Abstract
Huntington’s disease (HD) is an incurable and fatal hereditary neurodegenerative disorder of mid-life onset characterized by chorea, emotional distress, and progressive cognitive decline. HD is caused by an expansion of CAG repeats coding for glutamine (Q) in exon 1 of the huntingtin gene. Recent studies suggest that epigenetic modifications may play a key role in HD pathogenesis. Alterations of the epigenetic “histone code” lead to chromatin remodeling and deregulation of neuronal gene transcription that are prominently linked to HD pathogenesis. Furthermore, specific noncoding RNAs and microRNAs are associated with neuronal damage in HD. In this review, we discuss how DNA methylation, post-translational modifications of histone, and noncoding RNA function are affected and involved in HD pathogenesis. In addition, we summarize the therapeutic effects of histone deacetylase inhibitors and DNA binding drugs on epigenetic modifications and neuropathological sequelae in HD. Our understanding of the role of these epigenetic mechanisms may lead to the identification of novel biological markers and new therapeutic targets to treat HD.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-013-0206-5) contains supplementary material, which is available to authorized users.
Keywords: DNA methylation, Histone code, Chromatin remodeling, Noncoding RNA, microRNA, Huntington’s disease, Therapeutics
Introduction
Huntington’s disease (HD) is an autosomal dominant and fatal brain disorder characterized by chorea (uncoordinated movement), psychiatric symptoms, and cognitive decline. HD has a frequency as great as 10 cases per 100,000 and a 1–3 % new mutation rate [1]. HD occurs worldwide in all races and ethnic groups [2]. The frequency is highest in Venezuela along the shores of Lake Maracaibo. In the USA, there are 30,000 HD patients and another 200,000 are at genetic risk.
The first clinical description of HD and its hereditary nature was reported by George Huntington in1872 [3]. HD was initially regarded as a chronic encephalitis but in 1908 Jergelsma described the characteristic neuropathological alterations affecting the basal ganglia that are now accepted as the essential patho-anatomical feature of HD [4, 5]. The most striking neuropathological changes of HD are gross atrophy of the neostriatal nuclei, the caudate nucleus, and putamen accompanied by marked neuronal loss and astrogliosis [6–8]. Interestingly, there is a selective pattern of neuronal vulnerability and topographic susceptibility, and not all striatal neurons are equally affected. Medium-sized spiny neurons are most severely affected at the earliest stage of disease, whereas intrinsic locally arborizing aspiny striatal interneurons are relatively spared [8–14].
In 1993, the Huntington’s Disease Collaborative Research Group reported that a previously unknown “interesting transcript 15” on human chromosome 4 is mutated in HD patients and genetically linked to HD [15]. This HD-related lethal gene was named huntingtin (htt). The mutation of the htt gene was found to be an expansion of the wild-type htt allele that normally contains 15–35 CAG triplets in exon 1 to 36 or more repeats. HD is related to other neurodegenerative diseases, such as spinal and bulbar muscular atrophy (also known as Kennedy’s disease) and spinocerebellar ataxias (SCAs), which are caused by similar trinucleotide CAG repeat mutations. Spinal and bulbar muscular atrophy is caused by androgen receptor gene mutations, while SCA1 is caused by the expansion of a polyglutamine tract within the SCA1 gene product ataxin-1 [16, 17]. Interestingly, affected individuals with greater numbers of CAG repeats exhibit younger age of onset and there is a significant inverse relationship between age of onset and CAG repeat number in HD.
Htt is a ubiquitously expressed cytoplasmic protein found heterogeneously in neurons throughout the brain. Since the discovery of the htt gene, a number of hypothetical pathologic mechanisms have been suggested, but a direct pathway from the genetic mutation to neuronal degeneration has not been established. The exact function of htt remains unknown, but involvement in intracellular transport, autophagy, transcription, mitochondrial function, and signal transduction have been posited. Mutant htt (mthtt) inhibits fast axonal transport and destabilizes microtubules within the cell [18, 19]. Both normal and mutant alleles are expressed in heterozygous HD. While mutant htt protein is toxic and triggers the pathologic cascades of the disease through a “gain of function”, the deletion of the normal htt gene is also fatal, suggesting that the function of normal htt is important in survival [20–27]. It is well established that mutant htt and its proteolytic fragments engage in pathologic protein–protein interactions, contributing to alterations of cellular pathways those make neurons more susceptible to generic stresses, eventually leading to neuronal damage and death [28]. Mutant htt interactomes involve transcriptional dysregulation, mitochondrial dysfunction, pro-apoptotic signaling, oxidative injury, excitotoxicity, inflammatory reactions, and malfunctioning proteolysis. Despite significant progress towards understanding disease mechanisms over the more than 140 years since Dr. Huntington’s initial report, no treatment is currently available to prevent the onset, or to delay the insidious and relentless course of HD [29].
The term “epigenetics” was introduced by Dr. Waddington to explain biological events that are not described by genetic principals [30]. Since then, epigenetics has evolved and is now defined as the field of study connecting genotype to phenotype in the absence of altered DNA sequence [31]. In this paradigm, epigenetics is a very fruitful field to explore features and mechanisms underlying the temporal and spatial control of gene activity regulated by processes beyond DNA sequence mutation [32]. Epigenetic modifications encompass an array of molecular modifications to both DNA and chromatin, including regulation of noncoding RNAs (ncRNAs). In general, many genes contain DNA methylation sites (CpG islands) in their promoters [33]. Therefore, marked hypo- or hyper-DNA methylation may account for significant aspects of the molecular and pathogenic complexity of human genomes. A growing body of evidence suggests that alterations of epigenetic modifications constitute a basic molecular mechanism contributing to HD pathogenesis (Fig. 1). Understanding epigenetic mechanisms may therefore provide important insights leading to the identification of new biological markers and novel therapeutics to treat HD [34]. To this end we will provide a brief overview of recent findings related to alterations of DNA methylation, histone modification, and ncRNAs linked to HD pathogenesis, and discuss modulation of epigenetic components by therapeutic compounds and approaches to treat HD.
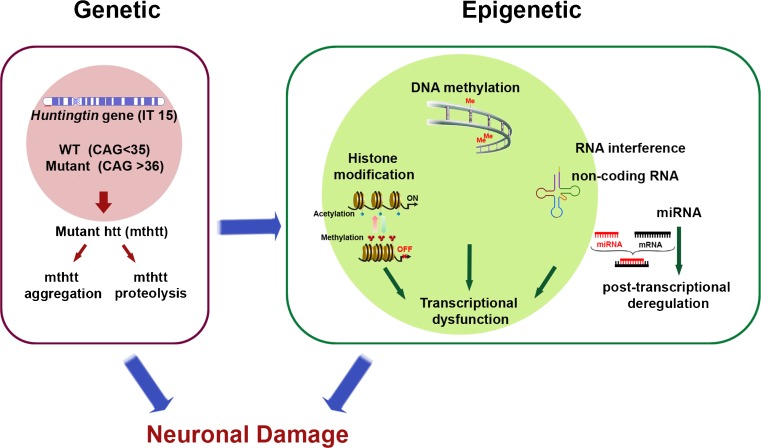
Fig. 1.
Alterations in epigenetic modifications are linked to the pathogenesis of Huntingdon’s disease (HD). Genetic mutation of the huntingtin (htt) gene (known as interesting transcript 15 (IT15)) leads to epigenetic alterations in neurons. DNA methylation is altered in the promoter region of neuronal genes in HD. Altered gene transcription in HD is associated with post-translational modifications of histone and abnormal nucleosomal dynamics. Neuronal gene expression is turned on (active) or off (silenced) depending on the dynamic status of histone acetylation versus methylation, respectively. Changes in noncoding RNA (ncRNA) and microRNA (miRNA) activity can deregulate gene expression at the transcriptional and post-transcriptional levels. Both aggregates and fragments of mutant htt (mthtt) may cause significant epigentic alterations that lead to synaptic and, ultimately, neuronal damage and loss in HD. The mechanisms by which these pathogenic insults trigger epigenetic modifications remain to be determined. WT = wild-type
Epigenetic Modifications in HD
DNA Methylation in HD
DNA methylation is a fundamental epigenetic modification that regulates gene expression and orchestrates changes in multiple genes. DNA methylation is a covalent addition of a methyl group to the number 5 carbon of the cytosine pyrimidine ring via DNA methyltransferase (DNMT) activity. DNMTs catalyze the transfer of a methyl group to single-stranded DNA using S-adenosyl methionine as the methyl donor. The recognition sequence for the mammalian DNMT is relatively conserved, with nearly all cytosine methylation occurring on 5-C-p-G-3 (CpG) [33, 35, 36]. DNA methylation typically occurs in CpG islands that are present in the 5′-untranslated regions (UTRs) of gene promoters. So far, 4 DNMTs are identified in mammals: DNMT1, DNMT2, DNMT3A, and DNMT3B. While DNMT1 is ubiquitously and most abundantly expressed in mammalian cells, the other DNMTs are differentially expressed in a cell type-specific manner. DNMT1 plays a key role in maintaining methylation in somatic cells, and loss of DNMT1 leads to nuclear disorganization, increased histone acetylation, and cell death [37–41]. DNA methylation affects the transcription of genes in 2 ways. First, methylated DNA physically impedes the binding of transcription factors to the gene. Second, and likely more important, methylated DNAs are occupied by single methyl-CpG-binding domain proteins (MBDs). Methylation stability is further maintained by the binding of MeCP1 to methylated regions of DNA. MeCP1 interacts with MBD2 and MeCP1/MBD2 complex, and, in conjunction with CDK2AP1 (Doc1), recruits other epigenetic components to the locus, such as nucleosome remodeling and histone deacetylase (NuRD) complex and other chromatin remodeling proteins that can modify histones, thereby forming compact and inactive heterochromatin [42, 43]. Consequently, DNA methylation in gene promoter regions results in gene inactivation/silencing or activation in a gene context-dependent manner (Fig. 1). Even though DNA methylation is the most studied epigenetic mechanism to date, epigenetic changes in terms of DNA methylation in HD are just beginning to be explored [44]. If DNA methylation is altered in HD it could affect many different aspects of gene expression because it is a highly conserved process.
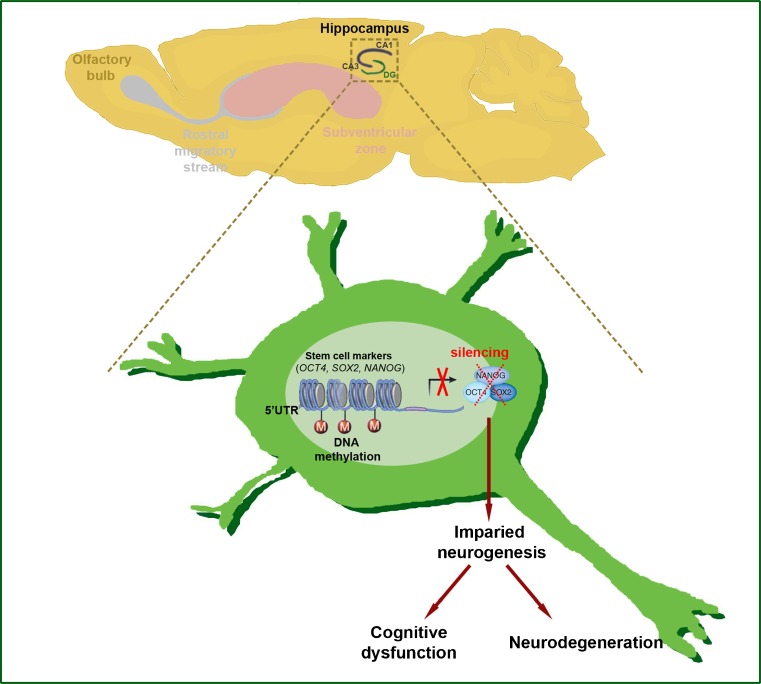
Altered DNA methylation has recently been found in HD patients and HD transgenic mice [44–46]. Interestingly, Ng et al. [44] showed that the promoter regions of the Ap-1, Sox2, Pax6, and Nes genes are highly methylated, and that expression levels of these genes are significantly reduced. Because these genes are directly involved in neurogenesis, DNA methylation-dependent impairment of hippocampal neurogenesis may be casually and mechanistically linked to cognitive decline in HD (Fig. 2). The DNA methylation pattern of these genes needs to be verified in HD patients in future studies.
Fig. 2.
DNA methylation deregulates neurogenesis in Huntingdon’s disease (HD). Regional-specific neural stem and progenitor cells turn into mature neurons of central nervous system by the process of neurogenesis. In HD, 5′-untranslated region (UTR) promoters of stem cell-related genes (octamer-binging transcription factor 4 (OCT4), SRY (sex determining region Y)-box 2 (SOX2), and Nanog homeobax (Nanog)) are methylated and neurogenesis is affected. Impaired neurogenesis results in cognitive dysfunction and could be an important epigenetic marker of neurodegeneration in HD
It has been shown that the adenosine A (2A) receptor (A2AR) is markedly reduced in HD and it has been implicated as a potential therapeutic target, but regulation of the A2AR gene (ADORA2A) expression is not known in HD [45]. A recent study shows that while the reduction of A2AR is correlated with increased levels of 5-methylcytosine in the 5′-UTR region of ADORA2A gene in the striatum of HD patients, A2AR down-regulation is correlated with a reduction of 5-hydroxymethylcytosine in the 5′-UTR region of the ADORA2A gene in HD transgenic mice [46]. Even though DNA methylation is differentially modulated between species, the evident DNA methylation of ADORA2A gene suggests additional molecular and pathological mechanisms possibly relevant to HD. Guanine methylation is less well studied than 5-cytosine methylation in HD models. Thomas et al. [47] have found that 7-methylguanosine (7-MG) levels are significantly changed in cytoplasmic and nuclear DNA in the human HD brain and animal models compared with controls [47]. Collectively, the above studies suggest that DNA methylation is one of the epigenetic alterations contributing to HD pathogenesis. However, how mutant htt triggers DNA methylation and which DNMTs and DNA methylation maintenance factors are directly or indirectly involved in abnormal epigenetic modifications in HD remains to be determined.
Histone Modifications and Chromatin Remodeling in HD
Histone modification is a second major epigenetic mechanism that has been widely studied. The association of histone proteins with DNA is affected by histone modifications that modulate the dynamic nature of chromatin fibers [48]. In general, gene expression is regulated by two components that act in concert: the binding of transcriptional activators and repressors, and the alteration of chromatin structure governed by histone modification and chromatin remodeling. Chromatin remodeling is a dynamic and highly regulated process that occurs through interactions between DNA, RNA, and histone proteins in the nucleus [49, 50]. A basic unit of chromatin is the nucleosome, which consists of core histone proteins (H2A, H2B, H3, and H4) that form an octamer around which DNA (147 base-pairs) winds tightly. When the nucleosome is assembled to a distinct compact histone/DNA conformation called heterochromatin, the result is gene silencing. In contrast, when the nucleosme is lightly packed and assumes a more relaxed structure called euchromatin, this relatively open chromatin region is associated with active gene transcription [51, 52] (Fig. 1). Chromatin organization is reversibly subjected to numerous post-translational modifications (PTMs) of histone proteins, collectively known as the “histone code”, that directly affects the plasticity of chromatin structure [53]. Amino (N)-terminal tails of core histones are strongly basic and contain specific amino acid residues that serve as sites for several PTMs, including acetylation, methylation, phosphorylation, and ubiquitylation [32, 54]. These covalent PTMs determine the “histone code”. For example, acetylation of histone H3K9 residue corresponds to transcriptionally active chromatin (euchromatin) that promotes transcription. In contrast, hypermethylation of histone H3K9 residue contributes to transcriptionally inactive chromatin (heterochromatin) and represses transcription [32]. PTMs of histone molecules are catalyzed by various enzymes, including histone acetyltransferase (HAT), histone deacetylase (HDAC), histone methyltransferase (HMT), and histone demethylase. In this context, the pattern of histone modifications is regulated and maintained through the balancing action of chromatin-modifying enzymes that add and remove modifications to histone tails in response to cellular signals. Importantly, histone modifications, such as hypo-acetylation and hypermethylation, have been identified in HD patients, HD animal models, and HD cell line models [53–56]. Interestingly, the hypo-acetylation of histone is correlated with down-regulations of genes in HD, and therapeutic inhibition of HDAC restores the acetylation level of histone and improves the neuropathology and the motor symptom in HD [56–61].
HAT Dysfuction in HD
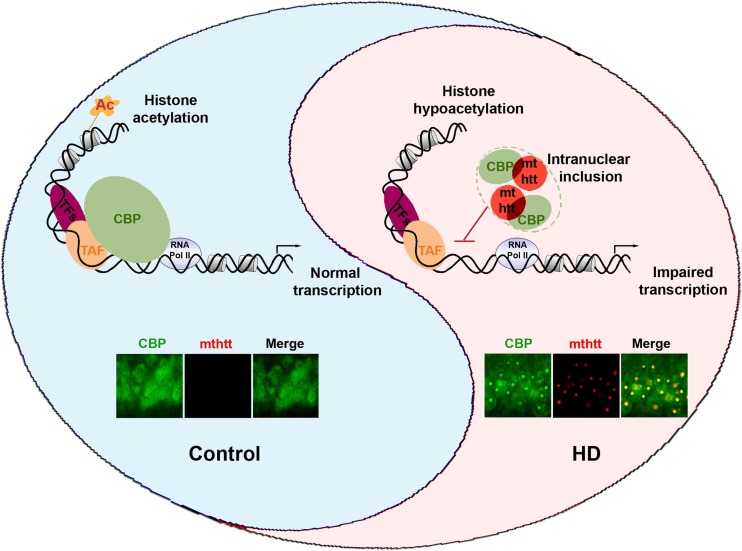
Cyclic adenosine monophosphate response element-binding protein (CREB) binding protein (CBP) functions as a HAT and a transcriptional cofactor. CBP plays a role as a HAT in acetylating histones that contribute to transcription by remodeling the chromatin structure [62]. CBP also interacts with diverse transcription factors and with components of the RNA polymerase II complex, thereby acting as a co-activator or repressor of transcription. A loss of CBP function interferes with transcription by inhibiting recruitment of the basal transcription machinery to the promoter and by altering the acetylation level of histones and chromatin structure in neurons (Fig. 3) [55, 63]. Importantly, it has been known for more than a decade that sequestration of CBP by mthtt leads to neuronal transcriptional dysfunction [21, 57]. The polyglutamine stretches in mthtt interact physically with CBP and block its transcriptional co-activator function, as well as intrinsic CBP HAT activity [50, 64]. Accordingly, the sequestration of CBP protein by mthtt expression causes the hypermethylation and hypo-acetylation of histone proteins, and the subsequent transcriptional dysfunction of neurons in HD [56, 59, 61, 65, 66]. These specific interactions and transcriptional dysfunction are attributable to pathological epigenetic modifications [51]. Korzus et al. [63] found that the stabilization of short-term memory into long-term memory is impaired in transgenic mice expressing CBP that lacks HAT activity, whereas acquisition of new information and short-term memory is spared. Concurrent with these findings, p300 (a CBP homologue) mutant mice lacking carboxy-terminal HAT and activation domains have impaired long-term recognition memory and contextual fear memory [67]. Moreover, Oliveira et al. [67] demonstrated that p300 is required for certain forms of memory, and that the HAT and carboxy-terminal domains play critical roles. The molecular dysfunction of CBP may therefore be linked to cognitive dysfunction in HD.
Fig. 3.
A scheme illustrates how mutant huntingtin (mthtt) contributes to cyclic adenosine monophosphate response element-binding protein (CREB) binding protein (CBP) dysfunction in Huntingdon’s disease (HD). In normal conditions, CBP maintains the acetylation status of histone through histone acetyltransferase activity and regulates the initiation of transcription by interacting with transcriptional complexes in a gene context-dependent manner. In HD, mthtt sequestrates CBP in nuclear inclusions (aggregate formation) and disrupts CBP-dependent histone modification and transcription. Consequently, imbalanced transcription and altered chromatin remodeling leads to neuronal damage resulting in cognitive dysfunction and other symptoms in HD. The confocal images show that colocalization of mthtt (red) and CBP (green) is found in nuclear inclusions of the hippocampus of HD (R6/2) mouse. TFs = transcription factors; TAF = TATA-binding protein (TBP)-associated factor; RNA Pol II = RNA polymerase II
Alteration of HMT in HD
Because abnormally increased histone methylation occurs concurrently with altered histone acetylation related to CBP dysfunction, our group hypothesized that CBP may have effects independent of HAT activity that contribute to chromatin remodeling [68]. We discovered an alternative mechanism of histone methylation associated with mono-allelic deletion of CBP that is regulated independently from HAT activity by induction of erythroblast transformation-specific (ETS)-related gene (ERG)-associated protein with Drosophila Su(var)3-9 and ‘Enhancer of zeste’ proteins (SET) domain [68]. We hypothesized that CBP represses the expression of SETDB1 gene and maintains an appropriate level of trimethylated histone H3K9 (H3K9me3) in neurons. A loss of CBP function leads to elevated SETDB1 gene expression and H3K9 hypermethylation. Indeed, the condensation of H3K9me3-dependent heterochromatin structure has been shown to be a prominent pathological feature of HD. Our group found that the levels of SETDB1 protein and histone H3K9me3 are elevated in striatal neurons of HD patients and HD transgenic animal models [66]. These data suggest that neuronal levels of SETDB1 and H3K9me3 may be predictive markers of nucleosomal dysfunction in HD [32]. Consistent with this, a recent study confirmed that altered gene transcription in HD is directly associated with H3K9me3-mediated chromatin remodeling [69] (Fig. 4). Altered expression of HMT and elevated H3K9me3 are linked to upstream transcriptional deregulation in both animal models of HD and in HD patients [65, 66, 70]. Thus, the increase of H3K9me3 level has been correlated with the formation of large constitutive heterochromatin domains and is thought to promote gene silencing in both global and local repression of transcription, including CHRM1 [68, 71].
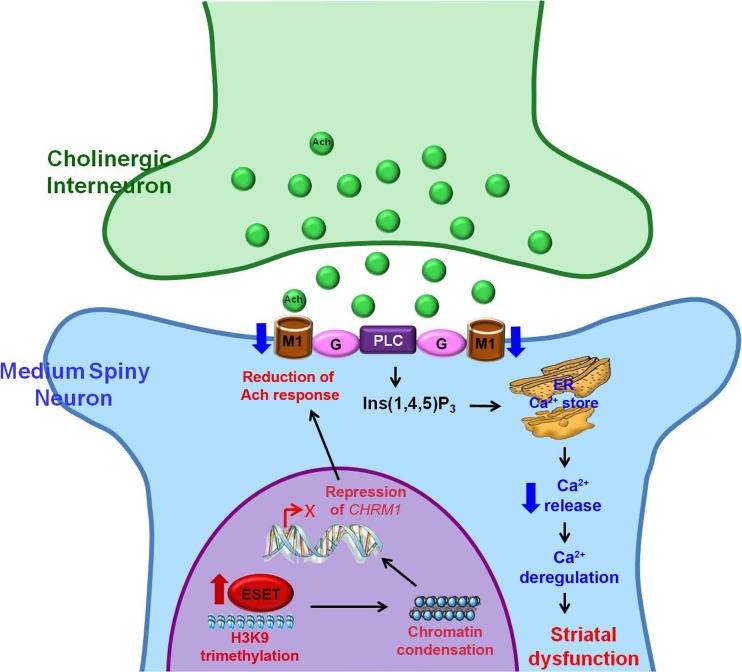
Fig. 4.
A scheme represents an epigenetic mechanism that abnormal activity of histone H3K9-specific methyltransferase (ESET) leads to synaptic failure and striatal dysfunction in Huntingdon’s disease (HD). ESET-induced and H3K9me3-mediated heterochromatin condensation results in the repression of the CHRM1 gene and subsequent reduction of CHRM1 protein in medium spiny neurons (MSNs). Down-regulation of CHRM1 fails to respond to acetylcholine (Ach) from cholinergic interneurons and to transduce the G-protein-coupled intracellular Ca2+-dependent signaling pathway, which affects on the synaptic function of MSNs. Consequently, deregulation of CHRM1-dependent striatal synaptic function contributes to neurodegeneration in HD. This figure is reproduced from [69]. PLC = phospholipase C; Ins(1,4,5)P3 = inositol 1,4,5-triphosphate; ER = endoplasmic reticulum
We recently discovered a novel epigenetic pathway whereby H3K9me3-dependent heterochromatin condensation leads to transcriptional deregulation of muscarinic acetylcholine (Ach) receptor 1 (CHRM1) by occupying its promoter in HD striatal cells [69]. CHRM1 promotes phosphatidylinositol hydrolysis and intracellular Ca2+ mobilization, while CHRM2 is coupled negatively to adenylate-cyclase activity [72–75]. The conventional wisdom is that loss of cholinergic receptor function directly results in synaptic dysfunction through inadequate intracellular signal transductions in neurons. The CHRM1 is highly expressed in the striatum of control brain, but is down-regulated in HD striatum [72, 76, 77]. Despite the deregulation of striatal cholinergic system has been suggested in the pathophysiology of HD, the cellular mechanisms underlying the epigenetic regulation of CHRM1 expression in striatal neurons are unknown. Furthermore, because Ach plays a key role in the regulation of striatal output by influencing the activity of gamma-aminobutyric acid-ergic medium spiny neurons, the interaction of Ach with pre- and post-synaptic CHRMs is pivotal to modulate the striatal activity (Fig. 4). Accordingly, the net effect of gamma-aminobutyric acid-ergic medium spiny neurons is dependent upon the expression type and location of the muscarinic receptors [78, 79]. Consequently, as CHRM1 is a major muscarinic receptor transducing intracellular Ca2+ signaling, the reduced CHRM1 protein levels result in the deregulation of intracellular Ca2+ release from endoplasmic reticulum in response to Ach in HD striatal cells [69] (Fig. 4).
Why the alteration of HMT and HAT activity turns into a pathologically catastrophic event under neurodegenerative conditions, despite the yin and yang balance of HAT and HMT activity is essential and critical in maintaining neuronal transcriptional and synaptic activity, is unknown. Probably, at first, the acute neuronal stresses may induce HMT and HAT, and lead to histone modifications and gene expression as a stress coping mechanism, and it should be a reversible reaction. However, in a chronic stress condition, the repetitive histone modifications by HMT and HAT may erroneously lose plasticity, convert epigenetic signals irreversibly, and contribute to neuronal damage and neurodegeneration.
Alteration of ncRNAs in HD
Several types of ncRNAs such as microRNAs (miRNAs), small interfering RNAs (siRNAs), ribosomal RNA, transfer RNA, small nucleolar RNA, and piwi-interacting RNA do not directly encode proteins. Investigations over the last decade have shed light on novel functions of noncoding small RNAs that control genetic pathways. Short (~22 nucleotides) miRNAs are involved in gene expression at the post-transcriptional level. They can target consensus RNA-binding motifs and induce degradation and translational repression. Their processing is initiated by RNA polymerase II as primary miRNAs. Drosha is a miRNA processor and is essential for miRNA maturation. Drosha cleaves primary miRNA to precursor-miRNAs (pre-miRNAs) in the nucleus [80]. Exportin-5 facilitates the exit of pre-miRNAs and Dicer removes the loop of pre-miRNAs to produce the mature miRNA duplex. One strand of the duplex combines with the RNA-induced silencing complex [81]. MiRNA machinery (microprocessor), including Drosha and DGCR8, participate in processing of miRNAs for the proper execution of gene expression programs [80]. They specifically target, cleave, or degrade messenger RNA (mRNA) and regulate its expression by inhibiting the consequent translation of target mRNAs into proteins.
A growing body of evidence shows that alterations of miRNAs are linked to HD pathogenesis. Johnson et al. [82] found that the level of neuronal specific miRNAs is decreased, while the level of target mRNAs is inversely elevated in murine models of HD and HD patients [81]. Lee et al. [83] performed miRNA arrays on the striatum and showed that miRNAs (e.g., miR-22, miR-29c, miR-128, miR-132, miR-138, miR-218, miR-222, miR-344, and miR-674) are down-regulated, and the levels of Drosha, a nuclear microprocessor, are correspondingly decreased in both transgenic HD R6/2 mice at 10 weeks of age and YAC128 mice at 12 months of age, respectively. These studies suggest additional mechanisms through which deregulation of miRNA biogenesis may contribute to post-transcriptional malfunction in HD [83, 84]. Furthermore, a recent study by Ghose et al. [85] suggests that mthtt decreases the expression of miR-125b and miR-15. As miR-125b and miR-15 are known to negatively regulate the expression of p53, these down-regulated miRNAs lead to increased levels of p53 which, in turn, decrease nuclear factor kappa B (NFκB)/p65 expression (RelA/NFκB), NFκB activity, and miR-146a expression. p53 interacts with mthtt aggregates and also induces nuclear and mitochondria-mediated neuronal damage in HD [86]. The cross-talk between miRNAs and transcription factors suggests another layer of mechanisms in HD pathogenesis. How mthtt triggers the abnormal biogenesis of miRNAs via microprocessors-dependent pathway is a topic for further study.
Therapeutic Approaches for Epigenetic Modifications in HD
Epigenetic modifications are reversible, while genetic mutations are irreversible. Therefore, from a therapeutic perspective, epigenetic components and modifications are a strong candidate of drug targets. Small compounds can dynamically modulate the status of DNA methylation and remodel the structure of chromatin through inhibition of DNA methylation and post-translational modifications of histone molecules in HD. By the same token, mutation of the htt allele may be correctable by noncoding small RNAs. In this context, the development of such drug agents that realign the epigenetic balance and subsequently improve HD-related epigenetic deficits is a main interest in HD-related research.
HDAC Inhibitors
Histone acetylation is regulated through the concerted activities of HAT and HDACs [51, 87]. It is widely believed that HAT activity acetylates on lysine residues in the histone tails and transforms intra- and/or internucleosomal structure locally and results in increased DNA transcription [88]. In contrast, recruitment of HDACs to DNA alters chromatin structure and inhibits transcription. Accordingly, HDAC inhibitors can promote either transcription activation or suppression by relaxing DNA conformations in a gene context-dependent manner. HDAC inhibitors have been preclinically tested in many neurodegenerative conditions, including animal models of HD, amyotrophic lateral sclerosis, and multiple sclerosis [55, 56, 65, 66, 89, 90]. Suberoylanilide hydroxamic acid and sodium butyrate are the first compounds that provide the efficacy of HDAC inhibition and histone modification in HD transgenic mice [91, 92]. These pioneering studies ignited the rationale for beginning clinical testing of HDAC inhibitors in humans with HD. Enhancement of memory formation is found in mice treated with sodium butyrate or suberoylanilide hydroxamic acid, or subjected to genetic knockout of the HDAC2 gene [92]. In the case of CBP deficiency and HAT deletion mutant animal models, HDAC inhibitors also improve memory and behavioral symptoms. In cell models of HD, polyglutamine decreases histone acetylation, and HDAC inhibitors have been shown to reduce polyglutamine-induced toxicity [93]. HDAC inhibitors also improve the phenotypes of transgenic Drosophila and mouse models of HD [57, 60, 65, 92, 94]. A number of HDAC inhibitors are currently under development as therapeutics to target neurodegenerative diseases.
Of the five classes of HDAC inhibitors, sodium butyrates are the most developed for clinical use, and their bioavailability in the central nervous system is known. The toxicity of sodium butyrate is low, and it is well tolerated in both human and animal studies [95–97]. Sodium butyrate modulates epigenetic histone modifications, improves motor performance and neuropathologic sequelae, and significantly extends survival of transgenic HD (R6/2) mice [94].
Phenylbutyrate is metabolized to phenylacetate that posseses HDAC inhibitory activity and shows high levels of brain bioavailability [98]. Phenylbutyrate is a feasible compound for testing in HD because it is Food and Drug Administration-approved and there is a preponderance of pharmacokinetic, toxicity, and available dosing information available. Phenylbutyrate remodels chromatin structures and improves the motor deficits and neuropathological phenotype observed in HD mice [65]. Given the potential benefit of HDAC inhibitors on the overall HD phenotype (mortality and neuropathology) in animal models, they have been applied in clinical trials for HD patients. A multicenter, double-blind, placebo-controlled study with open-label follow-up to determine the safety and tolerability of phenylbutyrate in patients with HD has been completed [99, 100]. As the potency of phenylbutyrate is very low and high doses are required in patients, it is not an ideal therapeutic compound for HD patients.
Thomas et al. [60] reported that a novel pimelic diphenylamide HDAC inhibitor 4b (HDACi 4b) ameliorates the disease phenotype and transcriptional abnormalities in HD transgenic (R6/2) mice [60]. HDACi 4b treatment effectively restored acetylation of histone H3 and corrected mRNA expression levels in HD mice. HDACi 4b also significantly improved body weight and several parameters of motor function, and ameliorated cognitive decline in N171-82Q transgenic mice [101]. Interestingly, HDACi 4b treatment modulated gene networks involving post-translational modification, including protein phosphorylation and ubiquitination pathways. Furthermore, activation of inhibitor of kappaB kinase by HDACi 4b contributed to phosphorylation, acetylation, and clearance of the Htt protein through the ubiquitin-proteasomal and autophagy pathways. In addition, the Thomas group has examined the selectivity and biological effects of HDACi 4b and related compounds against class I and class II HDACs to determine whether they restored expression of deregulated genes in HD mice and rescued disease effects in cell line and Drosophila models [102]. HDAC inhibitors targeting HDAC3 and HDAC1 ameliorated mthtt-induced eye and neurodegeneration in Drosophila and improved mthtt-elicited metabolic deficits in STHdhQ111 striatal cells. HDACi 4b and 136, 2 compounds showing high potency for inhibiting HDAC3, were most effective in reversing the expression of genes relevant to HD. These findings suggest that HDACi 4b possesses beneficial biological effects and efficacy that may be applicable to the treatment of HD patients. Taken together, it is encouraging that HDAC inhibitors improve phenotypes by either upregulating survival genes that are repressed in HD or by repressing prodeath genes that are elevated in HD [103]. However, the underlying precise mechanisms whereby HDAC inhibitors modulate neuronal function remain to be investigated.
DNA-binding Drugs
Anthracyclines, including mithramycin A and chromomycin A3, are potent DNA intercalating agents (Table 1). Mithramycin is isolated from Streptomyces argillaceus and has been used to treat Paget’s disease, hypercalcemia in malignancy, and several types of cancer [104–110]. Chromomycin is isolated from Streptomyces griseus [104]. Mithramycin and chromomycin A show anti-tumor properties by inhibiting the replication and the transcription process of cells. They are known to selectively modulate gene expression/transcription by blocking transcription activators and repressors that bind to guanine-cytosine (G-C)-rich regions of gene promoters [105, 111, 112]. The neuroprotectiive properties of mithramycin and chromomycin are correlated with their ability to inhibit DNA binding of the transcription factors Sp1 and Sp3 to their cognate G–C box in response to oxidative stress or DNA damage [113]. Interestingly, mithramycin action involves epigenetic compensation by reducing pericentromeric heterochromatin condensation, which ameliorates the clinical and neuropathological phenotype and extends survival of transgenic HD (R6/2) mice [56, 66]. Mithramycin A represses SETDB1/ESET expression and normalizes levels of H3K9me3, which are increased in R6/2 [56]. These findings support the role of epigenetic alterations in the R6/2 HD model and suggest that DNA-binding drugs, such as mithramycin, act by partially restoring perturbed histone modifications. Similarly, chromomycin restores the balance of methylation and acetylation of histone H3K9 towards greater acetylation in N171-82Q and R6/2 mice [70]. As a result, chromomycin landscapes transcriptionally active chromatin packaging and improves HD-related deficits and disease phenotype. Mithramycin and chromomycin are anticancer chemotherapeutics, but have not been used chronically owing to dose-related toxicity in humans. It is therefore important to develop safer DNA-binding drugs that may ultimately be helpful to HD patients.
Table 1.
Therapeutic regulation of epigenetic components in Huntington’s disease (HD)
| Drug/material | Epigenetic targets/function | Disease model | Effect | Clinical trial | Reference(s) |
|---|---|---|---|---|---|
| Histone and nucleosome | |||||
| Suberoyl bis-hydroxamic acid (SAHA) | Class I and II HDACs (predominantly class I)/inhibition | htt ex1p polyQ expanded flies | Inhibited neurodegeneration and impaired transport restoration | [57] | |
| 3-NP induced mouse model | Sp1 acetylation and abrogate oxidative stress | [128] | |||
| R6/2 transgenic mouse model | Increased histone acetylation, improved behavior, and increased neuronal survival | [91] | |||
| R6/2 transgenic mouse model | Decreased levels of HDAC2 and HDAC4 and ameliorated HD phenotype | [92] | |||
| Sodium butyrate | Class I and II HDACs/inhibition | R6/2 transgenic mouse model | Inhibited Htt aggregation, improved behavior, and increased survival | [94] | |
| Phenylbutyrate | Class I and II HDACs/inhibition | N171–82Q transgenic mouse | Increased survival, decreased gross brain, and neuronal atrophy | [65] | |
| HD patient | Improved cognitive function and behavior | Phase 2 | [99] | ||
| HD patient | Decreased symptoms related to HD genetic abnormalities | Phase 2 | [100] | ||
| Trichostatin A | Class I and II HDACs/inhibition | Striatal cells derived from HdhQ109 mice | Impaired transport restoration | [129] | |
| 4b | Class I HDACs/inhibition | R6/2 transgenic mouse model | Ameliorated HD phenotype | [62, 101, 102] | |
| Mithramycin | Nucleosome, ESET/transcription inhibition | R6/2 transgenic mouse model | Decreased histone methylation, and increased neuronal function and survival | [56] | |
| R6/2 transgenic mouse model | Improved behavior and neuropathological phenotype | [66] | |||
| N171–82Q and R6/2 transgenic mouse model | Improved nucleosomal dynamics, behavior, and neuropathological phenotype | [70] | |||
| Chromomycin | Nucleosome/transcription inhibition | N171–82Q and R6/2 transgenic mouse model | Improved nucleosomal dynamics, behavior, and neuropathological phenotype | [70] | |
| DNA methylation | |||||
| 5-azacytidine | DNA methyltransferase/inhibition | No study is yet available in HD model | Activated differential gene at the level of cellular division | [130] | |
| RNA interference | |||||
| AAV-shRNA | mRNA/inhibition | N171–82Q transgenic mouse | Less inclusion/improved stride length and behavior | [117] | |
| AAV1/2-HD70-injected rat | Less Fluoro-jade, more NeuN-positive cells and less feet-slipping | [118] | |||
| rAAV5-shRNA | mRNA/inhibition | R6/1 transgenic mouse model | Less nuclear inclusion/delayed onset of rear paw clasping | [119] | |
| HD190QG mouse model | Fewer aggregates | [120] | |||
| Adenovirus-shRNA | mRNA/inhibition | R6/2 transgenic mouse model | Fewer aggregates | [121] | |
| Lentivirus-shRNA | mRNA/inhibition | Lentivirus-HD171-82Q-injected mouse | Less inclusion and more DARPP32, NeuN-positive cells | [122] | |
| AAV-miRNA | mRNA/inhibition | CAG140 knock-in mouse model | Reduced toxicity compared with AAV-shRNA | [123] | |
| N171–82Q transgenic mouse | Improved behavior and prolonged lifespan | [122] | |||
| siRNA-Cholesterol | mRNA/inhibition | AAV1/8-HD400aa-100Q-injected mouse | Less inclusion and behavior | [125] | |
| ss-siRNA | mRNA/inhibition | HdhQ150 knock-in mouse model | Inhibited mutant htt expression | [126] | |
shRNA small hairpin RNA; AAV adeno-associated virus; miRNA microRNA; siRNA small interfering RNA; ss-siRNA single stranded siRNA; HDAC histone deacetylase; ESET ERG-associated protein with SET domain; mRNA messenger RNA; htt huntingtin; polyQ polyglutamin; 3-NP 3-nitropropionic acid; NeuN neuronal nuclei (known as Feminizing Locus on X-3, Fox-3, or Hexaribonucleotide Binding Protein-3); DARPP32 dopamine- and cAMP-regulated neuronal phosphoprotein 32
RNA Interference and Noncoding Small RNAs
Because HD is caused by a mutation at exon1 of htt gene, gene therapy to eliminate the expression of lethal gene (mthtt) using ncRNAs has been proposed and actively investigated over the last few years [114]. Wild-type htt protein functions as a survival factor that is indispensable for neuronal function, while mthtt protein is neurotoxic. A major question in gene therapy is how to nullify the mutant allele only without affecting the expression of wild-type allele because most HD patients posses one heterozygous mutant htt allele and one wild-type htt allele at the htt locus, (Fig. 5). Given this, the development of nucleic acid therapy by noncoding small RNAs could be an ideal approach to selectively silence mutant htt [115, 116]. Harper et al. [117] showed that RNA interference using adeno-associated virus-small hairpin RNA (shRNA) improves motor deficits and neuropathological phenotypes in a transgenic (N171-82Q) mouse model of HD. Most RNA interference studies using adenovirus-shRNA, lentivirus-shRNA, adeno-associated virus-miRNA, and cholesterol-conjugated siRNA have shown a reduction of mthtt aggregates, improvement of motor behavior, and reduced neuropathological sequelae (Table 1) [118–125].
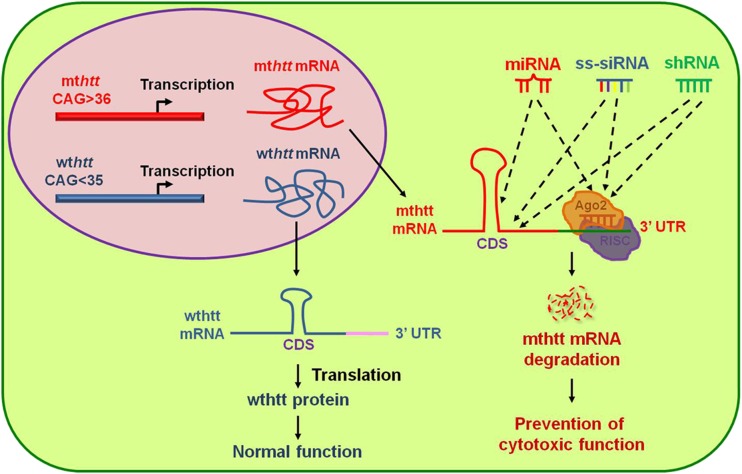
Fig. 5.
Therapeutic strategies using noncoding RNAs [microRNA (miRNA), small hairpin RNA (shRNA), and single-stranded small interfering RNA (ss-siRNA)] to nullify mutant huntingtin (mthtt) expression. The mthtt gene encodes cytotoxic mutant htt protein while wild-type htt (wthtt) expresses htt protein that functions in vesicle trafficking and neuronal survival. The development of nucleic acid therapy by non-coding small RNAs is an ideal approach to selectively silence mthtt without affecting the expression of the wild-type allele. miRNAs, ss-siRNAs, and shRNAs can target either coding sequence (CDS) or the 3-untranslated region (UTR) of mthtt messenger RNA (mRNA). Consequently, they participate in mRNA degradation through Argounate (Ago) and the RNA-induced silencing complex (RISC)-dependent pathway [114, 126]
Antisense nucleotides and siRNAs with central mismatch have been tested. Yu et al. [126] reported that single-stranded siRNAs (ss-siRNAs) are 100-fold more potent than unmodified RNA and 30-fold more allele-specific inhibitors of mthtt expression. Interestingly, chemically modified mismatched bases in ss-siRNAs mimic miRNA targeting and optimally distinguish mutant htt from wild-type htt alleles. Intraventricular infusion of ss-siRNA selectively nullifies the expression of mutant htt allele in an animal model of HD [126]. This study suggests that strategically designed ss-siRNAs could play a role through Argounate and the RNA-induced silecing complex-dependent pathway, and may be useful as an allele-specific drug for HD clinical trials (Fig. 5).
Conclusion and Future Perspectives
Dysfunction of epigenetic components and alteration of epigenetic modifications are closely linked to HD pathogenesis and may also be useful therapeutic targets for treating HD. The disruption of transcriptional homeostasis through DNA methylation, histone methylation/acetylation, and miRNA biogenesis is associated with a number of pathological mechanisms in HD. The use of noncoding small RNA-based therapy targeting mthtt in particular may be a successful therapeutic strategy [114]. Approaches using, noncoding small RNA-based therapeutic interventions need improved target specificity for the mthtt allele, efficacy, and region-specific delivery. It remains to be determined whether epigenetic alterations are a fundamental aspect of HD pathogenesis. At the present time it is clear that DNA methylation, histone methylation, acetylation status, and transcription cofactors are important markers directly or indirectly associated with transcriptional abnormalities in HD [32, 47, 127]. Further mechanistic studies are required to discover whether epigenetic alterations play a key role as a cause of disease progress in HD [30, 31]. It will be important to determine whether the correlation of CAG triplet repeat length with disease severity in HD patients parallels the severity of epigenetics abnormalities. Continued research identifying and clarifying the role of epigenetic factors will likely provide new insights into HD pathogenesis and open new directions for biomarkers research and therapeutics.
Electronic supplementary material
(PDF 499 kb)
Acknowledgments
This study was supported by NIH NS 067283–02 (H.R.), WCU Neurocytomics Program Grant (800–20080848) (H.R.), and SRC Grant (2010-0029-403) (H.R.) from National Research Foundation, and Flagship Grant (H.R.) from KIST.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.Myers RH, MacDonald ME, Koroshetz WJ, et al. De novo expansion of a (CAG)n repeat in sporadic Huntington’s disease. Nat Genet. 1993;5:168–173. doi: 10.1038/ng1093-168. [DOI] [PubMed] [Google Scholar]
- 2.Kremer BP, Goldberg S, Andrew J, et al. A worldwide study of the Huntington’s disease mutation the sensitivity and specificity of measuring CAG repeats. New Engl J Med. 1994;330:1401–1406. doi: 10.1056/NEJM199405193302001. [DOI] [PubMed] [Google Scholar]
- 3.Huntington G. On chorea. Med Surg Rep (Philadelphia) 1872;317–321.
- 4.Jergelsma G. Nue anatomische befunde bei paralysis agitans und bei chronischer progressive chorea. Neurol Centralbl. 1908;27:995–996. [Google Scholar]
- 5.Bruyn GW, Bots GTAM, Dom R. Huntinton’s chorea: Current neuropathological status. In: Chase TN, Wexler NS, Barbeau A, editors. Huntington’s disease. Advances in neurology. New York: Raven Press; 1979. pp. 83–93. [Google Scholar]
- 6.Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington’s disease. J Neuropath Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Kowall NW, Ferrante RJ. Huntinton’s disease. In: Markesbery WR, editor. Neuropathology of dementing disorders. New York: Oxford University Press; 1998. pp. 219–256. [Google Scholar]
- 8.Hersch SM, Rosas HD, Ferrante RJ. Neuropathology and pathophysiology of Huntington’s disease in movement disorders. In: Koller W, editor. Neurologic principles and practice. New York: McGraw-Hill; 2004. pp. 503–526. [Google Scholar]
- 9.Ferrante RJ, Kowall NW, Beal MF, Richardson EP, Jr, Bird ED, Martin JB. Selective sparing of a class of striatal neurons in Huntington’s disease. Science. 1985;230:561–563. doi: 10.1126/science.2931802. [DOI] [PubMed] [Google Scholar]
- 10.Ferrante RJ, Kowall NW, Richardson EP, Jr, Bird ED, Martin JB. Topography of encephalin, substance P and acetylcholinesterase staining in Huntington’s disease striatum. Neurosci Lett. 1986;71:283–288. doi: 10.1016/0304-3940(86)90634-8. [DOI] [PubMed] [Google Scholar]
- 11.Ferrante RJ, Kowall NW, Beal MF, Martin JB, Bird ED, Richardson EP., Jr Morphologic and histochemical characteristics of a spared subset of striatal neurons in Huntington’s disease. J Neuropathol Exp Neurol. 1987;1:12–27. doi: 10.1097/00005072-198701000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Ferrante RJ, Kowall NW, Richardson EP., Jr Proliferative and degenerative changes in striatal spiny neurons in Huntington’s disease: a combined study using the section-Golgi method and calbindin D28K immunocytochemistry. J Neurosci. 1991;11:3877–3887. doi: 10.1523/JNEUROSCI.11-12-03877.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graveland GA, Williams RS, DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington’s disease. Science. 1985;227:770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- 14.Kowall NW, Ferrante RJ, Martin JB. Patterns of cell loss in Huntington’s disease. TINS. 1987;10:24–29. [Google Scholar]
- 15.Huntington’s disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 16.Spada ARL, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 17.Orr HT, Chung MY, Banfi S, et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 18.Szebenyi G, Morfini GA, Babcock A, et al. Neuropathogenic forms of huntingtin and androgen receptor inhibit fast axonal transport. Neuron. 2003;40:41–52. doi: 10.1016/s0896-6273(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 19.Trushina E, Dyer RB, Badger JD, 2nd, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross CA. Intranuclear neuronal inclusions: a common pathogenic mechanism for glutamine-repeat neurodegenerative diseases? Neuron. 1997;19:1147–1150. doi: 10.1016/s0896-6273(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 21.Nucifora FC, Jr, Sasaki M, Peters MF, et al. Interference by huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 22.Cattaneo E. Dysfunction of wild-type huntingtin in Huntington disease. News Physiol Sci. 2003;18:34–37. doi: 10.1152/nips.01410.2002. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Li M, Drozda M, et al. Depletion of wild-type huntingtin in mouse models of neurologic diseases. J Neurochem. 2003;87:102–106. doi: 10.1046/j.1471-4159.2003.01980.x. [DOI] [PubMed] [Google Scholar]
- 24.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med (Suppl.) 2004;10:S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 25.Mangiarini L, Sathasivam K, Seller M, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87:493–506. [DOI] [PubMed]
- 26.Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a corditional model of Huntington’s disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 27.Ordway JM, Tallaksen-Greene S, Gutekunst CA, et al. Ectopically expressed CAG repeats cause intranuclear inclusions and a progressive late onset neurological phenotype in the mouse. Cell. 1997;91:753–763. doi: 10.1016/s0092-8674(00)80464-x. [DOI] [PubMed] [Google Scholar]
- 28.Beal MF, Ferrante RJ. Experimental therapeutics in transgenic mouse models of Huntington’s disease. Nat Rev Neurosci. 2004;5:373–384. doi: 10.1038/nrn1386. [DOI] [PubMed] [Google Scholar]
- 29.Ryu H, Rosas HD, Hersch SM, Ferrante RJ. The therapeutic role of creatine in Huntington’s disease. Pharmacol Ther. 2005;108:193–207. doi: 10.1016/j.pharmthera.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Waddington CH. The epigenotype. Endeavour. 1942;1:18. [Google Scholar]
- 31.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 32.Sadri-Vakili G, Cha JH. Mechanisms of disease: Histone modifications in Huntington’s disease. Nat Clin Pract Neurol. 2006;2:330–333. doi: 10.1038/ncpneuro0199. [DOI] [PubMed] [Google Scholar]
- 33.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 34.Ryu H, Ferrante RJ. Emerging chemotherapeutic strategies for Huntinton’s disease. Expert Opin Emerg Drugs. 2005;10:345–363. doi: 10.1517/14728214.10.2.345. [DOI] [PubMed] [Google Scholar]
- 35.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: Evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis JD, Meehan RR, Henzel WJ, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 37.Chan MF, van Amerongen R, Nijjar T, Cuppen E, Jones PA, Laird PW. Reduced rates of gene loss, gene silencing, and gene mutation in Dnmt1-deficient embryonic stem cells. Mol Cell Biol. 2001;21:7587–7600. doi: 10.1128/MCB.21.22.7587-7600.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan G, Beard C, Chen RZ, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson M, Krassowska A, Gilbert N, et al. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milutinovic S, Brown SE, Zhuang Q, Szyf M. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J Biol Chem. 2004;279:27915–27927. doi: 10.1074/jbc.M312823200. [DOI] [PubMed] [Google Scholar]
- 41.Espada J, Ballestar E, Santoro R, et al. Epigenetic disruption of ribosomal RAN genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 2007;35:2191–2198. doi: 10.1093/nar/gkm118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 43.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 44.Ng CW, Yildirim F, Yap YS, et al. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc Natl Acad Sci U S A. 2013;110:2354–2359. doi: 10.1073/pnas.1221292110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villar-Menéndez I, Blanch M, Tyebji S, et al. Increased 5-methylcytosine and decreased 5-hydroxymethylcytosine levels are associated with reduced striatal A(2A)R levels in Huntington’s disease. Neuromolecular Med. 2013;15:295–309. doi: 10.1007/s12017-013-8219-0. [DOI] [PubMed] [Google Scholar]
- 46.Wood H. Neurodegenerative disease: Altered DNA methylation and RNA splicing could be key mechanisms in Huntington disease. Nat Rev Neurol. 2013;9:119. doi: 10.1038/nrneurol.2013.23. [DOI] [PubMed] [Google Scholar]
- 47.Thomas B, Matson S, Chopra V, et al. A novel method for detecting 7-methyl guanine reveals aberrant methylation levels in Huntington disease. Anal Biochem. 2013;436:112–120. doi: 10.1016/j.ab.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hake SB, Xiao A, Allis CD. Linking the epigenetic ‘language’ of covalent histone modifications to cancer. Br J Cancer. 2004;90:761–769. doi: 10.1038/sj.bjc.6601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marques SC, Oliveira CR, Outeiro TF, Pereira CM. Alzheimer’s disease: the quest to understand complexity. J Alzheimers Dis. 2010;21:373–383. doi: 10.3233/JAD-2010-100303. [DOI] [PubMed] [Google Scholar]
- 50.Chouliaras L, Rutten BP, Kenis G, et al. Epigenetic regulation in the pathophysiology of Alzheimer’s disease. Prog Neurobiol. 2010;90:498–510. doi: 10.1016/j.pneurobio.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 52.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 54.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 55.Alarcón JM, Malleret G, Touzani K, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 56.Ferrante RJ, Ryu H, Kubilus JK, et al. Chemotherapy for the brain: the antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington’s disease. J Neurosci. 2004;24:10335–10342. doi: 10.1523/JNEUROSCI.2599-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steffan JS, Bodai L, Pallos J, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 58.Igarashi S, Morita H, Bennett KM, et al. Inducible PC12 cell model Huntington’s disease shows toxicity and decreased histone acetylation. Neuroreport. 2003;14:565–568. doi: 10.1097/00001756-200303240-00007. [DOI] [PubMed] [Google Scholar]
- 59.Sadri-Vakili G, Bouzou B, Benn CL, et al. Histones associated with downregulated genes are hypo-acetylated in Huntington’s disease models. Hum Mol Genet. 2007;16:1293–1306. doi: 10.1093/hmg/ddm078. [DOI] [PubMed] [Google Scholar]
- 60.Thomas EA, Coppola G, Desplats PA, et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington’s disease transgenic mice. Proc Natl Acad Sci U S A. 2008;105:15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McFarland KN, Das S, Sun TT, et al. Genome-wide histone acetylation is altered in a transgenic mouse model of Huntington’s disease. PLoS One. 2012;7:e41423. doi: 10.1371/journal.pone.0041423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 63.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.West RL, Lee JM, Maroun LE. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer’s disease patient. J Mol Neurosci. 1995;6:141–146. doi: 10.1007/BF02736773. [DOI] [PubMed] [Google Scholar]
- 65.Gardian G, Browne SE, Choi DK, et al. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington’s disease. J Biol Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- 66.Ryu H, Lee J, Hagerty SW, et al. ESET/SETDB1 gene expression and histone H3(K9) trimethylation in Huntington’s disease. Proc Natl Acad Sci U S A. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oliveira AM, Wood MA, McDonough CB, Abel T. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learn Mem. 2007;14:564–572. doi: 10.1101/lm.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee J, Hagerty S, Cormier KA, et al. Monoallele deletion of CBP leads to pericentromeric heterochromatin condensation through ESET expression and histone H3(K9) methylation. Hum Mol Genet. 2008;17:1774–1782. doi: 10.1093/hmg/ddn067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee J, Hwang YJ, Shin JY, et al. Epigenetic regulation of cholinergic receptor M1 (CHRM1) by histone H3K9me3 impairs Ca(2+) signaling in Huntington’s disease. Acta Neuropathol. 2013;125:727–739. doi: 10.1007/s00401-013-1103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stack EC, Del Signore SJ, Luthi-Carter R, et al. Modulation of nucleosome dynamics in Huntington’s disease. Hum Mol Genet. 2007;16:1164–1175. doi: 10.1093/hmg/ddm064. [DOI] [PubMed] [Google Scholar]
- 71.Wu R, Terry AV, Singh PB, Gilbert DM. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol Biol Cell. 2005;16:2872–2881. doi: 10.1091/mbc.E04-11-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sugita S, Uchimura N, Jiang ZG, North RA. Distinct muscarinic receptors inhibit release of GABA and excitatory amino acids in mammalian brain. Proc Natl Acad Sci U S A. 1991;88:2608–2611. doi: 10.1073/pnas.88.6.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bonner TI, Buckley NJ, Young AC, Brann MR. Identification of a family of muscarinic acetylcholine receptor genes. Science. 1897;237:527–532. doi: 10.1126/science.3037705. [DOI] [PubMed] [Google Scholar]
- 74.Hulme EC, Birdsall NJM, Buckley NJ. Muscarinic receptor subtypes. Annu Rev Pharmacol Toxicol. 1990;30:633–673. doi: 10.1146/annurev.pa.30.040190.003221. [DOI] [PubMed] [Google Scholar]
- 75.Wess J. Molecular biology of muscarinic acetylcholine receptors. Crit Rev Neurobiol. 1996;10:66–69. doi: 10.1615/critrevneurobiol.v10.i1.40. [DOI] [PubMed] [Google Scholar]
- 76.Calabresi P, Centonze D, Gubellini P, Pisani A, Bernardi G. Acetylcholine-mediated modulation of striatal function. Trends Neurosci. 2000;23:120–126. doi: 10.1016/s0166-2236(99)01501-5. [DOI] [PubMed] [Google Scholar]
- 77.Cha JH, Kosinski CM, Kerner JA, et al. Altered brain neurotransmitter receptors in transgenic mice expressing a portion of an abnormal human Huntington disease gene. Proc Natl Acad Sci U S A. 1998;95:6480–6485. doi: 10.1073/pnas.95.11.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Z, Kai L, Day M, et al. Dopaminergic control of corticostriatal long-term synaptic depression in medium spiny neurons is mediated by cholinergic interneurons. Neuron. 2006;50:443–452. doi: 10.1016/j.neuron.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 79.Wilson CJ. Striatal D2 receptors and LTD: yes, but not where you thought they were. Neuron. 2006;50:347–348. doi: 10.1016/j.neuron.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 80.Han J, Pedersen JS, Kwon SC, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol Dis. 2008;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 83.Lee ST, Chu K, Im WS, et al. Altered microRNA regulation in Huntington’s disease models. Exp Neurol. 2011;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 84.Buckley NJ, Johnson R. New insights into non-coding RNA networks in Huntington’s disease. Exp Neurol. 2011;231:191–194. doi: 10.1016/j.expneurol.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 85.Ghose J, Sinha M, Das E, Jana NR, Bhattacharyya NP. Regulation of miR-146a by RelA/NFkB and p53 in STHdhQ111/HdhQ111 cells, a cell model of Huntington’s disease. PLos One. 2011;6:e23837. doi: 10.1371/journal.pone.0023837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bae BI, Xu H, Igarashi S, et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron. 2005;47:29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 87.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 88.Fukuda H, Sano N, Muto S, Horikoshi M. Simple histone acetylation plays a complex role in the regulation of gene expression. Brief Funct Genomic Proteomic. 2006;5:190–208. doi: 10.1093/bfgp/ell032. [DOI] [PubMed] [Google Scholar]
- 89.Sugai F, Yamamoto Y, Miyaguchi K, et al. Benefit of valpronic acid in suppressing disease progression of ALS model mice. Eur J Neurosci. 2004;20:3179–3183. doi: 10.1111/j.1460-9568.2004.03765.x. [DOI] [PubMed] [Google Scholar]
- 90.Camelo S, Iglesias AH, Hwang D, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;164:10–21. doi: 10.1016/j.jneuroim.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 91.Hockly E, Richon VM, Woodman B, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci U S A. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mielcarek M, Benn CL, Franklin SA, et al. SAHA decreases HDAC2 and 4 levels in vivo and improves molecular phenotypes in the R6/2 mouse model of Huntington’s disease. PLos One. 2011;6:e27746. doi: 10.1371/journal.pone.0027746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McCampbell A, Taye AA, Whitty L, Penney E, Steffan JS, Fischbeck KH. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc Natl Acad Sci U S A. 2001;98:15179–15184. doi: 10.1073/pnas.261400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ferrante RJ, Kubilus JK, Lee J, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J Neurosci. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miller AA, Kurschel E, Osieka R, Schmidt CG. Clinical pharmacology of sodium butyrate in patients with acute leukemia. Eur J Cancer Clin Oncol. 1987;23:1283–1287. doi: 10.1016/0277-5379(87)90109-x. [DOI] [PubMed] [Google Scholar]
- 96.Daniel P, Brazier M, Cerutti I, et al. Pharmacokinetic study of butyric acid administered in vivo as sodium and arginine butyrate salt. Clin Chim Acta. 1989;181:255–263. doi: 10.1016/0009-8981(89)90231-3. [DOI] [PubMed] [Google Scholar]
- 97.Egorin MJ, Yuan ZM, Sentz DL, Plaosance K, Eiseman JL. Plasma pharmacokinetics of butyrate after intravenous administration of sodium, butyrate or oral administration of tributyrin or sodium butyrate to mice and rats. Cancer Chemother Pharmacol. 1999;43:445–453. doi: 10.1007/s002800050922. [DOI] [PubMed] [Google Scholar]
- 98.Dasgupta S, Zhou Y, Jana M, Banik NL, Pahan K. Sodium phenylacetate inhibits adoptive transfer of experimental allergic encephaplomyelitis in SJL/J mice at multiple steps. J immunol. 2003;170:3874–3882. doi: 10.4049/jimmunol.170.7.3874. [DOI] [PubMed] [Google Scholar]
- 99.Hogarth P, Lovrecic L, Krainc D. Sodium phenylbutyrate in Huntington’s disease: a dose-finding study. Mov Disord. 2007;22:1962–1964. doi: 10.1002/mds.21632. [DOI] [PubMed] [Google Scholar]
- 100.Ebbel EN, Leymarie N, Schiavo S, et al. Identification of phenylbutyrate-generated metabolites in Huntington disease patients using parallel liquid chromatography/electrochemical array/mass spectrometry and off-line tandem mass spectrometry. Anal Biochem. 2010;399:152–161. doi: 10.1016/j.ab.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jia H, Kast RJ, Steffan JS, Thomas EA. Selective histone deacetylase (HDAC) inhibition imparts beneficial effects in Huntington’s disease mice: implications for the ubiquitin-proteasomal and autophagy systems. Hum Mol Genet. 2012;21:5280–5293. doi: 10.1093/hmg/dds379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jia H, Pallos J, Jacques V, et al. Histone deacetylase (HDAC) inhibitors targeting HDAC3 and HDAC1 ameliorate polyglutamine-elicited phenotypes in model systems of Huntington’s disease. Neurobiol Dis. 2012;46:351–361. doi: 10.1016/j.nbd.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu Y, Chopra V, Chopra R, et al. Transcriptional modulator H2A histone family, member Y (H2AFY) marks Huntington disease activity in man and mouse. Proc Natl Acad Sci U S A. 2011;108:17141–17146. doi: 10.1073/pnas.1104409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Blanco G, Fu H, Mendez C, Khosla C, Salas JA. Deciphering the biosynthetic origin of the aglycone of the aureolic acid group of anti-tumor agents. Chem Biol. 1996;3:193–196. doi: 10.1016/s1074-5521(96)90262-8. [DOI] [PubMed] [Google Scholar]
- 105.Ralston SH. Pathogenesis and management of cancer associated hypercalcaemia. Cancer Surv. 1994;21:179–196. [PubMed] [Google Scholar]
- 106.Chakrabarti S, Bhattacharyya D, Dasgupta D. Structural basis of DNA recognition by anticancer antibiotics, chromomycin A3, and mithramycin: roles of minor groove width and ligand flexibility. Biopolymers. 2001;56:85–95. doi: 10.1002/1097-0282(2000)56:2<85::AID-BIP1054>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 107.Hescock H, Jr, Parker M, Wang TY, Ballinger R, Balducci L. Metastatic carcinoma of unknown primary: complete response to second-line treatment with plicamycin. Am J Med Sci. 1989;298:34–37. doi: 10.1097/00000441-198907000-00006. [DOI] [PubMed] [Google Scholar]
- 108.Prado L, Lombo F, Brana AF, Mendez C, Rohr J, Salas JA. Analysis of two chromosomal region adjacent to genes for a Type II polyketide synthase involved in the biosynthesis of the antitumor polyketide mithramycin in Streptomyces argillaceus. Mol Gen Genet. 1999;261:216–225. doi: 10.1007/s004380050960. [DOI] [PubMed] [Google Scholar]
- 109.Ryan WG. Treatment of Paget’s disease of bone with mithramycin. Clin Orthop. 1977;127:106–110. [PubMed] [Google Scholar]
- 110.Kennedy BJ. Mithramycin therapy in testicular cancer. J Urol. 1972;107:429–432. doi: 10.1016/s0022-5347(17)61046-2. [DOI] [PubMed] [Google Scholar]
- 111.Hagen G, Dennig J, Preiss A, Beato M, Suske G. Functional analyses of the transcription factor Sp4 reveal properties distinct from Sp1 and Sp3. J Biol Chem. 1995;270:24989–24994. doi: 10.1074/jbc.270.42.24989. [DOI] [PubMed] [Google Scholar]
- 112.Majello B, De Luca P, Suske G, Lania L. Differential transcriptional regulation of c-myc promoter through the same DNA binding sites targeted by Sp1-like proteins. Oncogene. 1995;10:1841–1848. [PubMed] [Google Scholar]
- 113.Chatterjee S, Zaman K, Ryu H, Conforto A, Ratan RR. Sequence-selective DNA binding drugs mithramycin A and chromomycin A3 are potent inhibitors of neuronal apoptosis induced by oxidative stress and DNA damage in cortical neurons. Ann Neurol. 2001;49:345–354. [PubMed] [Google Scholar]
- 114.Zhang Y, Friedlander RM. Using non-coding small RNAs to develop therapies for Huntington’s disease. Gene Ther. 2011;18:1139–1149. doi: 10.1038/gt.2011.170. [DOI] [PubMed] [Google Scholar]
- 115.Hu J, Matsui M, Gagnon KT, et al. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lombardi MS, Jaspers L, Spronkmans C, et al. A majority of Huntington’s disease patients may be treatable by individualized allele-specific RNA interference. Exp Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 117.Harper SQ, Staber PD, He X, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci U S A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Franich NR, Fitzsimons HL, Fong DM, Klugmann M, During MJ, Young D. AAV vector-mediated RNAi of mutant huntingtin expression is neuroprotective in a novel genetic rat model of Huntington’s disease. Mol Ther. 2008;16:947–956. doi: 10.1038/mt.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS, Mandel RJ. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington’s disease transgenic mice. Mol Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Machida Y, Okada T, Kurosawa M, Oyama F, Ozawa K, Nukina N. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem Biophys Res Commun. 2006;343:190–197. doi: 10.1016/j.bbrc.2006.02.141. [DOI] [PubMed] [Google Scholar]
- 121.Huang B, Schiefer J, Sass C, Landwehrmeyer GB, Kosinski CM, Kochanek S. High capacity adenoviral vector-mediated reduction of huntingtin aggregate load in vitro and in vivo. Hum Gene Ther. 2007;18:303–311. doi: 10.1089/hum.2006.160. [DOI] [PubMed] [Google Scholar]
- 122.Drouet V, Perrin V, Hassig R, et al. Sustained effects of nonallele-specific Huntingtin silencing. Ann Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 123.McBride JL, Boudreau RL, Harper SQ, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Boudreau RL, McBride JL, Martins I, et al. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Difiglia M, Sena-Exteves M, Chase K, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci U S A. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yu D, Pendergraff H, Liu J, et al. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Luthi-Carter R, Cha J-HJ. Mechanisms of transcriptional dysregulation in Huntington’s disease. Clin Neurosci Res. 2003;3:165–177. [Google Scholar]
- 128.Ryu H, Lee J, Olofsson BA, et al. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc Natl Acad Sci U S A. 2003;23:3597–3606. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dompierre JP, Godin JD, Charrin BC, et al. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Payao SL, Smith MD, Bertolucci PH. Differential chromosome sensitivity to 5-azacytidine in Alzheimer’ disease. Gerontology. 1998;44:267–271. doi: 10.1159/000022023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 499 kb)