34 cells [70]. RG108 (a designed small molecule isoform nonspecific Dnmt inhibitor) [94] and procainamide (a Dnmt1-specific inhibitor) [95] decreased significantly the amount of apoptosis in CPT-treated NSC34 neurons. RNAi using Dnmt3a-siRNA also decreased significantly the amount of NSC34 cell apoptosis induced by CPT [70].
Work in non-neural cells has shown that Dnmt overexpression can be cytotoxic. Overexpression of Dnmt1 and Dnmt3a in human and mouse tumor cell lines diminished clonogenicity and survival, while cells tolerated Dnmt3b overexpression [96]. In nonsmall cell lung cancer cell lines, activation of Dnmt1 can cause suppression of survivin gene expression and induce apoptosis [97]. Loss of survivin expression is associated with motor neuron degeneration in neonatal spinal cord organotypic slice cultures [98].
Based on different lines of experimental evidence using animal model and cell culture systems we have shown that motor neurons can engage epigenetic mechanisms to drive apoptosis, involving Dnmt up-regulation and increased DNA methylation [70]. Because the animal and cell culture paradigms were models of neuronal apoptosis, associations between Dnmts/DNA methylation and apoptosis can only be interpreted. Human cells with perturbations in DNA methylation linked to Dnmt3b abnormalities are hypersensitive to radiation-induced apoptotic and non-apoptotic cell death [99], so it is possible that DNA methylation can also regulate other forms of cell death, such as autophagy and programmed necrosis. We hypothesize that the neurodegeneration in human ALS, which is suspected to be some form of programmed cell death [92, 93], may be the result of abnormal epigenetic regulation. Epigenetic mechanisms involving DNA methylation might explain how individual variation in gene activity arising during development or from environmental and lifestyle factors is not genetically transmissible in sporadic forms of ALS.
DNA Methylation, Neurotransmission, Interneurons, and ALS
DNA methylation appears to function in synaptic plasticity [22, 71]. DNA methylation is a dynamic gene regulatory mechanism, as illustrated by some genes undergoing cyclical changes within minutes to hours [100]. Inhibition of DNA methylation can block long-term potentiation in acute hippocampal slices [22, 71]. Prolonged depolarization of cultured cortical neurons can cause demethylation of the gene promoter for the neurotrophin brain-derived neurotrophic factor [60]. Furthermore, spontaneous synaptic activity is regulated by DNA methylation that, in itself, changes in response to synaptic activity [101]. The finding that DNA methylation can regulate spontaneous miniature neurotransmission [101] is consistent with our finding that Dnmt3a is localized in axon terminals [70] and is pertinent to ALS. We have found deficits in spontaneous miniature and evoked inhibitory postsynaptic currents in motor neurons cultured from a transgenic mouse model of ALS [102]. Single-cell quantitative polymerase chain reaction revealed silencing of genes encoding glycine receptor channel subunits in motor neurons [102]. It is not yet clear whether the abnormality in ALS mouse motor neuron neurotransmission is autonomous or non-autonomous and related to primary abnormalities in spinal interneurons. Other studies of transgenic mouse models of ALS show that interneurons degenerate before motor neurons [91], and motor neurons become deficient in glycinergic innervation early in the course of the disease [103], leading to the hypothesis that ALS is initiated in interneurons [104]. This hypothesis has been supported by recent work modeling ALS in Drosophila [105] and in clinical studies of ALS patients [106]. Interneurons are interesting in the context of DNA methylation because they might be particularly sensitive to epigenetic modulation, as suggested in studies of prenatal stress in mice [107] and in the patterns of 5mC immunolabeling in human ALS cerebral cortex (Fig. 1). Their possible involvement in ALS pathogenesis would be consistent with non-autonomous mechanisms of degeneration for motor neurons [16, 91, 104, 108].
DNA Methylation, Skeletal Muscle, and ALS
Recent epidemiologic observations showing that professional soccer and football players, and Gulf War veterans have an increased risk of developing ALS have led to ideas about a potential association between physical activity and ALS, but this possibility has been disputed by indications that other genetic, environmental, and lifestyle factors, rather than physical activity per se, increase ALS susceptibility [109, 110]. It is pertinent, in this regard, that human ALS may be a form of metabolic disease with defects in energy metabolism manifesting as negative energy balance, hypermetabolism, and hyperlipidemia [111]. Evidence is accruing that DNA methylation can contribute to increased risk and development of metabolic disease by regulating the expression of genes that control body energy metabolism and glucose homeostasis [112].
Motor neuron non-autonomous mechanisms of disease in ALS also extend to the periphery because primary disease in skeletal muscle can cause motor neuron degeneration and ALS in transgenic mice [113]. Studies of human ALS support the concept that abnormalities in skeletal muscle are primary intrinsic mechanisms in the pathogenesis, rather than merely secondary changes related to denervation and neurogenic atrophy [114–116]. Evidence from monozygotic twins reveals that they have similar DNA methylation patterns in skeletal muscle when young, but markedly divergent patterns when older; thus, environmental and lifestyle factors have major epigenetic effect on skeletal muscle [12]. Elegant developmental experiments on mouse cells show that myoblasts are heterogeneous in their DNA methylation signatures, which then interact with environmental influences to confer muscle groups their distinct characteristics [117]. Skeletal muscle also has a high degree of plasticity that allows adaption to a variety of environmental and kinesiologic challenges, such as exercise [118], and acute exercise is associated with rapid gene promoter hypomethylation in skeletal muscle in human [119]. If skeletal muscle contributes to the etiology of ALS [113], then a variety of epigenetic mechanisms could contribute to the multifactorial disease process, and epigenetic modifying drugs could be used to target skeletal muscle peripherally, without the need for blood–brain barrier permeability, for the treatment of ALS.
Opening a New Door to ALS Therapy
New clinically relevant disease concepts, targets, and drug applications are necessary to move ALS therapy in new meaningful directions because status quo concepts and treatments are ineffective [2, 6]. DNA methylation is an exciting potential field relevant to human ALS in terms of identifying pathogenic mechanisms. If our hypothesis is correct regarding DNA methylation as a mechanism of disease in ALS, then new avenues for epigenetic therapeutics become available to ALS patients for testing in clinical trials (Table 1). DNA methylation can be modified pharmacologically, and by lifestyle intervention and rehabilitation programs [29, 120, 121]. Nucleoside analogs and Dnmt inhibitors are available (Table 1) to achieve DNA demethylation, and some DNA methylation modifying drugs are FDA-approved for human use. Nucleoside analogs are used widely as suicide inhibitors of Dnmts in treating neoplasia, but have dose-limiting toxicity and carcinogenic side effects [29, 120]. Blocking the catalytic activity of Dnmts with small molecules is another strategy. Procainamide and hydralazine are FDA-approved for the treatment of cardiac arrhythmia and hypertension owing to their sodium channel blocking activity and cyclic guanosine monophosphate modulation actions, respectively; however, they are now being tested as Dnmt inhibitiors in preclinical studies [120]. In addition to other actions, these drugs can bind CpG-rich sequences and act as partial competitive inhibitors of Dnmt1. Natural molecules (psammaplins) and rationally designed small molecules (RG108) are sound alternative approaches to modify DNA methylation. RG108 is an exciting drug because it has demethylating activity in vitro and in vivo in non-neuronal systems, and low toxicity—even at high concentrations [120]. We have shown the demethylating efficacy of RG108 in neuronal systems and its robust neuroprotective actions in cell and animal models of motor neuron degeneration [70], thus establishing aberrant DNA methylation as a new disease target in ALS and RG108 as a potential therapy.
Table 1.
DNA methylation inhibitor drugs possibly relevant to amyotrophic lateral sclerosis (ALS)
| Drug | Drug class | Mechanisms | Indications | Comments and concerns |
|---|---|---|---|---|
| 5-aza-cytidine (5-azaC) & 5-aza-2-deoxycytidine (5-azadC, decitabine) | Nucleoside analog prodrug, suicide inhibitor of Dnmt | DNA incorporation, DNA–Dnmt trapping, passive loss of DNA methylation | Hematological malignancies | Covalent DNA-protein trapping, carcinogenic, dose-limiting toxicity |
| Zebularine | Cytidine nucleoside analog | Inhibition of cytidine deaminase and Dnmt | Preclinical cancer chemoprevention | Lower toxicity than 5-azaC; orally active |
| Procainamide | Non-nucleoside, small molecule | Sodium channel blocker, Dnmt1 inhibitor | Cardiac arrhythmia | Long history of use in humans; off-target actions related to Na+ channel blocking, perhaps beneficial in ALS |
| Hydralazine | Non-nucleoside, small molecular | Increases cGMP, vasodilation, activates Hif1; direct Dnmt inhibition uncertain | Hypertension, preclinical for cancer chemotherapy | Long history of use in humans; can cause lupus-like syndrome; pleiotropic mechanisms of action |
| RG108 | Non-nucleoside, small molecule | Blocks noncovalently Dnmt active site, inhibition of Dnmt | Preclinical for cancer chemotherapy and ALS therapy | Shown to block DNA methylation accumulation in motor neurons and their degeneration; encouraging future |
| Psammaplin | Marine sponge bisulfide bromotyrosine antibiotic | Inhibits HDAC and Dnmt activities | Preclinical for cancer chemotherapy | Strong off-target cytotoxic effects in cultured human cells; limited in vivo data available |
Dnmt = DNA methyltransferase; cGMP = cyclic guanosine monophosphate; Hif1 = hypoxia‐inducible factor‐1.
Electronic supplementary material
(PDF 510 kb)
Acknowledgments
This work was supported by grants from the U.S. Public Health Service, National Institute of Neurological Disorders and Stroke (R01-NS065895 and R01-NS079348).
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N Eng J Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 2.Zoccolella S, Santamato A, Lamberti P. Current and emerging treatments for amyotrophic lateral sclerosis. Neuropsychiatr Dis Treat. 2009;5:577–595. doi: 10.2147/ndt.s7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisen A. Amyotrophic lateral sclerosis: a 40-year personal perspective. J Clin Neurosci. 2009;16:505–512. doi: 10.1016/j.jocn.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 4.Heath PR, Shaw PJ. Update on the glutamatergic neurotransmitter system and the role of excitotoxicity in amyotrophic lateral sclerosis. Muscle Nerve. 2002;26:438–458. doi: 10.1002/mus.10186. [DOI] [PubMed] [Google Scholar]
- 5.Martin LJ. Mitochondrial and cell death mechanisms in neurodegenerative diseases. Pharmaceuticals. 2010;3:839–915. doi: 10.3390/ph3040839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin LJ. Olesoxime, a cholesterol-like neuroprotectant for the potential treatment of amyotrophic lateral sclerosis. IDrugs. 2010;13:568–580. [PMC free article] [PubMed] [Google Scholar]
- 7.Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 8.Morris HR, Waite AJ, Williams NM, Neal JW, Blake DJ. Recent advances in the genetics of the ALS-FTLD complex. Curr Neurol Neurosci. 2012;12:243–250. doi: 10.1007/s11910-012-0268-5. [DOI] [PubMed] [Google Scholar]
- 9.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Chalabi A, Fang F, Hanby MF, et al. An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry. 2010;81:1324–1326. doi: 10.1136/jnnp.2010.207464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraga M, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Nat Acad Sci. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schymick JC, Talbot K, Traynor BJ. Genetics of sporadic amyotrophic lateral sclerosis. Hum Mol Gen. 2007;16:R233–R242. doi: 10.1093/hmg/ddm215. [DOI] [PubMed] [Google Scholar]
- 14.Turner BJ, Talbot K. Transgenics, toxicity and therapeutics in rodent models of mutant SOD1-mediated familial ALS. Prog Neurobiol. 2008;85:94–134. doi: 10.1016/j.pneurobio.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Redler RL, Dokholyan NV. The complex molecular biology of amyotrophic lateral sclerosis. Prog Mol Biol Trans Sci. 2012;107:215–262. doi: 10.1016/B978-0-12-385883-2.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 17.Qureshi IA, Mehler ME. Advances in epigenetics and epigenomics for neurodegenerative diseases. Curr Neurol Neurosci Rep. 2011;11:464–473. doi: 10.1007/s11910-011-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolffe AP, Matzke MA. Epigenetics: Regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 19.Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 20.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 21.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 1999;98:285-294. [DOI] [PubMed]
- 22.Levenson JM, Roth TL, Lubin FD, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 23.Curradi M, Izzo A, Badaracco G, Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol. 2002;22:3157–3173. doi: 10.1128/MCB.22.9.3157-3173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Yan L, Davidson N. DNA methylation in breast cancer. Endocr Relat Cancer. 2001;8:115–127. doi: 10.1677/erc.0.0080115. [DOI] [PubMed] [Google Scholar]
- 25.Luczak MW, Jagodzinski PP. The role of DNA methylation in cancer development. Folia Histochem Cytobiol. 2006;44:143–154. [PubMed] [Google Scholar]
- 26.Cheng X. Structure and function of DNA methyltransferases. Ann Rev Biophys Biomol Struct. 1995;24:293–318. doi: 10.1146/annurev.bb.24.060195.001453. [DOI] [PubMed] [Google Scholar]
- 27.Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase I: expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- 28.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 29.Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmcol Toxicol. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- 30.Mazin AL. Suicidal function of DNA methylation in age-related genome disintegration. Ageing Res Rev. 2009;8:314–327. doi: 10.1016/j.arr.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Brenner C, Fuks F. DNA methyltransferases: Facts, clues, mysteries. Curr Top Microbiol Immunol. 2006;301:45–66. doi: 10.1007/3-540-31390-7_3. [DOI] [PubMed] [Google Scholar]
- 32.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 33.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 34.Fatemi M, Pao M, Jeong S, et al. Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res. 2005;33:e176. doi: 10.1093/nar/gni180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 36.Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J Biol Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- 37.Varley KE, Gertz J, Bowling KM, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barres R, Osler ME, Yan J, et al. Non-CpG methylation of the PGP-1α promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 39.Ziller MJ, Muller F, Liao J, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2001;7:e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–260. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H, Coskun V, Tao J, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons in the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin S-G, Wu X, Li A, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nuc Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu SC, Zhang Y. Active DNA methylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Globisch D, Münzel M, Müller M, et al. Tissue distribution of 5-hydroxymethlycytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:31–39. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 48.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc Natl Acad Sci USA. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to from a complex at replication foci. Nature Genet. 2000;24:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 50.Espada J. Non-catalytic functions of DNMT1. Epigenetics. 2012;7:115–118. doi: 10.4161/epi.7.2.18756. [DOI] [PubMed] [Google Scholar]
- 51.Schaefer M, Lyko F. Solving the Dnmt2 enigma. Chromosoma. 2010;119:35–40. doi: 10.1007/s00412-009-0240-6. [DOI] [PubMed] [Google Scholar]
- 52.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 53.Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 54.Kangaspeska S, Stride B, Métivier R, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 55.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansen RS, Wijmenga C, Luo P, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klein CJ, Botuyan M-V, Wu Y, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winkelman J, Lin L, Schormair B, et al. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet. 2012;21:2205–2210. doi: 10.1093/hmg/dds035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Easwaran HP, Schermelleh L, Leonhardt H, Cardoso MC. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 2004;5:1181–1186. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinowich K, Hattori D, Wu H, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 61.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 2001;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 62.Endres M, Meisel A, Biniskiewicz D, et al. DNA methyltransferase contributes to delayed ischemia brain injury. J Neurosci. 2000;20:3175–3181. doi: 10.1523/JNEUROSCI.20-09-03175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan G, Beard C, Chen RZ, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hutnick LK, Golshani P, Namihira M, et al. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum Mol Genet. 2009;18:2875–2888. doi: 10.1093/hmg/ddp222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened life span. Dev Dyn. 2007;236:1663–1676. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- 66.Goto K, Numata M, Komura JI, et al. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- 67.Brooks PJ, Marietta C, Goldman D. DNA mismatch repair and DNA methylation in adult brain neurons. J Neurosci. 1996;16:939–945. doi: 10.1523/JNEUROSCI.16-03-00939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inano K, Suetake I, Ueda T, et al. Maintenance-type DNA methyltransferases is highly expressed in post-mitotic neurons and localized in the cytoplasmic compartment. J Biochem. 2000;128:315–321. doi: 10.1093/oxfordjournals.jbchem.a022755. [DOI] [PubMed] [Google Scholar]
- 69.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferase DNMT3A and DNMT3B in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 70.Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M, Martin LJ. Epigenetic regulation of motor neuron cell death through DNA methylation. J Neurosci. 2011;31:16619–16636. doi: 10.1523/JNEUROSCI.1639-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 72.LaPlant Q, Vialou V, Covington HE, 3rd, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feng J, Zhou Y, Campbell SL, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108:3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shmookler Reis RJ, Goldstein S. Mitochondrial DNA in mortal and immortal cells: Genome number, integrity, and methylation. J Biol Chem. 1983;258:9078–9085. [PubMed] [Google Scholar]
- 76.Pollack Y, Kasir J, Shemer R, Metzger S, Szyf M. Methylation pattern of mouse mitochondrial DNA. Nucleic Acids Res. 1984;12:4811–4824. doi: 10.1093/nar/12.12.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oates N, Pamphlett R. An epigenetic analysis of SOD1 and VEGF in ALS. Amyotroph Lateral Scler. 2007;8:83–86. doi: 10.1080/17482960601149160. [DOI] [PubMed] [Google Scholar]
- 78.Morahan JM, Yu B, Trent RJ, Pamphlett R. Are metallothionein genes silenced in ALS? Toxicol Lett. 2007;168:83–87. doi: 10.1016/j.toxlet.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 79.Yang Y, Gozen O, Vidensky S, Robinson MB, Rothstein JD. Epigenetic regulation of neuron-dependent induction of astroglial synaptic protein GLT1. Glia. 2010;58:277–286. doi: 10.1002/glia.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morahan JM, Yu B, Trent RJ, Pamphlett R. A genome-wide analysis of brain DNA methylation identifies new candidate genes for sporadic amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2009;10:418–429. doi: 10.3109/17482960802635397. [DOI] [PubMed] [Google Scholar]
- 81.Ladd-Acosta C, Pevsner J, Sabunciyan S, et al. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nishioka M, Shimada T, Bundo M, et al. Neuronal cell-type specific DNA methylation patterns of the Cacna1 gene. Int J Devl Neurosci. 2013;31:89–95. doi: 10.1016/j.ijdevneu.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 83.Figueroa-Romero C, Hur J, Bender DE, et al. Identification of epigenetically altered genes in sporadic amyotrophic lateral sclerosis. PLoS ONE. 2012;7:e52672. doi: 10.1371/journal.pone.0052672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ginsberg SD, Hemby SE, Mufson EJ, Martin LJ. Cell and tissue microdissection in combination with genomic and proteomic applications. In: Zaborsky L, Wouterlood FG, Lanciego JL, editors. Neuroanatomical tract-tracing 3: molecules, neurons, and systems. Singapore: Springer; 2006. pp. 109–141. [Google Scholar]
- 85.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xi Z, Zinman L, Moreno D, et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet. 2013;92:981–989. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hadano S, Hand CK, Osuga H, et al. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet. 2001;29:166–173. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
- 88.Yang Y, Hentati A, Deng HX, et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet. 2001;29:160–165. doi: 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
- 89.Labonte B, Suderman M, Maussion G, et al. Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry. 2012;69:722–731. doi: 10.1001/archgenpsychiatry.2011.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martin LJ, Chen K, Liu Z. Adult motor neuron apoptosis is mediated by nitric oxide and Fas death receptor linked to DNA damage and p53 activation. J Neurosci. 2005;25:6449–6459. doi: 10.1523/JNEUROSCI.0911-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martin LJ, Liu Z, Chen K, et al. Motor neuron degeneration in amyotrophic lateral sclerosis mutant superoxide dismutase-1 transgenic mice: mechanisms of mitochondriopathy and cell death. J Comp Neurol. 2007;500:20–46. doi: 10.1002/cne.21160. [DOI] [PubMed] [Google Scholar]
- 92.Martin LJ. Neuronal death in amyotrophic lateral sclerosis is apoptosis: possible contribution of a programmed cell death mechanism. J Neuropathol Exp Neurol. 1999;58:459–471. doi: 10.1097/00005072-199905000-00005. [DOI] [PubMed] [Google Scholar]
- 93.Martin LJ. p53 is abnormally elevated and active in the CNS of patients with amyotrophic lateral sclerosis. Neurobiol Dis. 2000;7:613–622. doi: 10.1006/nbdi.2000.0314. [DOI] [PubMed] [Google Scholar]
- 94.Brueckner B, Garcia Boy R, Siedlecki P, et al. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- 95.Lee BH, Yegnasubramanian S, Lin X, Nelson WG. Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem. 2005;280:40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Santourlidis S, Kimura F, Fischer J, Schulz WA. Suppression of clonogenicity by mammalian Dnmt1 mediated by the PCNA-binding domain. Biochem Cell Biol. 2004;82:589–596. doi: 10.1139/o04-099. [DOI] [PubMed] [Google Scholar]
- 97.Li HL, Ma AN. Induction of apoptosis of non-small cell lung cancer by a methylated oligonucleotide targeting surviving gene. Cancer Gene Therapy. 2010;17:441–446. doi: 10.1038/cgt.2009.82. [DOI] [PubMed] [Google Scholar]
- 98.Tolosa L, Mir M, Asensio VJ, et al. Vascular endothelial growth factor protects spinal cord motoneurons against glutamate-induced excitotoxicity via phosphatidylinositol 3-kinase. J Neurochem. 2008;105:1080–1090. doi: 10.1111/j.1471-4159.2007.05206.x. [DOI] [PubMed] [Google Scholar]
- 99.Narayan A, Tuck-Muller C, Weissbecker K, et al. Hypersensitivity to radiation-induced non-apoptotic and apoptotic death in cell lines from patients with the ICF chromosome instability syndrome. Mut Res. 2000;456:1–15. doi: 10.1016/s0027-5107(00)00103-2. [DOI] [PubMed] [Google Scholar]
- 100.Metivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 101.Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chang Q, Martin LJ. Glycine receptor channels in spinal motoneurons are abnormal in a transgenic mouse model of amyotrophic lateral sclerosis. J Neurosci. 2011;31:2815–2827. doi: 10.1523/JNEUROSCI.2475-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chang Q, Martin LJ. Glycinergic Innervation of motoneurons is deficient in amyotrophic lateral sclerosis mice: a confocal quantitative analysis. Am J Path. 2009;174:574–585. doi: 10.2353/ajpath.2009.080557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martin LJ, Chang Q. Inhibitory synaptic regulation of motoneurons: a new target of disease mechanisms in ALS. Mol Neurobiol. 2012;45:30–42. doi: 10.1007/s12035-011-8217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McGown A, McDearmid JR, Panagiotaki N, et al. Early interneuron dysfunction in ALS: Insights from a mutant sod1 zebrafish model. Ann Neurol. 2001;73:246–258. doi: 10.1002/ana.23780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Turner MR, Kiernan MC. Does interneuronal dysfunction contribute to neurodegeneration in ALS? Amyotroph Lateral Scler. 2012;13:245–250. doi: 10.3109/17482968.2011.636050. [DOI] [PubMed] [Google Scholar]
- 107.Matrisciano F, Tueting P, Dalal I, et al. Epigenetic modifications of GABAergic interneurons are associated with the schizophrenia-like phenotype induced by prenatal stress in mice. Neuropharmacology. 2013;68:184–194. doi: 10.1016/j.neuropharm.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McGeer PL, McGeer EG. Inflammatory processes in amyotrophic lateral sclerosis. Muscle Nerve. 2002;26:459–470. doi: 10.1002/mus.10191. [DOI] [PubMed] [Google Scholar]
- 109.Harwood CA, McDermott CJ, Shaw PJ. Physical activity as an exogenous risk factor in motor neuron disease (MND): A review of the evidence. Amyotroph Lateral Scler. 2009;10:191–204. doi: 10.1080/17482960802549739. [DOI] [PubMed] [Google Scholar]
- 110.Huisman MHB, Seelen M, de Jong SW, et al. Lifetime physical activity and the risk of amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 2013. doi:10.1136/jnnp-2012-304724. [DOI] [PubMed]
- 111.Dupuis L, Pradat P-F, Ludolph AC, Loeffler J-P. Energy metabolism in amyotrophic lateral sclerosis. Lancet Neurol. 2011;10:75–82. doi: 10.1016/S1474-4422(10)70224-6. [DOI] [PubMed] [Google Scholar]
- 112.Ribel-Madsen R, Fraga MF, Jacobsen S, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS ONE. 2012;7:e51302. doi: 10.1371/journal.pone.0051302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wong M, Martin LJ. Skeletal muscle-restricted expression of human SOD1 causes motor neuron degeneration in transgenic mice. Hum Mol Genet. 2010;19:2284–2302. doi: 10.1093/hmg/ddq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vielhaber S, Winkler K, Kirches E, et al. Visualization of defective mitochondrial function in skeletal muscle fibers of patients with sporadic amyotrophic lateral sclerosis. J Neurol Sci. 1999;169:133–139. doi: 10.1016/s0022-510x(99)00236-1. [DOI] [PubMed] [Google Scholar]
- 115.Echaniz-Laguna A, Zoll J, Ponsot E, et al. Muscular mitochondrial function in amyotrophic lateral sclerosis is progressively altered as the disease develops: A temporal study in man. Exp Neurol. 2006;198:25–30. doi: 10.1016/j.expneurol.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 116.Pradat P-F, Barani A, Wanschitz J, et al. Abnormalities of satellite cells function in amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2011;12:264–271. doi: 10.3109/17482968.2011.566618. [DOI] [PubMed] [Google Scholar]
- 117.Donoghue MJ, Sanes JR. All muscles are not created equal. Trends Genet. 1994;10:396–401. doi: 10.1016/0168-9525(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 118.Coffey AG, Hawley JA. The molecular bases of training adaptation. Sports Med. 2007;37:737–763. doi: 10.2165/00007256-200737090-00001. [DOI] [PubMed] [Google Scholar]
- 119.Barrès R, Yan J, Egan B, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15:405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 120.Nebbioso A, Carafa V, Benedetti R, Altucci L. Trials with ‘epigenetic’ drugs: an update. Mol Oncol. 2012;6:657–682. doi: 10.1016/j.molonc.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Baar K. Epigenetic control of skeletal muscle fiber type. Acta Physiol. 2010;199:477–487. doi: 10.1111/j.1748-1716.2010.02121.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 510 kb)