Abstract
This review highlights recent discoveries that have shaped the emerging viewpoints in the field of epigenetic influences in the central nervous system (CNS), focusing on the following questions: i) How is the CNS shaped during development when precursor cells transition into morphologically and molecularly distinct cell types, and is this event driven by epigenetic alterations?; ii) How do epigenetic pathways control CNS function?; iii) What happens to “epigenetic memory” during aging processes, and do these alterations cause CNS dysfunction?; iv) Can one restore normal CNS function by manipulating the epigenome using pharmacologic agents, and will this ameliorate aging-related neurodegeneration? These and other still unanswered questions remain critical to understanding the impact of multifaceted epigenetic machinery on the age-related dysfunction of CNS.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-013-0229-y) contains supplementary material, which is available to authorized users.
Key Words: Epigenetics, CNS, chromatin, neurodegeneration, aging, histone code, HDAC, DNA methylation
Introduction
The molecular mechanisms underlying the age-related decline in the function of the central nervous system (CNS) are not currently known, but recent work points toward a dysregulation of complex and interconnected pathways, which impose epigenetic control over cellular identity and/or specification. Epigenetics refer to a set of potentially self-perpetuating mechanisms that produce heritable alterations in the chromatin structure. These alterations, viewed historically in the context of cell division and early development, are aimed to choreograph the myriad cellular and molecular events in order to establish an “epigenetic landscape” [1] which drives phenotypic diversity amongst cell types that share a common genome. At first glance, the conceptual framework of epigenetics bears little relevance to the adult brain as the brain is composed of a large proportion of postmitotic and highly differentiated cells. However, recent explorations of the brain epigenome are providing unprecedented insights into the importance of specific epigenetic mechanisms in controlling gene expression, not only in early brain development, but in adult brain function as well, calling into place postmitotic epigenetic mechanisms. These epigenetic mechanisms serve as essential mediators that interface the extrinsic environment and the intrinsic genetic program that allows for a postmitotic “reprogramming process” of neuronal cells in order to achieve plasticity at many levels of the neural circuitry in response to environmental factors, for example diet [2] and experiential factors [3].
Currently known epigenetic mechanisms operating within the cells include post-translational modifications of histones (histone PTMs) and incorporation of histone variants; changes in DNA methylation and adenosine triphosphate-dependent chromatin remodeling; and the implementation of RNA interference pathways and nonprotein-coding RNAs (see [4] for a review). To drive gene transcription, all of these epigenetic components operate in high-performance collaborative fashion mediated by transcriptional factors (TF). Emerging sequencing technologies have made it possible for more sensitive, precise, and genome-wide scale measurements of epigenetic DNA and histone modifications, and for profiling of various noncoding RNAs and chromatin remodeling complexes.
In this review, we will discuss recent progress that points to an active role of epigenetic regulations in neurobiology. We will focus on our current understanding of the molecular basis of epigenetic signaling to chromatin, and will discuss examples of this signaling in context of neurogenesis, diseases of the CNS, and aging. Many reviews on epigenetic regulation have been published and we have not attempted a comprehensive coverage herein of this topic; rather, we have focused on a personal selection of epigenetic phenomena that are exciting at present or promise to be so in the future.
Epigenetics and Aging of CNS
An increasing body of literature indicates that a substantial reorganization of the brain epigenome occurs during postnatal development and aging, which make the hypothesis that aging is associated with epigenetic changes in the brain quite attractive. The time in life when brain aging begins is undefined. However, the finding that normal aging shares common molecular signatures with some neurological and chronic deteriorating psychiatric disorders, as exemplified by early-stage schizophrenia [5], suggest that some symptoms of aging, i.e., dementia, psychosis, and progressive neurodegeneration could be explained by the molecular mechanisms in common. This assumption offers a possibility of identifying early epigenetic components and mechanisms that may be engaged in preventive or detrimental age-related CNS functions. Importantly, the epigenetic concept of aging is also attractive because, in contrast to genetic mutations, epi-mutations (epigenetic markings) are now thought to be reversible through multitude of pharmacological interventions [6].
Both DNA methylation 5-positon of cytosine (5-mC) catalyzed by DNA methyltransferases and 5-hydroxymethylcytosine (5-hmC), which is converted from 5-mC by ten–eleven translocation proteins (for detailed reviews see[7] and [8]), play important roles in mammalian neurogenesis. Both an establishment and maintenance of DNA methylation are essential for normal development and function of the mammalian brain. The role of DNA methylation seems to be indispensible in maintaining neural stem cell self-renewal [9, 10], postnatal neurogenesis [11], and learning and memory, as well as in synaptic plasticity [12, 13]. Mutations of methyl-CpG binding protein 2, which binds to methylated DNA and acts as a transcriptional repressor or activator, causes Rett syndrome and related neurodevelopmental disorders [14, 15]. More interestingly, both loss and over-expression of methyl-CpG binding protein 2 not only affect the overall level of 5-hmC, but also modulate its distribution in the genome during mouse cerebellum development [16]. During the last several years, the DNA methylation signature was found to show certain dynamics associated with brain development and aging, suggesting that epigenetic changes in levels of 5-mC and 5-hmC could function as an intermediate step for the internal/external environmental regulation of the brain epigenome, which ultimately leads to gene expression changes. Age-related changes in DNA methylation/hydroxymethylation dynamics tend to occur more often outside of CpG islands, as shown for cerebellum [17] and prefrontal cortex [18], and seems to affect genes associated with cognitive impairment and stereotypic movement. Such age-related epigenetic drift could affect an individual's vulnerability to neurodegenerative disease.
In addition to changes in DNA methylation during development and aging, dynamic changes to the epigenetic landscapes of PTMs also occurs. An age-dependent histone PTMs drift mainly affects areas that can differentiate between open and repressive chromatin as demonstrated for the human prefrontal and cerebellar cortex [19], and hundreds of loci undergo substantial chromatin remodeling in cortical neurons between infancy and advanced age [20]. Interestingly, the brains of SAMP8 mice, which are prone to accelerated senescence and have learning and memory deficits, also show age-related drifts in histone PTMs [21]. These epigenetic drifts are defined by a loss of the histone methylation modifications associated with active gene expression, and a robust rise in the repressive histone methylation ([22]; Fig. 1 and discussed in detail in [23]), as well as a robust decline in histone acetylation [24]. We will discuss several of these important findings in great details later on. Cumulative evidence suggests that such age-related epigenetic drifts in the CNS could affect multiple interconnected entities, contributing to a decline in the signaling capacity of nerve cells by modifying neuronal and oligodendroglial gene transcription program, inflicting defects in axon myelination and many other molecular alterations that have been linked to various cognitive disorders of the adult brain with and without neurodegeneration [25–28].
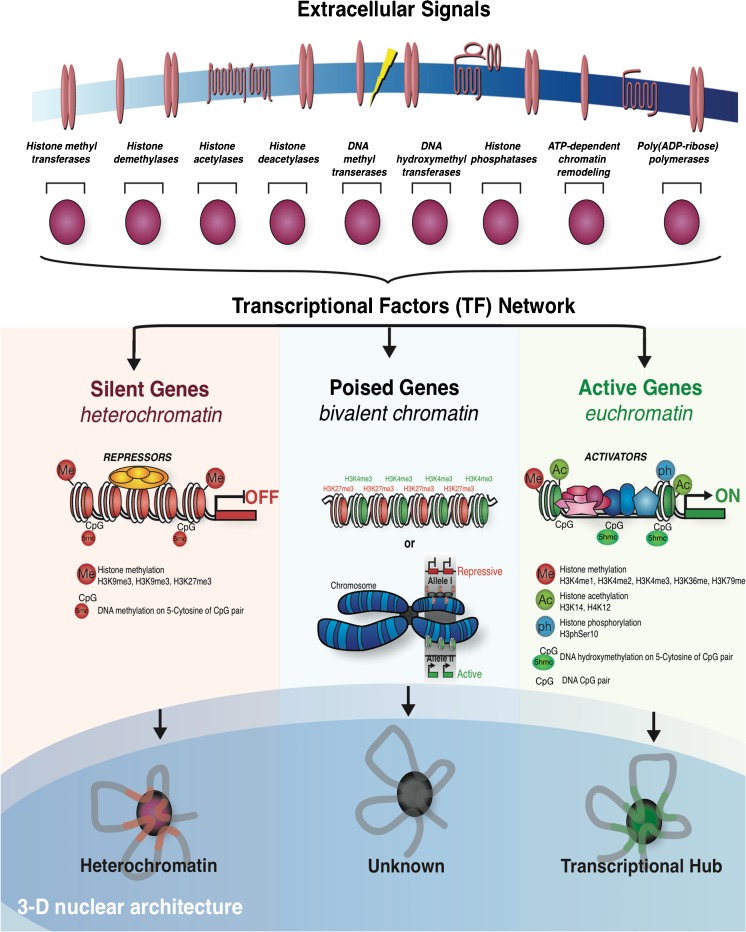
Fig. 1.
Dynamic genome architecture in the nuclear space: regulation of gene expression in 3 dimensions. Epigenetic response to extrinsic signals occurs through the transcriptional factors network. Chromosomes are organized into loose (euchromatin) and highly condensed (heterochromatin) domains. Heterochromatin is transcriptionally repressive, whereas euchromatin is transcriptionally permissive. Bivalent chromatin (middle) that poise genes for transcriptional activation are thought to be either superposition of active and silent epigenetic marks, or simply represent monoallelic gene expression. An 11-nm “beads-on-a-string” chromatin fiber is comprised of nucleosomal arrays and has an additional level of organization in the context of 3-dimensional (3D) nuclear volume (bottom). The high level of transcriptional regulation depends on the 3D organization and functional compartmentalization of the nucleus [42, 43]. In mammalian cells, transcription of genes that are concomitantly activated in response to extracellular stimuli or during cell differentiation occurs specifically at intranuclear foci enriched with active RNA polymerase II (RNAPII) [142]. These transcriptional hubs are known as transcription factories and, in addition to RNAPII, they often include transcription factors (TF) [41]. The distribution of DNA methylation and a small subset of post-translational histone marks represents different flavors of chromatin related either to gene activation (right) or gene repression (left). ATP adenosine triphosphate; ADP adenosine diphosphate; Me methylation; 5 m 5-methylcytosine; Ac acetylation; 5hm 5-hydroxymethylcytosine; ph phosphorylation
Combined, these findings leave no room for doubt that epigenetic influences within the CNS are, indeed, subject to dynamic changes throughout all periods of maturation and aging, and this process of epigenetic drift has important ramifications for the neurobiology of disease.
“Epigenetic Memory” Concept and CNS Dysfunction
Coordinated genome-wide gene activation and gene repression shapes functional cellular identities during development and aging of the CNS. Multiple levels of epigenetic regulation converge in the chromatin to institute a flexible, but tightly regulated, cellular expression program by balancing transcriptionally permissive, less condensed euchromatin, and highly condensed and often repressed heterochromatin (Fig. 1) [29]. Dysfunction in this balance is viewed as a major driving force of aging, suggesting that aging results in a progressive loss of this “epigenetic memory” of chromatin state in mature and progenitor (adult stem) CNS cells, thereby resulting in profound changes of the intrinsic properties of these cells due to global changes of gene expression or the ability of cells to respond to stimuli [30–35].
A molecular “epigenetic memory” of gene expression as a concept is not novel. It is a well characterized phenomenon defined by numerous studies of imprinting and inheritance of parental traits [36–38]. It is often viewed as a lifelong molecular information storage mechanism that is put in place during development, and is necessary for perpetuating the correct cellular phenotype during mitosis. In addition to imprinting, the epigenome can reflect a cell’s past history of activities of transcriptional enhancers [39]. The traditional view has been that once epigenetic marks (discussed below) have been laid down during development, they remain unchanged for the life of the organism. Recent work in the adult organism, however, has challenged this view. For example, published data indicate that epigenetic marks can be rapidly (within minutes) and transiently (less than 24 h) changed in order to provide dynamic regulation of gene transcription in the adult brain [40].
Very little is known about how epigenetic mechanisms regulate this dynamic and immensely complex gene transcription. While a great deal of effort has been given to the role of TFs that can either act as activators or as repressors, depending on their context or interacting partners, an emerging field of neuro-epigenetics is just starting to gather evidence implicating specific spatial and temporal chromatin states driving transcription in the CNS. These specific chromatin states are required not only for the TF interaction with cognate DNA sequence, but also function as operational modules indispensible for the establishment and maintenance of the specialized 3-dimensional (3D) nuclear architecture critical for supporting transcriptional outcomes [15, 41–44]. Although it was initially thought that TF dominate the epigenetic landscape as their overexpression can reprogram cell fates, it is apparent now that mechanisms of TF action cannot be clearly separated from the action of epigenetic machineries in altering chromatin states. TFs are likely to interact and act in complexes with enzymes that remodel chromatin, manipulate DNA methylation, and force deposition or exchange of histone variants. TF-mediated recruitment of such proteins could directly aid in a step-wise process that starts with distal and proximal enhancers that execute their influences on gene promoters and then continue in nonlinear fashion to form a complex network of intra- and extrachromosomal interaction to generate a wide variety of chromatin states [42, 44, 45].
For instance, new results suggest that the loss of capability that some cells within the CNS undergo for such rapid and dynamic change in gene expression may contribute to cognitive impairment in the aged brain. Finally, observations reported by Miller et al. [13] present a hypothesis that the “epigenetic memory” operation system controlling these dynamic transcriptional responses could be particularly vulnerable to the aging process. Next we will discuss the integral parts, or components, of the “epigenetic memory” paradigm.
Chromatin is a Major Interface Between Epigentic Mechanisms and the Gene Transcription Program
3D Nuclear Architecture and Gene Transcription
Application of novel methods for detection and characterization of local and long-range chromosomal contacts, combined with microscopy and computational modeling, have yielded more coherent insights into 3D chromatin architecture in relation to gene regulation (for a review see [46]). A large body of literature indicates that alterations in the 3D chromatin state in adult neurons are important for mediating various aspects of experience-dependent plasticity, such as learning and memory, stress response, and cognition, thus suggesting that it might be an important, integral component of aging.
It has become well accepted that the position of a gene within the 3D structure of the nucleus and assignment to particular nuclear compartments (heterochromatin vs euchromatin) has been shown to correlate with its transcriptional activity (shown in Fig. 1). Upon cellular proliferation, the position of the gene is established during early G1 upon chromosome decondensation [32]. Proximity to the nuclear envelope or peri-centromeric heterochromatin is generally associated with gene silencing [47–49]. The relative position of individual genes to subnuclear compartments or other genes within cellular nuclei can change at different stages of gene activation and/or cell differentiation. For instance, in neurons, TFIIIC binding within short interspersed nuclear elements mediates the rearrangement of nuclear architecture, possibly by coordinating the simultaneous expression of activity-dependent neuronal genes necessary for dendritic growth [50–52]. Mis-regulation of short interspersed nuclear elements has been shown to be associated with changes in the chromatin environment, and the aging of adult stem and somatic cells in humans [53, 54].
A gene and its regulatory elements can be relocated upon transcriptional activation into specific compartments. Both transgene and endogenous gene mobility has been reported upon binding of transcriptional activator [31, 55, 56]. For instance, large-scale chromatin reorganization and nuclear repositioning from the periphery to the center have been described for the Mash1 locus during neural induction [33]. Although an entire, relatively large (2-Mb) genomic segment was found to relocate at the onset of neurogenesis, this change of subnuclear positioning resulted in activation of some, but not all genes within the repositioned chromosomal domain. This suggests that induction of transcription requires not only a favorable 3D chromatin environment, but also a specific TF recruitment to a cognate DNA site.
Cellular differentiation leads to the restructuring of the chromatin accompanied by the change in the global nuclear architecture. For example, it has been shown that heterochromatic markers, such as HP-1 proteins, as well as heterochromatin-related histone modifications, change their localization from dispersed and very dynamic in embryonic stem cells to more concentrated distinct loci during cellular differentiation (for a review see [7]).
As differentiation advances, cells undergo global chromatin reorganization [57] leading to accumulation of more rigid heterochromatin, driven by the compaction of major satellite repeats [58] and the pericentric regions of some chromosomes, resulting in concentrated heterochromatic foci detectable upon cytological analysis [58]. Although chromatin tends to be more confined in differentiated cells, examples of dramatic changes on the nuclear architecture of postmitotic cells have been reported. Early studies of Purkinje cells (PCs) identified striking morphological changes associated with kinetochore [34] and pericentric heterochromatin [35] movements. In newly born mice, PCs are already in a postmitotic state, but are not yet terminally differentiated. Analysis of PC nuclei during postnatal development clearly demonstrates that nuclear architecture undergoes profound changes manifested by the movement in kinetochore regions of the chromosomes and pericentric heterochromatin away from nuclear membrane (periphery) towards nuclear center. In addition to obvious movements of pericentrometic heterochromatin, the nucleoli in PCs undergo dynamic changes in size and positioning [59]. Although the possible functional implications of these observations in postmitotic PCs cell is not explained at present, it can be anticipated that various levels of epigenetic regulation of chromatin can contribute to this developmental process and its malfunction can be an integral part of disease related pathologies and aging. The discovery that methyl-DNA binding protein MeCP-2 is required for induction of chromocenters clustering, and maturation of the nucleoli in developing neurons in vivo and in vitro further corroborates this statement [32].
Therefore, it is not surprising that nuclear architecture defects have been shown to correlate with manifestation of a number of human diseases, as well as aging. A striking example is provided by analysis of a leucine-rich repeat kinase 2 dominant mutation (G2019S; glycine-to-serine substitution at amino acid 2019) [60], which is associated with familial and sporadic Parkinson’s disease (PD), as well as the impairment of adult neurogenesis in mice [38]. It has been found that the gene mutation results in the aberrant cellular morphology of neural stem cells (NSCs). Abnormality in chromatin architecture was depicted in these experiments: the nuclei had acquire the pedal-like shapes during in vitro aging. The deformed NSC nuclear shape is accompanied by decreased abilities of clonal expansion, neural differentiation, and resistance to stresses, which partially explains PD syndrome [61]. These results demonstrate the importance of nuclear architecture in the pathology of the neurodegeneration, and reinforce the notion that epigenetic mechanisms responsible for the fine-tuning of 3D chromatin structure within the cells could be potential targets in therapeutic interventions.
Many aspects of epigenetic regulations discussed in this review have direct influence on the organization of the chromatin during development and aging of the CNS. Comprehending how the nuclear genomic organization is established and maintained to provide for neural cellular diversity, connectivity, and plasticity within CNS, and how it influences gene expression, might subsequently allow for a better understanding of CNS development, homeostasis, and aging.
Nucleosome Composition
Before we look into the epigenetic alteration occurring in CNS upon aging, it is important to discuss the basics of chromatin composition. Early work on the nuclear packaging of chromosomal DNA has defined the basic unit of the DNA/protein complex known as chromatin. This fundamental unit, the nucleosome, is comprised of 2 copies of the histone proteins H3, H4, H2A, and H2B, creating a bead-like structure. Then, 146 base pairs of DNA are wrapped around the surface of this structure formed by these core histone proteins. The linker histone, H1, binds the nucleosome at the entry and exit sites of the DNA wrapping around the complex thus locking the nucleosomal particle in place (Fig. 2a).
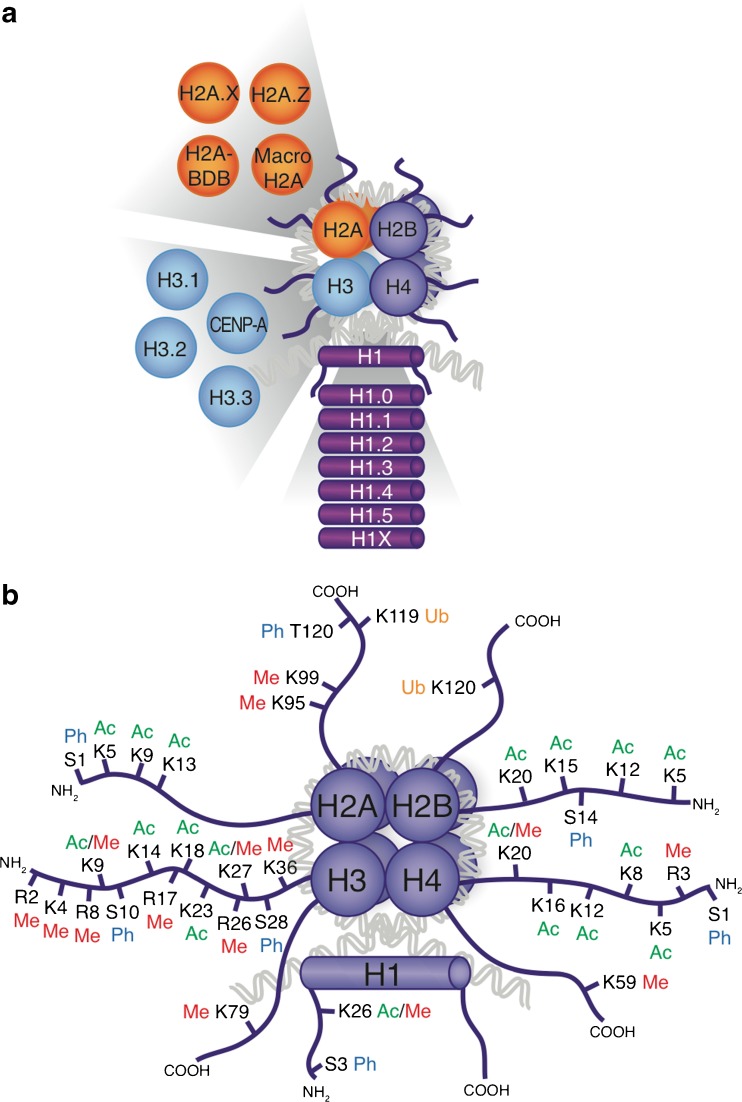
Fig. 2.
Histone post-translational modifications and variants. a Schematic drawing of a nucleosome with the 4 canonical histones (H2A, H2B, H3, and H4), the linker histone H1 and their variants. b Currently known covalent histone post-translational modifications PTMs are highlighted on the N- and C-terminal tails of each histone. Me = methylation; Ac = acetylation; Ub = ubiquitination; Ph = phosphorylation
Two of the most prevalent neurodegenerative diseases—Alzheimer’s disease (AD) and PD diseases—share the common feature: a normally soluble peptide (amyloid-beta) or protein (alpha-synuclein) aggregate into an ordered fibrillar structure. An interesting observation reported by Duce et al. [47] highlights the unexpected dynamics of the linker histone H1. This study demonstrated that histone H1 is localized within the cytoplasm of neurons and astrocytes from disease-affected areas. The pull-down experiment suggests that the strongest detected interacting protein to the amyloid-like motifs of amyloid-beta, alpha-synuczzlein and lysozyme is, in fact, histone H1. As histone H1 is known to be an integral part of chromatin within the nucleus, with a primary role of binding DNA that enters and exits from the nucleosome (Fig. 2a), and facilitating the shift in equilibrium of chromatin towards a more condensed form, depletion of the nuclear histone H1 upon onset of AD or PD might suggest a different degree of chromatin compaction within the cells with ordered fibrillar aggregates related to these diseases. Histone H1 and the members of this family can differentially affect the condensation of the chromatin fiber and its stability [62]. It is therefore reasonable to assume that this family of histones might play pivotal roles in chromatin organization during the aging of CNS. One explanation for these aforementioned intriguing observations relates to the possible role of histone chaperones in the mechanistic aspects of chromatin packaging.
Histone Chaperones
In general, a shift in the pool of histones available within the nucleus of the cell could potentially promote changes in the nuclear architecture and ultimately tip the balance within transcriptional programs. As a mediator of the external signals, chromatin is anything but static. Nucleosome unwrapping and disassembly events, which must occur during DNA replication, transcription, and DNA repair, can directly influence the state of chromatin compaction [63]. Several lines of evidence obtained in yeast, Drosophila, and humans indicate that chromatin undergoes disassembly during the onset of DNA double-strand break repair at the double-strand break sites. Importantly, recovery from DNA damage requires restoration of chromatin structure, which depends on re-deposition of the histones back onto the DNA by means of coordinated action of histone-chaperones and ATP-dependent chromatin remodeling complexes (reviewed in [64]). Therefore, if this “access, repair, restore” model of the chromatin dynamics is critical for the successful DNA repair and restoration of cellular function, then it is plausible that mis-regulation or interruption of the DNA damage-specific chromatin disassembly/reassembly pipeline could potentiate a deficiency in the nuclear histone composition, leading to a persistent DNA damage response and, ultimately, cellular senescence, as demonstrated in the human fibroblast models [65, 66]. Similarly, this deficiency may be an integral part of a chain of molecular events that lead to loss of plasticity within the CNS and physiological changes in neurodegenerative diseases.
Histone chaperones are proteins that can assemble histones and DNA into the nucleosome structure, as well as disassemble an intact nucleosome into its subcomponents to facilitate chromatin opening not only during DNA repair but also upon gene transcription [64]. The chaperones also function to provide guidance for histone folding, prevent histone aggregation, and mediate histone transport between the nucleus and cytoplasm [63, 67, 68]. A new study of activity-dependent gene transcription regulation has highlighted an indispensible role of the histone chaperone death-domain associated protein (DAXX) in cortical neurons [31]. Chromatin-associated protein DAXX [69] acts as a histone chaperone for the histone variant H3.3 [70, 71], and has the ability to interact with transcription factors and chromatin modifiers, which include histone deacetylases (HDACs), the histone acetyl-transferase cyclic adenosine monophosphate response element-binding protein (CREB) binding protein (CBP), and DNA methyltransferases [72–75]. New data demonstrate that DAXX interacts with histone H3.3 in neurons and regulates deposition of this histone at regulatory elements of activity-regulated genes upon membrane polarization. Loading of histone H3.3 by DAXX occurs during transcription through a mechanism involving a calcium-dependent phosphorylation switch. This event, together with the action of another chaperone, HIR histone cell cycle regulation defective homology A (HIRA), which loads H3.3 at the transcriptional start sites and gene body [76, 77], is apparently instrumental to activity-triggered chromatin changes in neurons. Recent analysis of somatic mutations associated with human pediatric glioblastoma revealed that mutations in H3.3–ATRX–DAXX chromatin remodeling pathway was strongly associated with alternative lengthening of telomeres and specific changes in gene expression profiles [77], suggesting that defects of the chromatin architecture underlie pediatric and young adult glioblastoma multiforme (GBM) pathogenesis.
Removed from the context of neurobiology, the roles of numerous histone chaperones have been intensively studied genetically and biochemically (see [64]); however, the molecular link between histone chaperone function and chromatin reconfiguration upon development and aging CNS has not yet been explored in full.
Histone Variants
In addition to the 4 canonical histone proteins (H2A, H2B, H3, and H4), many variant forms of histones exist in different organisms. H1.0, H1.1, H1.2, H1.3, H1.4, H1.5, and H1X are the variants of H1; H2A.X, H2A.Z, H2A-BDB, and Macro H2A are replacement variants for H2A; and H3.1, H3.2, H3.3, and centromere protein A (CENP-A) are variants of the core histone H3 (Fig. 2a; for review see [7] and [79]). The variants are usually present as single-copy genes that are unrestricted in their expression to the S phase, but are expressed throughout the entire cell cycle. Unlike major subtypes, the variant histone genes contain introns, and their transcripts are often polyadenylated. These features are considered important in the post-transcriptional regulation of these proteins [80]. Biochemical and structural studies have shown that nucleosomes containing histone variants have altered affinity, stability, and sequence preference [48, 49, 81]. For instance, in mammalian cells, variant nucleosomes containing H3.3/H2AZ are unstable, suggesting a more accessible state of chromatin marked by these nucleosome variants [82].
As chromatin maintenance in nondividing cells relies on the incorporation of replication-independent histone variants, it is likely that histone variants will greatly affect neuronal biology. In the 1980s, it was reported that the mouse brain, like testis and oocytes, expresses a unique set of histone variants [46]. Some variants exchange with pre-existing histones during development and differentiation, and are therefore referred to as replacement histones. In fact, several known in the scientific literature support the dependency of the basic aspects of neuronal function on the numerous histone variants (e.g., H2AX, H3.3, macroH2a, and H1.0) [31–33]. For instance, during brain maturation, histone H1.0 accumulates in both nerve and glial cells, and it is subjected to age-dependent deamination [34]. Overexpression of H1.0 can transiently block cell cycle progression in proliferating cells [35]. Indirect evidence for the role of histone H3.3 in the control of neural cell cycle regulation comes from recent reports demonstrating that mutations in H3.3 are linked to pediatric glioblastoma [61, 78, 83]. A further example includes the regulation of adult neurogenesis by gamma aminobutyric acid (GABA)-induced cascades, leading to phosphorylation of H2AX in NSCs, subsequently blocking NSC self-renewal [32] and therefore inhibiting neurogenesis. Interestingly, H2AX histone variants display progressive increases in expression with age in rat cortex [60] and ataxia telangiectasia mutated (ATM), the enzyme responsible for phosphorylation of H2AX, has been shown to be significant for nervous system development. ATM mutation causes pronounced and debilitating neurodegeneration (for a review see [38]).
However, beyond a very fundamental understanding of histone variant expression in the brain, little is known regarding the functions of these variants and factors that regulate their expression, either at a transcriptional or a post-transcriptional level. Recently, Lal et al. [84] have shown that the suppression of H2AX by microRNA mir-24 in terminally differentiated blood cells is responsible for the diminished DNA repair ability of these cells. It is still unclear if the same pathways are in place in the developing and adult CNS.
Currently, the majority of studies aimed at elucidating the functions of the histone variants are based on the correlation between the localization of the variant and the transcriptional activity of the locus, or on analyses of phenotypes associated with the loss of the variant. For example, H2Be is a histone variant that is exclusively expressed by olfactory sensory neurons in a replication-independent fashion [40]. Loss of H2Be alters olfactory function and gene expression despite the fact that H2Be knockout mice appear healthy and fertile. H2Be replaces the canonical H2B to the extent that it appears to be regulated by neuronal activity, from nondetectable levels in highly active neurons to almost complete replacement in neurons that are inactive. The complete replacement of H2B by H2Be promotes neuronal cell death, and genetic manipulation aimed at deleting H2Be significantly extends the lifespan of neurons. These interesting data uncover a novel chromatin-based mechanism for activity-dependent neuronal plasticity and suggest that histone variants participate in a pathway that shapes the cellular and molecular composition of the olfactory epithelium, based on signals from the external environment.
Therefore, if one considers all possible variations in nucleosomal composition, it is clear that a staggering variety of possible biological readouts of these diverse nucleosomes exist within the CNS. The full potential of this diversity and the functional relevance to the development of CNS and its aging attributes have not yet been fully explored or appreciated.
“Histone Code” Concept in Neurobiology: The Challenge of Complexity
In the framework of chromatin, nucleosomes can be 1) covalently modified by chromatin modifying complexes, providing a covalent histone tail and globular domain modifications, as shown in Fig. 2b, or 2) repositioned by chromatin remodeling complexes, causing an alteration of DNA–histone contacts. This occurs in a highly combinatorial and, sometimes, in a mutually exclusive fashion. Nucleosomal packaging and histone modifications dictate the different degrees of primary chromatin compaction, for example 6 nucleosomes per 11 nm in the euchromatic chromatin fiber vs 12–15 nucleosomes per 11 nm in heterochromain achieved by additional chromatin structural proteins [85]. The core histones are subjected to numerous and different PTMs, including acetylation, methylation, phosphorylation, poly-adenosine monophosphate-rybosylation, ubiquitination, and sumoylation (Fig. 2b) [86]. Different chromatin states are defined by combinatorial patterns in these histone modifications, and are often referred to as the “histone code” [87], as shown in Fig. 1. Each histone modification can induce or inhibit subsequent PTMs, and such cross-talk can operate on the same nucleosome or can be established between nucleosomes [88]. To add to this complexity, the chromatin structure is also influenced by effector or “reader” proteins that recognize single or multiple histone PTMs. (The example of repressive Polycomb group complex stepwise action on chromatin is shown later in Fig. 4 to illustrate the complexity and hierarchy of events deployed to establish silenced state of chromatin.) Moreover, this recognition can occur with PTMs on a single nucleosome, or several nucleosomes that can be either present on the same or different chromatin fibers (interchromosomal interactions). We will address some of these specific “writer” proteins at a later point. The majority of histone PTMs have shown to be reversible and could be mediated by multiple enzymatic machineries (Fig. 3). The balance between these enzymatic machineries responsible for the nucleosomal remodeling or establishment and maintenance of removal of histone PTMs significantly contributes to chromatin dynamics upon neurogenesis and are indispensable for driving cell type-specific biological outcomes in the CNS.
Fig. 4.
Combinatorial and sequential histone post-translational modifications (PTMs) deposition by Polycomb complexes. This graphical representation illustrates an example of combinatorial action of histone PTM writers, which add covalent PTMs to histone tails; readers, which recognize and bind histone PTMs; and erasers, which remove histone PTMs. Protein families associated with these steps are listed on the left. In this example, the PRC2 complex adds (writes) trimethylation on lysine 27 of histone H3. This is then recognized by the (reader) complex PRC1. The RING1A and RING1B subunits of PRC1 (writers) subsequently act to ubiquitinate lysine 199 of histone H2A. UTX-1 and JMJD3 can act to remove (erase) histone H3 K27 trimethylation. HMT = histone methyltransferase; HAT = histone acetyl transferase; ADP = adenosine diphosphate; SH2 = Src homology 2; FHA = forkhead-associated; HDMT = histone demethylase; HDAC = histone deacetylase; Me = methylation; Ub = ubiquitination
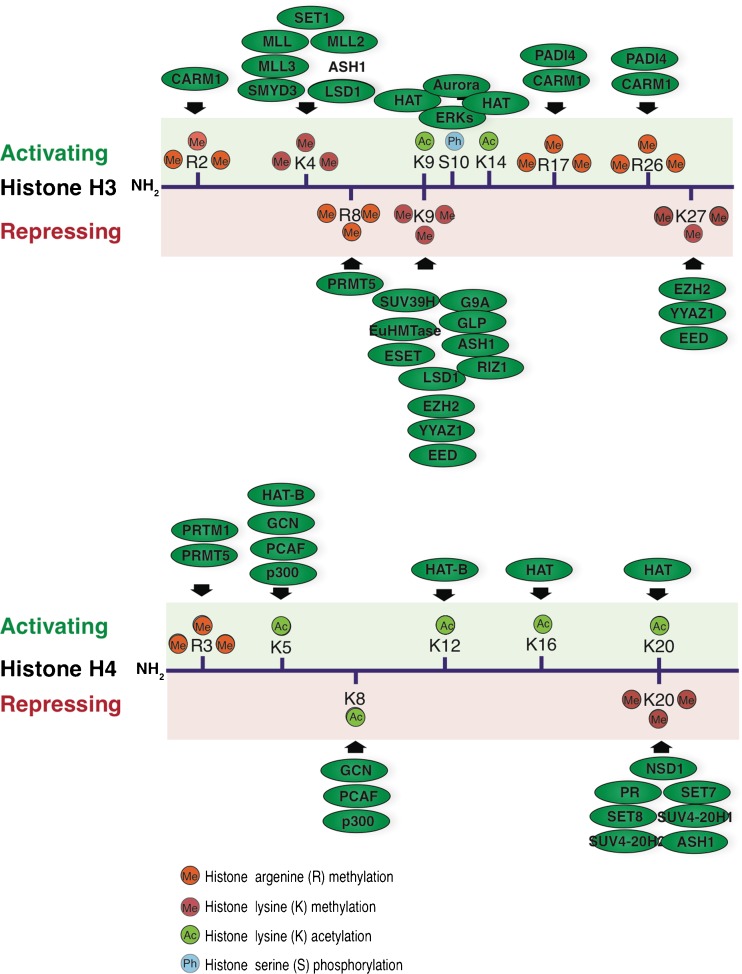
Fig. 3.
Post-translational modifications of N-terminus of histone H3 and H4 and enzymes responsible for their deposition. Relevance to the gene transcriptional activation or repression is indicated
It is vital to emphasize caution in interpretation of epigenetic data. Owing to the cellular heterogeneity of CNS tissues, one of the major challenges of investigating chromatin regulation is our current difficulty in defining individual chromatin states and remodeling activities, not only within specific functional regions of the brain, but also within specific neuronal subtypes (e.g., dopaminergic, cholinergic, glutamatergic, GABAergic, etc.), as well as between neurons and other cell types of the CNS (e.g., glia). These unique cell-type specific signatures, rather then averaged chromatin states across all of the cell types in the CNS, are highly likely to vary significantly in order to support unique transcriptional outputs and cellular functions.
Extracellular and intracellular signaling cascades can communicate with nucleosome remodelers and histone chaperones, histones and DNA, therefore modifying enzymes to alter the chromatin structure (Fig. 1). Such stimuli-induced epigenetic alterations play pivotal roles in normal CNS functions, and can persist long after the initiating signal has gone. Indeed, epigenetic changes can remain throughout the lifetime of an organism and can even be transmitted to subsequent generations. In the brain, one of the most heterogeneous tissues, the complexity of epigenetic responses to the signaling is likely more complex than anticipated. Different brain areas react differently to the same environmental challenge. For instance, whereas chronic treatment with the antidepressant imipramine decreases HDAC5 levels in the hippocampus, the same treatment increases HDAC5 levels in the N-Acetyl cysteine (NAc) [43, 45]. To add to this complexity, even within the same brain area, different cell types also react differently to identical environmental stimuli. This is illustrated by the case of GABAergic and dopaminergic neurons, which are have been to be characterized by DNA hyper- and hypomethylation, respectively, in the brain of schizophrenic patients [42]. The last, but not least, important notion is that this multitude of epigenetic responses is apparently sex-dependent (reviewed in detail in [44] and [45]).
Much progress has been made in recent years in an extensive search for these specific chromatin states at the molecular, cellular, and neural network levels. However, the greater goal is to know whether, or under what circumstances, chromatin modifications can be stably maintained and propagated, and to pursue an identification of chromatin alterations in the process of neuronal plasticity and long-lasting changes in brain function.
Chromatin Landscape Influences on CNS Function
Chromatin landscape alteration is emerging as a major consequence of signaling events. How do signaling cascades communicate with writer, eraser, and reader proteins? Histone phosphorylation offers an attractive theory as there is a direct link between the regulation of gene expression through the histone code and the activity of protein kinases and protein phosphatases involved in cellular signaling.
Histone Phosphorylation
Among many histone PTMs (Fig. 2b and Fig. 3), phosphorylation on serine 10 (phSer10) of histone H3 offers the best example of a direct link between signal transduction and histone modifications [89]. H3 Ser10 residue seems to constitute a converging site of multiple intracellular pathways and kinases. Mitogenic phosphorylation of H3 in response to epidermal growth factor or 12-O-tetradeconyl-phorbol-13-acetate involves the mitogen-activated protein kinase (MAPK) cascade and the consequent activation of the extracellular-regulated kinases (ERKs). The MAPK-activated RSK-2 was identified as an epidermal growth factor-induced histone H3 kinase [90]. In addition, the same Ser10 site can also be phosphorylated by the mitogen- and stress-induced kinases MSK-1 and MSK-2 [91]. Mitogenic stimulation induces rapid, transient phosphorylation of histone H3 [90, 92, 93], which has been associated with transcriptional activation of immediate–early genes (IEGs) [94]. Such Ser10 residue phosphorylation occurs in only a small subset of H3 molecules [90]. Interestingly, H3phSer10 could be also observed at metaphase as a hallmark chromatin condensation at mitosis. In this case phosphorylation is provided by a kinase of the Aurora family and all H3 molecules are phosphorylated during mitosis [95].
In the mammalian nervous system, chromatin remodeling via H3 phosphorylation has been reported to occur in response to a light stimulation of clock neurons within the suprachiasmatic nucleus, and after GABA stimulation of neurons within the supraoptic nucleus. The light stimulation induces the ERK pathway [96, 97], phosphorylation of CREB [98], and a robust induction of IEG transcription in the CNS [96]. The activation of IEGs has suggested to be important for genomic events that control long-term changes within the nervous system, thus making H3phS10 an interesting candidate for further investigation of molecular underpinning of these long-term changes [99, 100]. The kinetics of histone H3 phosphorylation parallels the induction of phosphorylation of the ERKs. Double-labeling experiments using specific phospho-antibodies clearly demonstrated that these two events are concomitantly occurring in the same neurons, correlating with the temporal induction of the gene expression of IEGs, such is c-fos, MKP-1, and MKP-3. Published data provide evidence of profound chromatin remodeling in hippocampal neurons in response to various signaling systems. For instance, systemic injection of the agonists for the dopamine (DA), muscarinic acetylcholine (mACh) and glutamate (GLU) receptors results in a specific, intense phosphorylation of Ser10 of H3 in different subfields of the hippocampus, thus likely reflecting the intrinsic physiological differences among neurons of the various regions and the selective cellular distribution of the individual receptors. It is therefore an attractive possibility that induced levels of MPK-1 and MPK-3 phosphatases could also modulate the phosphorylation state of histone H3, while controlling the levels of phosphorylated ERKs. This could suggest a possible feedback mechanism after stimulation of DA, mACh, and GLU receptors. To date, however, there is no information about Ser10H3 phosphatases related to transcriptional regulation and there is no evidence that phosphatases of the MKP class could dephosphorylate the H3 substrate.
Similar to in vivo data, evidence of a causal link between ERK activation, H3 phosphorylation, and IEG transcriptional activation was also obtained in in vitro models using cultured cells—once again indicating that the MAPK signal transduction pathway plays a central role in H3 phosphorylation [90, 91, 96].
It is important to emphasize that ERK activation is crucial in different models of learning and memory (reviewed in [101]). ERKs have been recognized as biochemical signal integrators and molecular coincidence detectors for coordinating responses to extracellular signals in neurons [102]. An intriguing possibility is that ERK signal integration does not simply sum up signals, but rather allows synergistic effects, at least in some cases. If ERK/MAPK signaling pathways constitute a key set of molecules integrating signals in the hippocampus, than a likely scenario emerging from published data identifies histone-H3 phosphorylation as yet another biochemical signal integrator downstream of ERKs, which is capable of coordinating responses of extracellular signals in hippocampal neurons with gene transcription. The aged brain is seemingly less plastic owing to increased chromatin condensation characterized by accumulating epigenetic silencing modifications, which shut down the expression of genes that are important for learning and memory. In this context, the histone H3phSer10 and silencing histone PTMs seem to be mutually exclusive. The role of histone H3phSer10 as a biochemical signal integrator in aging and neurodegeneration awaits further investigation.
Histone Acetylation
Histone acetylation/deacetylation is one of the major epigenetic processes that control gene transcription [103, 104]. It is achieved by opposing and balancing within the cells the action of particular “writers” called histone acetyl transferases (HATs) and “erasers”, the histone deacetylases (HDACs) (Figs 3 and 4). Pharmacological HDAC inhibition induces an increase in acetylation levels of histone proteins, causing chromatin to conform more openly to the point that transcriptional factors and RNAP II interact with DNA to modulate transcription (reviewed in [105]). Moreover, histone acetylation and HDAC inhibitors (HDACi) have shown to be involved in the control of apoptosis, cell survival, differentiation, energy metabolism, and response to internal and external environmental factors (see [106] and [107] for a review).
While the effects of HATs and HDACs within the neural epigenome have been traditionally studied in the context of early developmental and heritable cognitive disorders, recent studies point to aberrant histone acetylation status as a key mechanism underlying acquired inappropriate alterations of genome structure and function in postmitotic neurons during the aging process. As several HATs are vital in the regulation of neuronal plasticity and memory formation, chromatin acetylation status can be compromised in the CNS as a result of the aging process. A number of differing epigenetic abnormalities pertaining to histone acetylation have been reported in AD, Huntington’s disease (HD) and PD.
For instance, one of the theories for mutant huntingtin toxicity in the onset of HD is based on the finding that the polyglutamine repeat region binds to the acetyl-transferase domain of two HATs: CBP and p300/CBP-associated factor [45, 50]. It has been proposed that such interaction sequesters these HATs away from chromatin, causing a reduction in global H3 and H4 acetylation levels and, as a result, altering gene expression patterns. This further supports the notion of HD as a disease of aberrantly reduced histone acetylation, and treatment with various HDACi has been shown to rescue histone acetylation levels and to improve neurodegeneration and pathological symptoms in cellular, Drosophila, and mouse models [45, 54].
Unlike HD, accumulated evidence in AD is somewhat contradictory, implicating both histone hyper- and hypo-acetylation in AD and AD-related pathologies. Numerous experiments suggest that AD could be a disease of aberrantly increased histone acetylation. It has been reported that during the processing of the membrane precursor amyloid precursor protein (APP), an intracellular fragment is released into the cytosol. In vitro, the product of processing interacts with the HAT TIP60 through the protein Fe65, and the resulting complex acts to enhance gene transcription [108, 109]. Intriguingly, it is possible that the nuclear fraction of Fe65 could be involved not only in the regulation gene transcription, but also in DNA repair processes within the cells. It has been suggested that this complex may also play a role in histone H4 acetylation required for DNA repair, which is an interesting notion considering that the amount of DNA double-strand breaks are increased in AD and AD models [110, 111]. Furthermore, presenilin1 (PS1) was shown to play an inhibitory role on the HAT CBP through proteasomal degradation, and mutations in PS1 found in hereditary AD were shown to result in aberrantly high CBP activity [112]. Data demonstrate that overexpression of HDAC1 could protect against p25/Cdk5-mediated DNA damage and neurotoxicity. Experimental evidence also suggests that lentiviral overexpression of SIRT1 in the hippocampus of p25 transgenic mice confers significant protection against neurodegeneration by rescuing neurons from p25/Cdk5-mediated neurotoxicity [113], and acts in a similar fashion to a further transgenic mouse model of AD (APP/PS1) [114] via mechanisms involving p53 and α-secretase, respectively. However, it is not clear whether, in this case, SIRT1 also had a deacetylating effect on histones. In comparing these results, a substantial body of evidence suggests that any inhibition of HDACs can be protective and beneficial in AD. For example, APP overexpression in cultured cortical neurons leads to H3 and H4 hypo-acetylation. This coincides with decreased level of CBP [115]. Furthermore, both general and class I-selective HDACi have been shown to ameliorate cognitive defects in transgenic AD mouse harboring hereditary AD mutation [116]. Such apparent inconsistency may be due to brain heterogeneity, and appears to conflict because it represents a superposition of context-dependent epigenetic events that co-occur depending on brain area, cell type, or even in the same cell at different loci, as discussed before.
In recent years, epigenetic components linking PD pathology to histone acethylation/deacetylation have also been detected. For example, nuclear α-synuclein was found to directly bind to histones in Drosophila [117]. Alpha-synuclein localizes to the nucleus and presynaptic nerve terminals and mediates neurotoxicity in the nucleus. Targeting of alpha-synuclein to the nucleus promotes toxicity, whereas cytoplasmic sequestration is protective in both cell culture and transgenic Drosophila. The study describes the reduced levels of histone acetylation in a Drosophila model of PD and suggests histone hypo-acetylation may be causally involved in PD pathology [117]. Supporting this notion, inhibition of SIRT2 has been shown to rescue neurotoxicity in Drosophila [118]. It is not known whether this also affected the histone level. However, the effects of HDACi on PD remain unclear. In some clinical reports, it has been suggested that there might be a cause-and-effect relationship between exposure to the HDACi, valproate, and the occurrence of Parkinsonism. However, most of these reports focus on the direct effect of HDACi on neurons, with insufficient consideration for the role of microglia in neuroprotection. Recently emerged data have demonstrated the immunomodulatory effects of HDACi on microglia. It was found that different classes of the HDACi, such as valproate, Trichostatin A, and sodium butyrate, induce microglial apoptosis via the mitochondria-related pathway. The apoptosis is preceded by the enhancement of acetylation of histone H3 and a robust decrease in LPS-produced pro-inflammatory responses, thus protecting dopaminergic neurons from damage in mesencephalic neuron–glia cultures [119]. Contrary to this, treatment of dopaminergic cell lines with the HDACi Trichostatin A exacerbated PD-related neurotoxic damage [120]. Collectively, these findings caution against over-interpretation of the HDACi data, particularly with the notion that substrates, other than histones, may be more critically involved in HDAC/HAT-mediated action within the CNS.
Histone Lysine Methylation
There are probably up to 100 histone methyltransferases (HMT) and demethylases encoded in the human genome, and many of these enzymes are defined by functional domains outside the catalytic sites that are thought to contribute to target specificity and genomic occupancy patterns (some of these methyltransferases are listed in Fig. 3). Unfortunately, the mechanisms of altered histone methylation and HMT activity, which may be key to achieving a greater understanding of CNS plasticity, have not been fully investigated. There are two types of histone methylation: arginine (R) and lysine (K) methylation, both of which have important functions in gene activation and repression (shown in Fig. 3). It is known that that histone lysine (K) methylation has a longer half-life then acetylation [121] or phosphorylation [122], and, like other histone PTMs, defines chromatin states and function. Different degrees (mono-, di-, and trimethylation) of the specific histone lysine has been implicated in transcriptional regulation, silencing, and enhancer functions (Fig. 3) (for review see [123]). In this review, we will focus only on histone H3 and its Lys-9 (H3K9) methylation, discussing very sparse evidence of its role in the stress response, neurodegeneration, and memory consolidation.
Increased histone H3K9 and H3K27 methylation is a well established mechanism of gene silencing, resulting in transcriptional repression. These PTMs participate in the formation of facultative and constitutive heterochromatin, and are used by specific “reader” complexes that reinforce genome stability ([7] and Fig. 4). It has been shown that single, traumatic stressor, acute restrain stress produced a substantial elevation in H3K9me3 in the hippocampus, a highly plastic brain region particularly susceptible to the effects of the environmental stress and learning [124]. Hunter et al. [125] have demonstrated that that stress-induced increases in H3K9me3 levels in the hippocampus are strongly targeted to retrotransposable elements and rapidly silence their expression. The same group have shown that acute stress decreased global levels of the H3K27me3 mark and had no effect on the levels of the H3K4me3 methylation mark [126]. It is important to note that transposable elements constitute an order of magnitude larger fraction of the genome than protein-coding genes [51, 127]. New findings suggest that transposable elements are important inducers of genomic instability [128], cellular senescence [53, 54], and neuro-inflamation [129]. Not less important, it has been reported that the capacity of some of those elements to regulate gene transcription under normal or stressed conditions could also be beneficial [51, 52, 55, 130, 131]. On this background, the data from stress-induced H3K9me3 accumulation in retrotransposal elements might represent an additional level of epigenetic regulation in the context of hippocampal function, suggesting, therefore, that epigenetic chromatin remodeling of a portion of the genome containing retrotransposons may have both beneficial and detrimental effects, and affect spatial memory, anxiety, and age-related neurodegeneration.
Analysis of HD brains revealed an excessive H3K9 trimethylation in neocortex and caudate nucleus [132]. Data also demonstrate expression of Ets-related gene (ERG)-associated protein with SET domain, (ESET/ KMT1C/SETDB1) is markedly increased in HD patients and in transgenic R6/2 HD mice. Interestingly, ESET occupancy in neuronal chromatin is highly restricted, and may be confined to <0.75 % of annotated genes. ESET is a HMT that specifically targets HK9 site with a trimethyl group [133]. Excessive H3K9 trimethylation together with increased expression of histone variant macro-H2A1 have been also observed in blood and brain tissue of striatum and frontal cortex from individuals with HD [134]. As histone H3K9 methylation is broadly associated with repressive chromatin remodeling, these changes, in conjunction with additional effects in nucleosomal density and stability, may contribute to the decreased transcriptional activity that has been observed in the striatum of people with HD disease. In particular, the decreased expression of the neurotrophic factor BDNF, dopamine receptors, and components of the MAPK signaling cascade were suggested to be targets for this event [135]. However, mice with a genetic ablation of kruppel-associated boxes (KRAB)-associated protein 1, Kap1 (also known as TRIM28/TIF1b/KRIP1), encoding the ESET binding partner [136], show increased anxiety and deficits in cognition and memory [137], and overexpression of ESET in adult forebrain neurons results in changes in motivational and effective behaviors [138]. These findings further speak to the context-dependent action of ESET and complex interpretation of results related to the therapeutic potential of modulation of H3K9 methylation.
The aged brain differs drastically from its younger counterpart and seems to be less plastic. Such lack of plasticity seems to be characterized by accumulating epigenetic silencing modifications, which shut down the expression of genes that are important for learning and memory. Data obtained from learning and memory animal models have indicated that a distortion of combinatorial action of DNA methylation and histone methylation (discussed elsewhere [139, 140]) might account for cognitive decline during aging. Several data imply a role of H3K9me2 and its “writers” Lysine(K) methyltransferase (KMT) in long-term memory (LTM) formation. Recently, G9a/GLP were found to play a role in memory consolidation [141]. G9a/G9a-like protein (GLP) complex has been implicated in diverse processes, including transcriptional silencing, heterochromatin formation, and DNA methylation [133, 142, 143]. G9a/GLP-mediated transcriptional silencing in the hippocampus and entorhinal cortex (EC) during memory consolidation is regulated by deposition of H3K9me2. Data demonstrate that inhibition of the GLP/G9a activity in EC, but not in hippocampus, results in enhanced memory consolidation by lifting the repression of memory permissive genes [141]. Such differential regulation of the genes in the hippocampus and EC highlights the function of G9a/GLP in molecular cross-talk between two regions of the brain. For instance, analysis of G9a/GLP inhibition in the EC resulted in the down-regulation of the normally observed increases in H3K9me2 levels, while further elevating H3K4me3 and H3K9ac levels in the EC during memory consolidation. Moreover, in the hippocampus, G9a/GLP blockade in the EC further increased H3K9me2 and H3K4me3 regulation. These results underscore two important concepts. First is that in the CNS, histone modifications do not occur in isolation; rather, their combinatorial effects mediate the transcriptional signature of genes within brain regions, as well as across brain regions that are necessary for LTM formation. Second, KMTs are important regulators of chromatin structure in the adult CNS that serve to coordinate both dynamic and persistent gene expression changes in several memory-related brain regions that are critical for LTM formation and storage.
Conclusions
Overall, the studies we have briefly discussed in this review are very exciting as they suggest the importance of chromatin organization in maintaining transcriptional homeostasis in the adult brain. The staggering complexity of epigenetic regulation within the CNS holds the key not only to the puzzle of brain development, but also opens the new conceptual exploration of the epigenetic molecular mechanisms underlying adult brain function, such as learning, environmental adaptation, neuronal survival, and aging. The elucidation of details of epigenetic underpinning promises to have important implications for novel advances in neurobiology. We strongly believe that one day it will offer novel targets for combating human diseases, potentially leading to new diagnostic and therapeutic avenues.
Electronic supplementary material
(PDF 1224 kb)
Acknowledgments
YZ is a Buck Institute Research Fellow, and experiments from VVL laboratory are supported by NIH R21 AG043921 and Buck Institute Trust funds. We apologize to our colleagues for omission of so many important research contributions owing to the space constraints of this review. We thank J. Jenkins and L. Hankock for help with the manuscript preparation. We acknowledge members of the Lunyak laboratory for helpful discussions. No real or perceived conflict of interest is declared. Full conflict of interest disclosures are available in the electronic supplementary material for this article.
Required Author Forms Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.Waddington CH. The strategy of the genes; a discussion of some aspects of theoretical biology. London: Allen & Unwin; 1957. [Google Scholar]
- 2.Waterland RA, Jirtle RL. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 4.Qureshi IA, Mehler MF. Long non-coding RNAs: Novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics 2013. [DOI] [PMC free article] [PubMed]
- 5.Tang B, Chang WL, Lanigan CM, Dean B, Sutcliffe JG, Thomas EA. Normal human aging and early-stage schizophrenia share common molecular profiles. Aging Cell. 2009;8:339–342. doi: 10.1111/j.1474-9726.2009.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narayan P, Dragunow M: Pharmacology of epigenetics in brain disorders. British journal of pharmacology (2010) 159(2):285–303. [DOI] [PMC free article] [PubMed]
- 7.Tollervey JR, Lunyak VV. Epigenetics: Judge, jury and executioner of stem cell fate. Epigenetics. 2012;7:823–840. doi: 10.4161/epi.21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delatte B, Fuks F. Tet proteins: On the frenetic hunt for new cytosine modifications. Brief Funct Genomics. 2013;12:191–204. doi: 10.1093/bfgp/elt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 10.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases DNMT3a and DNMT3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 11.Wu H, Coskun V, Tao J, et al. DNMT3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng J, Zhou Y, Campbell SL, et al. Dnmt1 and dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller CA, Gavin CF, White JA, et al. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in x-linked mecp2, encoding methyl-CPG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 15.Lunyak VV, Burgess R, Prefontaine GG, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 16.Szulwach KE, Li X, Li Y, et al. 5-hmc-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, Pan Q, Lin L, et al. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum Mol Genet. 2012;21:5500–5510. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Numata S, Ye T, Hyde TM, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stadler F, Kolb G, Rubusch L, et al. Histone methylation at gene promoters is associated with developmental regulation and region-specific expression of ionotropic and metabotropic glutamate receptors in human brain. J Neurochem. 2005;94:324–336. doi: 10.1111/j.1471-4159.2005.03190.x. [DOI] [PubMed] [Google Scholar]
- 20.Cheung I, Shulha HP, Jiang Y, et al. Developmental regulation and individual differences of neuronal h3k4me3 epigenomes in the prefrontal cortex. Proc Natl Acad Sci U S A. 2010;107:8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang CM, Tsai SN, Yew TW, Kwan YW, Ngai SM. Identification of histone methylation multiplicities patterns in the brain of senescence-accelerated prone mouse 8. Biogerontology. 2010;11:87–102. doi: 10.1007/s10522-009-9231-5. [DOI] [PubMed] [Google Scholar]
- 22.Lu T, Pan Y, Kao SY, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 23.Jakovcevski M, Akbarian S. Epigenetic mechanisms in neurological disease. Nat Med. 2012;18:1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peleg S, Sananbenesi F, Zovoilis A, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 25.Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct hdac(s) to treat cognitive disorders. Trends Pharmacol Sci. 2010;31:605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Erraji-Benchekroun L, Underwood MD, Arango V, et al. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry. 2005;57:549–558. doi: 10.1016/j.biopsych.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 27.Copray S, Huynh JL, Sher F, Casaccia-Bonnefil P, Boddeke E. Epigenetic mechanisms facilitating oligodendrocyte development, maturation, and aging. Glia. 2009;57:1579–1587. doi: 10.1002/glia.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ginsberg SD. Expression profile analysis of brain aging. In: Riddle DR (ed.) Brain aging: Models, methods, and mechanisms. CRS press. Boca Raton, FL, 2007, chaper 7, pp: 159–189.
- 29.Patterton D, Wolffe AP. Developmental roles for chromatin and chromosomal structure. Dev Biol. 1996;173:2–13. doi: 10.1006/dbio.1996.0002. [DOI] [PubMed] [Google Scholar]
- 30.Bonisch C, Hake SB. Histone H2A variants in nucleosomes and chromatin: More or less stable? Nucleic Acids Res. 2012;40:10719–10741. doi: 10.1093/nar/gks865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michod D, Bartesaghi S, Khelifi A, et al. Calcium-dependent dephosphorylation of the histone chaperone daxx regulates h3.3 loading and transcription upon neuronal activation. Neuron. 2012;74:122–135. doi: 10.1016/j.neuron.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernando RN, Eleuteri B, Abdelhady S, Nussenzweig A, Andang M, Ernfors P. Cell cycle restriction by histone h2ax limits proliferation of adult neural stem cells. Proc Natl Acad Sci U S A. 2011;108:5837–5842. doi: 10.1073/pnas.1014993108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scaturro M, Nastasi T, Raimondi L, Bellafiore M, Cestelli A, Di Liegro I. H1(0) rna-binding proteins specifically expressed in the rat brain. J Biol Chem. 1998;273:22788–22791. doi: 10.1074/jbc.273.35.22788. [DOI] [PubMed] [Google Scholar]
- 34.Lindner H, Sarg B, Hoertnagl B, Helliger W. The microheterogeneity of the mammalian h1(0) histone. Evidence for an age-dependent deamidation. J Biol Chem. 1998;273:13324–13330. doi: 10.1074/jbc.273.21.13324. [DOI] [PubMed] [Google Scholar]
- 35.Brown DT, Alexander BT, Sittman DB. Differential effect of h1 variant overexpression on cell cycle progression and gene expression. Nucleic Acids Res. 1996;24:486–493. doi: 10.1093/nar/24.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bantignies F, Cavalli G. Cellular memory and dynamic regulation of polycomb group proteins. Curr Opin Cell Biol. 2006;18:275–283. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at lys-9 is an early mark on the x chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 38.McKinnon PJ. Ataxia telangiectasia: New neurons and ATM. Trends Mol Med. 2001;7:233–234. doi: 10.1016/s1471-4914(01)02035-4. [DOI] [PubMed] [Google Scholar]
- 39.Hon GC, Rajagopal N, Shen Y, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet 2013; 45:1198–1206. [DOI] [PMC free article] [PubMed]
- 40.Santoro SW, Dulac C. The activity-dependent histone variant h2be modulates the life span of olfactory neurons. eLife. 2012;1:e00070. doi: 10.7554/eLife.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Develop. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 42.Edelman LB, Fraser P. Transcription factories: Genetic programming in three dimensions. Curr Opin Genet Dev. 2012;22:110–114. doi: 10.1016/j.gde.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 44.Sutherland H, Bickmore WA. Transcription factories: Gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 45.Xu M, Cook PR. The role of specialized transcription factories in chromosome pairing. Biochim Biophys Acta. 2008;1783:2155–2160. doi: 10.1016/j.bbamcr.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 46.Pina B, Suau P. Core histone variants and ubiquitinated histones 2a and 2b of rat cerebral cortex neurons. Biochem Biophys Res Commun. 1985;133:505–510. doi: 10.1016/0006-291x(85)90935-0. [DOI] [PubMed] [Google Scholar]
- 47.Duce JA, Smith DP, Blake RE, et al. Linker histone h1 binds to disease associated amyloid-like fibrils. J Mol Biol. 2006;361:493–505. doi: 10.1016/j.jmb.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 48.Thakar A, Gupta P, Ishibashi T, et al. H2a.Z and h3.3 histone variants affect nucleosome structure: Biochemical and biophysical studies. Biochemistry. 2009;48:10852–10857. doi: 10.1021/bi901129e. [DOI] [PubMed] [Google Scholar]
- 49.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants h3.3 and h2a.Z. Genes Develop. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crepaldi L, Policarpi C, Coatti A, et al. Binding of tfiiic to sine elements controls the relocation of activity-dependent neuronal genes to transcription factories. PLoS Genet. 2013;9:e1003699. doi: 10.1371/journal.pgen.1003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lunyak VV, Atallah M. Genomic relationship between sine retrotransposons, pol iii-pol ii transcription, and chromatin organization: The journey from junk to jewel. Biochem Cell Biol 2011; 89:495–504. [DOI] [PMC free article] [PubMed]
- 52.Lunyak VV. Boundaries. Boundaries…Boundaries??? Curr Opin Cell Biol. 2008;20:281–287. doi: 10.1016/j.ceb.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Geesman GJ, Hostikka SL, et al. Inhibition of activated pericentromeric sine/alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle. 2011;10:3016–3030. doi: 10.4161/cc.10.17.17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Cecco M, Criscione SW, Peckham EJ, et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell. 2013;12:247–256. doi: 10.1111/acel.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lunyak VV, Prefontaine GG, Nunez E, et al. Developmentally regulated activation of a sine b2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 56.Hu Q, Kwon YS, Nunez E, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and lsd1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Surani MA. Reprogramming of genome function through epigenetic inheritance. Nature. 2001;414:122–128. doi: 10.1038/35102186. [DOI] [PubMed] [Google Scholar]
- 58.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Solovei I, Grandi N, Knoth R, Folk B, Cremer T. Positional changes of pericentromeric heterochromatin and nucleoli in postmitotic Purkinje cells during murine cerebellum development. Cytogenet Genome Res. 2004;105:302–310. [Google Scholar]
- 60.Bosch A, Suau P. Changes in core histone variant composition in differentiating neurons: The roles of differential turnover and synthesis rates. Eur J Cell Biol. 1995;68:220–225. [PubMed] [Google Scholar]
- 61.Fontebasso AM, Liu XY, Sturm D, Jabado N. Chromatin remodeling defects in pediatric and young adult glioblastoma: A tale of a variant histone 3 tail. Brain Pathol. 2013;23:210–216. doi: 10.1111/bpa.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wan LB, Bartolomei MS. Regulation of imprinting in clusters: Noncoding rnas versus insulators. Adv Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]
- 63.Tyler JK. Chromatin assembly. Cooperation between histone chaperones and atp-dependent nucleosome remodeling machines. Eur J Biochem. 2002;269:2268–2274. doi: 10.1046/j.1432-1033.2002.02890.x. [DOI] [PubMed] [Google Scholar]
- 64.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopez MF, Tollervey J, Krastins B, et al. Depletion of nuclear histone h2a variants is associated with chronic DNA damage signaling upon drug-evoked senescence of human somatic cells. Aging. 2012;4:823–842. doi: 10.18632/aging.100507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Das C, Tyler JK, Churchill ME. The histone shuffle: Histone chaperones in an energetic dance. Trends Biochem Sci. 2010;35:476–489. doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Itoh T, Ausio J, Katagiri C. Histone h1 variants as sperm-specific nuclear proteins of rana catesbeiana, and their role in maintaining a unique condensed state of sperm chromatin. Mol Reprod Dev. 1997;47:181–190. doi: 10.1002/(SICI)1098-2795(199706)47:2<181::AID-MRD9>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 69.Yang X, Khosravi-Far R, Chang HY, Baltimore D. Daxx, a novel fas-binding protein that activates jnk and apoptosis. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein daxx is a novel histone chaperone involved in the replication-independent deposition of h3.3. Genes Develop. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. DAXX is an h3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. DAXX and histone deacetylase ii associate with chromatin through an interaction with core histones and the chromatin-associated protein DEK. J Cell Sci. 2002;115:3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- 73.Kuo HY, Chang CC, Jeng JC, et al. Sumo modification negatively modulates the transcriptional activity of CREB-binding protein via the recruitment of DAXX. Proc Natl Acad Sci U S A. 2005;10247:16973–16978. doi: 10.1073/pnas.0504460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Puto LA, Reed JC. Daxx represses relb target promoters via DNA methyltransferase recruitment and DNA hypermethylation. Genes Develop. 2008;22:998–1010. doi: 10.1101/gad.1632208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salomoni P, Khelifi AF. DAXX: Death or survival protein? Trends Cell Biol. 2006;16:97–104. doi: 10.1016/j.tcb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 76.Goldberg AD, Banaszynski LA, Noh KM, et al. Distinct factors control histone variant h3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elsaesser SJ, Allis CD. HIRA and DAXX constitute two independent histone h3.3-containing predeposition complexes. Cold Spring Harb Symp Quant Biol. 2010;75:27–34. doi: 10.1101/sqb.2010.75.008. [DOI] [PubMed] [Google Scholar]
- 78.Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone h3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 79.Kamakaka RT, Biggins S. Histone variants: Deviants? Genes Develop. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- 80.Old RW, Woodland HR. Histone genes: Not so simple after all. Cell. 1984;38:624–626. doi: 10.1016/0092-8674(84)90256-3. [DOI] [PubMed] [Google Scholar]
- 81.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 82.Jin C, Zang C, Wei G, et al. H3.3/h2a.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gessi M, Gielen GH, Hammes J, et al. H3.3 g34r mutations in pediatric primitive neuroectodermal tumors of central nervous system (CNS-PNET) and pediatric glioblastomas: Possible diagnostic and therapeutic implications? J Neurooncol. 2013;112:67–72. doi: 10.1007/s11060-012-1040-z. [DOI] [PubMed] [Google Scholar]
- 84.Lal A, Pan Y, Navarro F, et al. Mir-24-mediated downregulation of h2ax suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bassett A, Cooper S, Wu C, Travers A. The folding and unfolding of eukaryotic chromatin. Curr Opin Genet Dev. 2009;19:159–165. doi: 10.1016/j.gde.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 86.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 87.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 88.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell 2010;142:682-685. [DOI] [PMC free article] [PubMed]
- 89.Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone h3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 90.Sassone-Corsi P, Mizzen CA, Cheung P, et al. Requirement of rsk-2 for epidermal growth factor-activated phosphorylation of histone h3. Science. 1999;285:886–891. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]
- 91.Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC. The nucleosomal response associated with immediate-early gene induction is mediated via alternative map kinase cascades: Msk1 as a potential histone h3/hmg-14 kinase. EMBO J. 1999;18:4779–4793. doi: 10.1093/emboj/18.17.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mahadevan LC, Willis AC, Barratt MJ. Rapid histone h3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell. 1991;65:775–783. doi: 10.1016/0092-8674(91)90385-c. [DOI] [PubMed] [Google Scholar]
- 93.Chadee DN, Hendzel MJ, Tylipski CP, et al. Increased ser-10 phosphorylation of histone h3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J Biol Chem. 1999;274:24914–24920. doi: 10.1074/jbc.274.35.24914. [DOI] [PubMed] [Google Scholar]
- 94.Nowak SJ, Corces VG. Phosphorylation of histone h3 correlates with transcriptionally active loci. Genes Develop. 2000;14:3003–3013. doi: 10.1101/gad.848800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Crosio C, Fimia GM, Loury R, et al. Mitotic phosphorylation of histone h3: Spatio-temporal regulation by mammalian aurora kinases. Mol Cell Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- 97.Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate map kinase activation in the suprachiasmatic nuclei. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- 98.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mperiod promoters by creb-dependent signaling and clock/bmal1 activity. Proc Natl Acad Sci U S A. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goelet P, Castellucci VF, Schacher S, Kandel ER. The long and the short of long-term memory—a molecular framework. Nature. 1986;322:419–422. doi: 10.1038/322419a0. [DOI] [PubMed] [Google Scholar]
- 100.Wisden W, Errington ML, Williams S, et al. Differential expression of immediate early genes in the hippocampus and spinal cord. Neuron. 1990;4:603–614. doi: 10.1016/0896-6273(90)90118-y. [DOI] [PubMed] [Google Scholar]
- 101.Selcher JC, Nekrasova T, Paylor R, Landreth GE, Sweatt JD. Mice lacking the erk1 isoform of map kinase are unimpaired in emotional learning. Learn Mem. 2001;8:11–19. doi: 10.1101/lm.37001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rosenblum K, Futter M, Jones M, Hulme EC, Bliss TV. Erki/ii regulation by the muscarinic acetylcholine receptors in neurons. J Neurosci. 2000;20:977–985. doi: 10.1523/JNEUROSCI.20-03-00977.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 104.Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology 2013. [DOI] [PMC free article] [PubMed]
- 105.Camelo S, Iglesias AH, Hwang D, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin a ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;164:10–21. doi: 10.1016/j.jneuroim.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 106.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 107.Pirooznia SK, Elefant F. Targeting specific hats for neurodegenerative disease treatment: Translating basic biology to therapeutic possibilities. Front Cell Neurosci. 2013;7:30. doi: 10.3389/fncel.2013.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cao X, Sudhof TC. A transcriptionally [correction of transcriptively] active complex of app with fe65 and histone acetyltransferase tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 109.Minopoli G, Gargiulo A, Parisi S, Russo T. Fe65 matters: New light on an old molecule. IUBMB Life. 2012;64:936–942. doi: 10.1002/iub.1094. [DOI] [PubMed] [Google Scholar]
- 110.Adamec E, Vonsattel JP, Nixon RA. DNA strand breaks in Alzheimer's disease. Brain Res. 1999;849:67–77. doi: 10.1016/s0006-8993(99)02004-1. [DOI] [PubMed] [Google Scholar]
- 111.Kim D, Frank CL, Dobbin MM, et al. Deregulation of HDAC1 by p25/CDK5 in neurotoxicity. Neuron. 2008;60:803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marambaud P, Wen PH, Dutt A, et al. A cbp binding transcriptional repressor produced by the PS1/epsilon-cleavage of n-cadherin is inhibited by PS1 FAD mutations. Cell. 2003;114:635–645. doi: 10.1016/j.cell.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 113.Kim D, Nguyen MD, Dobbin MM, et al. Sirt1 deacetylase protects against neurodegeneration in models for alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Donmez G, Wang D, Cohen DE, Guarente L. Sirt1 suppresses beta-amyloid production by activating the alpha-secretase gene adam10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Rouaux C, Jokic N, Mbebi C, Boutillier S, Loeffler JP, Boutillier AL. Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J. 2003;22:6537–6549. doi: 10.1093/emboj/cdg615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ricobaraza A, Cuadrado-Tejedor M, Perez-Mediavilla A, Frechilla D, Del Rio J, Garcia-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an alzheimer's disease mouse model. Neuropsychopharmacology. 2009;34:1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- 117.Kontopoulos E, Parvin JD, Feany MB. Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet. 2006;15:3012–3023. doi: 10.1093/hmg/ddl243. [DOI] [PubMed] [Google Scholar]
- 118.Outeiro TF, Kontopoulos E, Altmann SM, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of parkinson's disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 119.Chen PS, Wang CC, Bortner CD, et al. Valproic acid and other histone deacetylase inhibitors induce microglial apoptosis and attenuate lipopolysaccharide-induced dopaminergic neurotoxicity. Neuroscience. 2007;149:203–212. doi: 10.1016/j.neuroscience.2007.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Y, Wang X, Liu L, Wang X. HDAC inhibitor trichostatin A-inhibited survival of dopaminergic neuronal cells. Neurosci Lett. 2009;467:212–216. doi: 10.1016/j.neulet.2009.10.037. [DOI] [PubMed] [Google Scholar]
- 121.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. J Biol Chem. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Barth TK, Imhof A. Fast signals and slow marks: The dynamics of histone modifications. Trends Biochem Sci. 2010;35:618–626. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 123.Peter CJ, Akbarian S. Balancing histone methylation activities in psychiatric disorders. Trends Mol Med. 2011;17:372–379. doi: 10.1016/j.molmed.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Crosio C, Heitz E, Allis CD, Borrelli E, Sassone-Corsi P. Chromatin remodeling and neuronal response: Multiple signaling pathways induce specific histone h3 modifications and early gene expression in hippocampal neurons. J Cell Sci. 2003;116:4905–4914. doi: 10.1242/jcs.00804. [DOI] [PubMed] [Google Scholar]
- 125.Hunter RG, Murakami G, Dewell S, et al. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci U S A. 2012;109:17657–17662. doi: 10.1073/pnas.1215810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal h3 histone methylation by acute and chronic stress. Proc Natl Acad Sci U S A. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the encode pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kazazian HH, Jr, Goodier JL. Line drive. Retrotransposition and genome instability. Cell. 2002;110:277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- 129.Perron H, Lang A. The human endogenous retrovirus link between genes and environment in multiple sclerosis and in multifactorial diseases associating neuroinflammation. Clin Rev Allergy Immunol. 2010;39:51–61. doi: 10.1007/s12016-009-8170-x. [DOI] [PubMed] [Google Scholar]
- 130.Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: Sine RNAs regulate mRNA production. Curr Opin Genet Dev. 2010;20:149–155. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Muotri AR, Marchetto MC, Coufal NG, et al. L1 retrotransposition in neurons is modulated by mecp2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ryu H, Lee J, Hagerty SW, et al. Eset/setdb1 gene expression and histone h3 (k9) trimethylation in huntington's disease. Proc Natl Acad Sci U S A. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rea S, Eisenhaber F, O'Carroll D, et al. Regulation of chromatin structure by site-specific histone h3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 134.Hu Y, Chopra V, Chopra R, et al. Transcriptional modulator h2a histone family, member y (h2afy) marks huntington disease activity in man and mouse. Proc Natl Acad Sci U S A. 2011;108:17141–17146. doi: 10.1073/pnas.1104409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stack EC, Del Signore SJ, Luthi-Carter R, et al. Modulation of nucleosome dynamics in huntington's disease. Hum Mol Genet. 2007;16:1164–1175. doi: 10.1093/hmg/ddm064. [DOI] [PubMed] [Google Scholar]
- 136.Ayyanathan K, Lechner MS, Bell P, et al. Regulated recruitment of hp1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes Develop. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jakobsson J, Cordero MI, Bisaz R, et al. Kap1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron. 2008;60:818–831. doi: 10.1016/j.neuron.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 138.Jiang Y, Jakovcevski M, Bharadwaj R, et al. SETDB1 histone methyltransferase regulates mood-related behaviors and expression of the nmda receptor subunit NR2B. J Neurosci. 2010;30:7152–7167. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Penner MR, Roth TL, Chawla MK, et al. Age-related changes in arc transcription and DNA methylation within the hippocampus. Neurobiol Aging. 2011;32:2198–2210. doi: 10.1016/j.neurobiolaging.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Peters AH, Schubeler D. Methylation of histones: Playing memory with DNA. Curr Opin Cell Biol. 2005;17:230–238. doi: 10.1016/j.ceb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 141.Gupta-Agarwal S, Franklin AV, Deramus T, et al. G9a/glp histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J Neurosci. 2012;32:5440–5453. doi: 10.1523/JNEUROSCI.0147-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shinkai Y, Tachibana M. H3k9 methyltransferase G9a and the related molecule GLP. Genes Develop. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1224 kb)