Abstract
The acetylation of histone and non-histone proteins controls a great deal of cellular functions, thereby affecting the entire organism, including the brain. Acetylation modifications are mediated through histone acetyltransferases (HAT) and deacetylases (HDAC), and the balance of these enzymes regulates neuronal homeostasis, maintaining the pre-existing acetyl marks responsible for the global chromatin structure, as well as regulating specific dynamic acetyl marks that respond to changes and facilitate neurons to encode and strengthen long-term events in the brain circuitry (e.g., memory formation). Unfortunately, the dysfunction of these finely-tuned regulations might lead to pathological conditions, and the deregulation of the HAT/HDAC balance has been implicated in neurological disorders. During the last decade, research has focused on HDAC inhibitors that induce a histone hyperacetylated state to compensate acetylation deficits. The use of these inhibitors as a therapeutic option was efficient in several animal models of neurological disorders. The elaboration of new cell-permeant HAT activators opens a new era of research on acetylation regulation. Although pathological animal models have not been tested yet, HAT activator molecules have already proven to be beneficial in ameliorating brain functions associated with learning and memory, and adult neurogenesis in wild-type animals. Thus, HAT activator molecules contribute to an exciting area of research.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-013-0204-7) contains supplementary material, which is available to authorized users.
Keywords: HAT activator molecule, Lysine acetylation, CREB-binding protein, Learning and memory, Adult neurogenesis, Neurodegenerative diseases
Introduction
DNA is packed in the nucleus of mammalian cells in association with histones, non-histone proteins, and RNA in a highly organized structure known as chromatin. A nucleosome is a single unit of chromatin comprising 146 base pairs of DNA wrapped around histone octamers. The histone N-terminal tails are subjected to various post-translational modifications, including acetylation, methylation, phosphorylation, ubiquitination, and sumoylation [1, 2]. Histone acetyltransferase (HAT) catalyzes the acetylation of core histones through the addition of an acetyl group from the pseudo-substrate acetyl coenzyme A (acetyl-CoA) to the lysine residue on the ε-amino group on the N-terminal of histones. Apart from histone, HATs also have many non-histone protein substrates, suggesting a new nomenclature, KAT for lysine (K) acetyl transferase, replacing the term HAT [3]. However, in the following sections, we will use HAT to avoid possible confusion. Another class of enzymes, histone deacetylases (HDACs), catalyzes deacetylation through the hydrolysis of an acetyl moiety from the lysine residue.
Although the functional implications of each of these modifications have not been fully elucidated, studies have shown that it is difficult to identify the dynamic acetyl mark(s) that specifically respond to a pathway from the overall pre-existing marks that maintain the global chromatin structure. In terms of function, lysine acetylation initiates molecular processes leading to two biochemical consequences: 1) the recruitment of coactivator complexes through conserved domains, such as bromodomains; and 2) the participation of co-repressor complexes through HDACs. Together, these changes will affect chromatin structure, leading to further functional consequences. Indeed, strict regulation of the dynamic balance between acetylation and deacetylation is critical for various cellular physiological phenomena, such as proliferation, differentiation, cell growth and gene expression. Most recently, the acetylation balance has been implicated in intracellular pH regulation [4].
Alterations in the acetylation balance have been shown in neurologic disorders and neurodegenerative diseases [5–7]. Despite decoding chromatin language in pathological conditions or cognitively deficient states, HDAC inhibitors (HDACi) might represent a good therapeutic option, as these compounds induce a histone hyperacetylated state that might compensate for acetylation deficits. HDACi-based therapeutic strategies were first applied to models of polyglutamine (polyQ) diseases, such as Huntington’s [8, 9] and Kennedy’s [10], and then subsequently revealed to be successful to some extent in several other animal models of neurodegenerative disease [5, 6, 11, 12]. Importantly, this strategy has also been successfully applied in animals subjected to experimental brain damage [13, 14] and recently shown to improve memory functions in aged mice [15]. HDACi are the only molecules used for increasing cellular acetylation, and two of these molecules have been approved for clinical use in cutaneous T cell lymphoma [16]. However, caveats remain as to the specificity and the molecular and cellular mechanisms underlying the activity of these inhibitors in neurons, whereby long-term application might be detrimental.
In this review, we will briefly describe different HAT families and the regulation, function, and important role of these molecules in the cell fate specification of neural stem cells. As enzymes, HATs are druggable, and few HAT activator molecules have been developed. We will also discuss the mechanisms for HAT regulation and impairment in neurodegenerative diseases and neuronal plasticity to understand the benefits of therapeutic strategies based on reactivating acetylation using HAT activators versus HDAC inhibitors. Moreover, we will describe two interesting studies concerning HAT activator molecules.
Although lysine acetylation is best characterized in the context of nuclear histones, combinatorial approaches and proteomic analyses have revealed almost 2,000 acetylated proteins in the cell that are involved in a broad range of cellular functions [17, 18]. Thus, the potential effect of HAT activation or impairment on nonhistone targets will also be reviewed.
HATs
Families of HATs
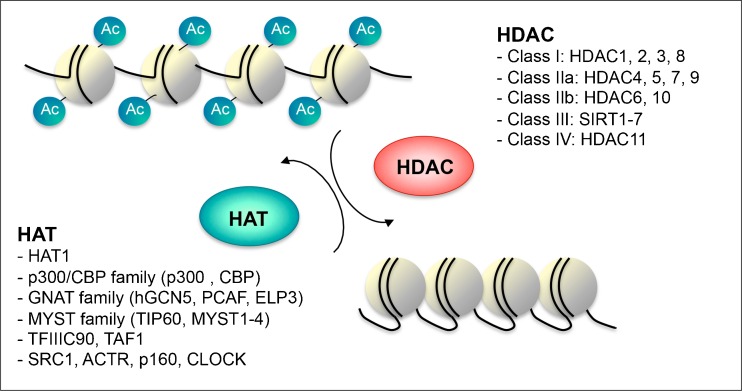
HATs catalyze the acetylation of lysine residues, which has been accepted as an important epigenetic marker. Acetylation occurs on both histone and nonhistone proteins, with an estimated 2,000–4,000 acetylated proteins and 15,000 acetylation sites in animal tissues [19]. The majority of the functional attributes of acetylation have been revealed as a result of several decades of research on histone acetylation, with recent evidence of nonhistone protein acetylation. In the context of chromatin, histones undergo acetylation to yield a more relaxed chromatin conformation resulting from hydrophobicity, a net change in the overall charge, and reduced electrostatic interactions. Acetylated histones recruit bromodomain-containing proteins, which are primarily transcription factors and cofactors, further enhancing gene expression [20]. HATs are highly conserved from yeasts to humans. Figure 1 summarizes the different HAT and HDAC families. These enzymes are broadly classified into cytoplasmic type A and nuclear type B HATs. Nuclear HATs are subclassified into 5 major families: 1) Gcn5-related N-acetyltransferases (GNAT), 2) p300/cyclic adenosine monophosphate response element-binding protein (CREB) binding protein (CBP), 3) MOZ, yeast YBF2, SAS2, and TIP60 (MYST), 4) transcription factor-related HATs, and 5) nuclear receptor-associated HATs. GCN5, a member of the GNAT family was the first identified acetyltransferase. The major members of the GNAT family are general control of amino acid synthesis 5 (Gcn5); elongation protein 3 (ELP3); p300/CBP-associated factor (PCAF); chromodomain on chromosome Y protein; establishment of cohesion 1; establishment of cohesion 1 homolog 1; establishment of cohesion 1 homolog 2 (a paralog of establishment of cohesion 1 homolog 1); ADA-two-A containing 2; and mechanosensory abnormal 17 [21]. GNATs comprise a conserved acetyltransferase domain, a bromodomain that recognizes acetylated lysine residues, and a transcription coactivator ADA2-binding domain. The 2 paralogs of p300/CBP family are transcriptional coactivators p300 and CBP. The 5 principal protein interaction domains of p300 and CBP are the nuclear receptor interaction domain; the CREB and myeloblastosis (MYB)-interaction domain; the kinase inducible binding domain (KIX), which binds to the Ser-133-phosphorylated kinase inducible domain region of CREB; the cysteine/histidine regions (CH1 and CH3); and the interferon response-binding domain, which is also the steroid receptor coactivator 1 interacting domain of CBP [22]. p300/CBP also possesses an acetyltransferase domain, bromodomain, and a plant homeodomain-type zinc finger motif. The members of the MYST family of acetyltransferases, including TAT interacting proteins 60 (Tip60), monocytic leukaemia zinc-finger protein (MOZ), males absent on the first (MOF), monocytic leukaemia zinc-finger protein related factor (MORF) and human acetylase binding to ORC1 (HBO1), have a 240 amino acid-long MYST region with a canonical acetyl-CoA-binding site and a C2HC-type zinc finger motif. Most of the MYST family acetyltransferases are characterized by the presence of a chromodomain, involved in protein–protein interactions. Structural alignments suggest these chromodomains to be slightly different than the canonical polycomb associated chromodomains [23].
Fig. 1.
Histone acetyletransferase (HAT) and histone deacetylase (HDAC) families. Illustration of chromatin conformation according to the HAT/HDAC balance. The acetylation levels of nucleosome histone tails, at lysine residues, are determined through the interplay between acetylation and deacetylation mechanisms engaged respectively through HATs and HDACs enzymes. The different families and classes of enzymes are noted. Ac = Acetyl; CBP = cyclic adenomonophosphate response element-binding (CREB) binding protein; GNAT = Gcn5-related N-acetyltransferases; hGCN5 = human general control of amino acid synthesis protein 5-like 2; PCAF = p300/CBP-associated factor; ELP3 = elongation protein 3; MYST = MOZ/YBF2/SAS2/TIP60; TIP60 = TAT interacting proteins 60; TFIIIC90 = transcription factor IIIC 90kDa; TAF1 = TATA Box Binding Protein-Associated Factor, SRC1 = steroid receptor coactivator 1; ACTR = SRC-3, steroid receptor coactivator/AIB-1, Activated In Breast cancer-1/TRAM-1, thyroid hormone receptor activator molecule 1/ NCOA3, nuclear receptor coactivator 3
HAT activities have been identified in several proteins associated with transcription activation through hormonal signals. This family of HATs is classified as nuclear receptor coactivators, which include steroid receptor coactivator-1, SRC-3, steroid receptor coactivator/AIB-1, Activated In Breast cancer-1/ TRAM-1, thyroid hormone receptor activator molecule 1/ NCOA3, nuclear receptor coactivator 3 (ACTR), and transcriptional intermediary factor 2 (reviewed in [24]). These enzymes are evolutionarily related to HATs, which interact with p300. The transcription factor-related HATs include TATA box binding protein (TBP)-associated factor TAFII250 and TFIIIC. TAFII250 is a subunit of the general transcription factor transcription initiation factor II D (TFIID). The general function of TFIIIC involves the initiation of the transcription promoter complex through DNA binding and the recruitment of TBP-containing TFIIIB and RNA polymerase III [25]. At least three subunits of TFIIIC (TFIIIC110 and TFIIIC90) harbor HAT activity in vitro [26]. However, this classification should be reconsidered, as several nuclear HATs exhibit nuclear cytoplasmic shuttling, and cytoplasmic lysine acetylation has been associated with important physiological outcomes [27].
HAT Function and Regulations
Acetylation plays a central role in numerous biological processes, including gene expression [28, 29], chromatin dynamics, cell-cycle progression [30–33], DNA repair [34], and apoptosis. HATs are regulated through various mechanisms, including auto-acetylation, signaling pathways-induced modifications, and cellular metabolites.
Auto-acetylation
Auto-acetylation involves the regulation of enzyme activity through the acetylation lysine residues, typically resulting in increased catalytic activity. The first reported acetyltransferase to undergo auto-acetylation is Tip60. Lysine 327 in the active site of the MYST domain is conserved amongst all the MYST proteins and acts as a key regulatory marker for enzyme activity upon acetylation. The intra- or intermolecular auto-acetylation of p300/CBP-associated factor (PCAF) specifically targets 5 lysine residues in the nuclear localization signal. Auto-acetylated PCAF localizes to the nucleus and activates gene transcription. Deacetylated PCAF is located primarily in the cytoplasm of apoptotic cells, suggesting a role for PCAF auto-acetylation in the regulation of cellular processes [35]. Similarly, p300 and CBP activity is also regulated through auto-acetylation [36]. The auto-acetylation of p300 induces a structural conformational change, leading to the exposure of substrate-binding regions. p300 auto-acetylation results in the acetylation of approximately 12 lysine residues in the unique activation loop of p300. Thus, the acetylation status of this enzyme facilitates the compartmentalization and induction enzymatic activity [36]. To maintain acetylation homeostasis, the nicotinamide adenine dinucleotide (+)-dependent HDAC sirtuin 2 (SIRT2) regulates p300 auto-acetylation [37]. HDAC3 deacetylates PCAF [38] and SIRT1 deacetylates Tip60 [39].
HAT Function of CBP
The HAT function of CBP can be regulated by different signaling pathways affecting its phosphorylation status by nuclear calcium or cyclic adenosine monophosphate pathways [40, 41], or its arginine methylation level by the coactivator-associated arginine methyltransferase I [42, 43]. Such modifications affect the enzymatic activity.
Cellular Metabolites
Cellular metabolites, such as nicotinamide adenine dinucleotide (+) and acetate, also act as regulators of acetylation. These compounds modulate HDACs; however, the direct roles of cellular metabolites on HATs have not been thoroughly investigated [4, 44].
HATs During Neural Development
Among the HATs, CBP/p300 are crucial enzymes in development; mutations and deletions of either enzyme causes Rubinstein–Taybi syndrome, characterized by mental disability, among other features [45]. Several studies have described a role for CBP/p300 in neural development. In mice, the mono-allelic abrogation or loss of the enzymatic activity of either enzyme results in embryonic lethality observed as early as E9.0 to E11.5 in a dominant fashion, consistent with the early stages of neural development, with impaired hematopoiesis, angiogenesis, heart, lung, and intestine organogenesis [46–49]. Interestingly, an analysis of the distribution of the p300 messenger RNA (mRNA) during mouse development showed the elevated expression in the neural tissue, suggesting the involvement of this enzyme in neural development. Consistently, knocking out CBP or p300 leads to an open neural tube, potentially reflecting defects in twist (without changes in twist expression) or other transcription factors, such as paired box 3 (PAX3) and activating enhancer binding protein 2 (AP2), thereby affecting neural development [49, 50]. Neural tube closure typically begins at E8.5 and spreads along neural folds; however, in CBP-null embryos, different types of neural tube closure defects are observed. Hemorrhaging in the telencephalon and mesencephalon, reflecting defective vasculature, has also been observed and hypothesized as the primary cause of embryonic death [50].
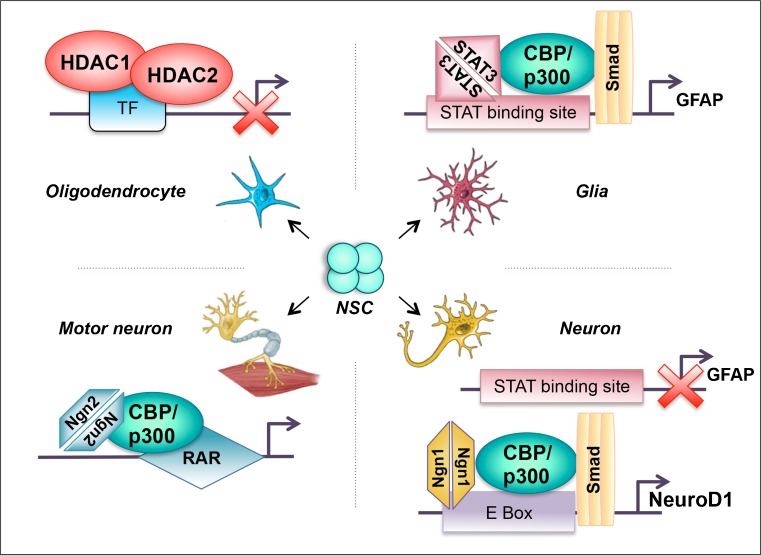
At the molecular level, HATs have been implicated in the cell fate specification of neuronal stem cells (NSC) (Fig. 2). p300 bridges signal transducers and activators of transcription 3 and mothers against decapentaplegic homolog 1 (Smad1), leading to synergistic astrocyte differentiation from neural progenitors [51]. CBP/p300 complexes with the transcription factor neurogenin 2 and the retinoic acid receptor to induce histone acetylation and transcriptionally active chromatin during motor neuron specification [52]. The closely related basic helix-loop-helix (bHLH) factor neurogenin 1 (Ngn1) represses gliogenesis through the sequestration of the CBP–Smad1 complex away from astrocyte differentiation genes, such as glial fibrillary acidic protein (GFAP), during neurogenesis [53]. CBP/p300 and Smad1, separately or together, subsequently associate with neurogenin at neural-specific promoters, such as NeuroD, thereby promoting neuronal differentiation [53]. The post-translational modifications of CBP and p300 also regulate the functions of these enzymes, and the phosphorylation of CBP through the atypical protein kinase C (aPKCζ) plays a critical role in controlling glial and neuronal differentiation [54].
Fig. 2.
Role of acetylation in different lineage determination. The neural stem cells (NSCs) exist in a niche, which can be differentially modulated to specific neuronal lineages. A differential recruitment of specific transcription factors (TF) to the same acetyltransferases determine specific neural cell fates from the NSCs. Cyclic adenomonophosphate response element-binding (CREB) binding protein (CBP)/p300 histone acetyletransferases (HATs) interact with STAT and SMAD activating glial fibrillary acidic protein (GFAP) expression, thus specifying the glial lineage. Increased expression of neurogenin (Ngn1) titrates this complex, thus leading to the release of STAT, blocking GFAP expression. The new Ngn1–CBP/p300–SMAD complex subsequently binds to the E box elements, which results in a neuron cell type due to the activation of NeuroD1 expression [53]. CBP/p300 when bound to retinoic acid receptor (RAR) and neurogenin 2 (Ngn2) leads to a differentiation of the motor neuron cells. The deacetylases histone deacetylases (HDACs) HDAC1 and HDAC2 act as a general repressor, blocking the transcription factor and thereby resulting in oligodendrocyte specification
Among the other families of HATs, the role of Gcn5 in neural development has also been documented, as the acetyltransferase activity and sufficient expression of this enzyme are required for neural tube closure in mice [55, 56]. Surprisingly, while p300 HAT mutants exhibit defects in multiple organ systems, the effects of mutating Gcn5 are restricted to the neural tissue, implicating the specificity of Gcn5 acetyltransferase activity, with potential redundancy with PCAF in other tissues [55]. Gcn5 also plays HAT-independent roles during development, and the complete loss of this protein leads to early embryonic lethality after gastrulation [57, 58]. Gcn5, as a member of the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex, accelerates neural dysfunction characteristic of polyQ-Atxn7, which causes spinocerebellar ataxia type 7 [59]. At the transcriptional level, knocking out Gcn5 in NSCs leads to changes in gene expression in a pattern similar to the Myc knockout (KO). This functional overlap indicates that Gcn5/Myc coregulate transcriptional programs in NSCs, a process required for stem cell proliferation and normal brain growth [60]. Interestingly, the KO of both CBP/p300 and Gcn5 leads to neural tube closure defects, reminiscent of knocking out other transcriptional regulators, such as p53, suggesting the complex, but overlapping, functions of various factors in neural tube closure.
While the effects of knocking out other proteins of the Tip60/NuA4 complex have been studied previously, the function of the MYST family of HATs in neural development remains unknown. However, the role of the interaction between Tip60 and ATXN1 in spinocerebellar ataxia type 1 pathogenesis, involving cerebellar degeneration, implicates a role for these HATs in neural development [61].
Though the roles of different HATs in neural development have been well-studied or at least indicated, further extensive studies will be required to provide a more detailed picture of the role of histone acetylation in the regulation of transcriptional programs during neural development.
Because acetylation plays a significant role during neural development, it would be intuitively obvious that deacetylation through HDACs also plays a role in modulating cell differentiation during neural development. Particularly, HDAC1 and HDAC2 play a role in oligodendrocyte differentiation through crosstalk with the canonical Wnt signaling pathway, mediated through transcription factor 7-like 2 (TCF7L2) [62] (Fig. 2). The inhibition of HDAC1 and HDAC2 causes the developmental plasticity of precursors, leading to differentiation into other cell types [63, 64].
Druggability of HAT Domains
An imbalance in acetylation homeostasis leads to a plethora of disease conditions; hence, the modulation of the activity of HATs could be an efficient method for therapeutics. Several domains of HAT, which play essential roles in the cell, can be targeted using small molecule modulators. These targets include 1) HAT domains, 2) auto-acetylation residues/regions, and 3) bromodomains, which recognize acetylation marks. These can be targeted and modulated using a small molecule or peptide approach. Several groups worldwide are designing synthetic or natural chemical probes that target epigenetic modifications. Small-molecule chemical probes have recently contributed to advances in the search for epigenetic drugs. The first reported HAT inhibitor, Lys-CoA, specifically inhibited p300 HAT activity [65]. Extracts from plants with medicinal value have served as therapeutics in the medieval world and present an excellent platform to develop new-generation therapeutics. The first reported HAT inhibitor from natural sources was obtained from Garcinia indica. Garcinol from G. indica nonspecifically inhibits p300 and PCAF HAT activity, but, recently, this inhibitor has shown great promise as a potent anticancer drug [66]. Owing to the specific focus of this article, we shall confine the review to HAT activators and their therapeutic potential.
Known HAT Activator Molecules (Structure and Their Possible Mechanism of Action)
The use of small molecules to modulate enzyme function is an emerging concept, and various molecules (synthetic and natural) have been identified with strong therapeutic potential against numerous diseases, including cancer, AIDS, diabetes, depression, and neurodegenerative conditions [7, 67, 68]. HAT activators are a class of small-molecule modulators that activate enzyme activity. The first reported and the most characterized HAT activator is N-(4-chloro-3-trifluoromethyl-phenyl)-2-ethoxy-6-pentadecyl-benza-mide (CTPB), derivatized using anacardic acid as a synthon [69, 70]. Anacardic acid can be isolated from cashew nut shell liquid, and this compound acts as an inhibitor of p300 and PCAF HAT activity. In contrast, CTPB can efficiently activate p300 HAT activity in a concentration-dependent manner, but does not alter PCAF HAT activity. In vitro CTPB induces the maximum activation of p300 HAT activity to ~4-fold at a 200–275 μM concentration. Interestingly, CTPB could also enhance p300 HAT-dependent transcription from a chromatin template in vitro. The mechanism of activation through CTPB and its derivative N-(4-chloro-3-trifluoromethyl-phenyl)-2-ethoxy-benzamide (CTB) was explored using surface-enhanced Raman spectroscopy, indicating a conformational change upon activator binding, which possibly recruits more acetyl-CoA and enhances auto-acetylation. Furthermore, the location of –CF3 and –Cl at the para position of the benzamide ring of CTPB and CTB is critical for HAT activation [71]. As CTPB and CTB are hydrophobic in nature, these compounds might form micelle-like structures in aqueous solution and directly bind to the hydrophobic pockets of p300. As most of the hydrophobic pockets of p300 are present in proximity to the HAT domain, the –CF3 and –Cl regions of these molecules could bind to the amide groups of the α-helix and β-sheets of p300 HAT domain and alter its conformation. In need of a more stable and potent activator of HATs, TTK21 was synthesized from salicylic acid. TTK21 enhances p300 and CBP acetylation in vitro and, upon conjugation with a carbon nanosphere (CSP), could enter the nucleus of the neural-derived cells, SHSY-5Y, and cross the blood–brain barrier (BBB), inducing histone H2B and H3 acetylation in specific regions of the mouse brain [72] (described in detail in the section ‘Potential use of HAT Activator as Therapeutic Option’).
Another small molecule with strong HAT activation ability towards p300/CBP is nemorosone, a derivative of benzophenone. Nemorosone is a polyisoprenylated benzophenone derivative that bears a C(1) acyl group and an adjacent C(8) quaternary center without any secondary cyclization. Thermodynamic studies using surface plasmon resonance indicated that nemorosone has high affinity towards p300 protein (0.25 μM), and this interaction is associated with a low kinetic dissociation constant (3.2 × 10-4 s-1), suggesting a hardly dissociable complex with a stable tertiary structure. Furthermore, the major region of nemorosone, which interacts with p300, was identified as a hydroxylated enol involving C(2), C(3), and C(4) [73]. Efforts to isolate small-molecule activators using the basic backbone structure of anacardic acid are currently underway. In a recent study, pentadecylidenemalonate was prepared, which surprisingly showed mixed inhibitor/activator properties towards HATs, as this chemical inhibited p300/CBP HAT activity and activated PCAF HAT activity [74, 75]. Pentadecylidenemalonate/SPV106 could cross mammalian cell membranes and enhance histone H3 acetylation. Moreover, this chemical is also the only reported PCAF activator.
HAT Impairment in Neurodegenerative Disorders
Neurodegenerative diseases, such as Huntington’s (HD), Parkinson’s (PD), Alzheimer’s (AD), or amyotrophic lateral sclerosis (ALS), are characterized by a progressive loss of neuronal structure and function that leads to neuronal death. In these diseases, impairments of HAT activity, histone, and nonhistone protein acetylation have been reported, associated with a decrease of cognitive functions and/or motor impairments. In this section, we will describe how HAT function is impaired in different pathologies (e.g., nuclear delocalization, enzymatic activity inhibition, etc.), how this dysregulation will contribute to the overall pathogenic process, and how maintaining adequate HAT/HDAC homeostasis in the cell is particularly important for neuronal survival [76–78], and how HATs might constitute targets for new therapeutic strategies. Table 1 summarizes the substrates of HATs and their potential links to neurological disorders.
Table 1.
Histone acetyltransferases (HATs), their substrates, and potential links to neurological diseases. Summary of the different HATs for which a potential relationship to pathological conditions or mechanisms has been documented. The associated modifications of histone and non-histone substrates induced through HAT impairments and potentially implicated in these diseases are also reported. The model (cellular, animal, or human) and the brain tissues in which modifications have been observed are indicated
| HATs | Substrates | Models | Related pathologies or mechanisms | |
|---|---|---|---|---|
| CBP | Histone | H3, H4 | In vitro, cellular and Drosophila lines | HD [8] |
| H3 | HdhQ7/Q111 mice (hipp) | HD [88] | ||
| H3, H4 | Cellular and C57BL/6J mice (striatum, substantia nigra) | PD [111] | ||
| H3 | SOD1 G86R mice (spinal cord) | ALS [76, 126] | ||
| H3 | EID1 trangenic mice (hipp) | AD [157] | ||
| H2A, H2B >> H3, H4 | Cbp +/−; cKO CBP mice (hipp) | LTM [199, 204] | ||
| H2B >> H3, H4 | Adenoviral CBP inhibition (hipp) | LTP and LTM [205] | ||
| Non-Histone | p53 | Cellular and R6/2 mice (striatum) | HD [9] | |
| UBF-1 | Cellular | HD [92] | ||
| Htt | Cellular, mice (cotex, striatum), Caenorhabditis elegans models, and human brain | HD [91] | ||
| α-tubulin | Human brain | HD [94] | ||
| p53 | Cellular | Axon regeneration [137, 138] | ||
| p53 | EID1 transgenic mice (hipp) | AD [157] | ||
| Tau | Cellular and human brain | AD [162] | ||
| P300 | Histone | H3 | EID1 transgenic mice (hipp) | AD [157] |
| Non-histone | p65 | Cellular, transgenic mice (substantia nigra) | PD [112] | |
| p53 | EID1 transgenic mice (hipp) | AD [157] | ||
| Tau | Cellular and human brain | AD [162] | ||
| ELP3 | Non-histone | α-tubulin | Cellular | ALS [128] |
| Bruchpilot (ELKS) | Drosophila lines | ALS [132] | ||
| PCAF | Histone | H3, H4 | In vitro, cellular, and Drosophila lines | HD [8] |
| / | / | Drosophila lines | HD [86] | |
CBP = cyclic adenomonophosphate response element-binding (CREB) binding protein; ELP3 = elongation protein 3; PCAF = p300/CBP-associated factor; UBF-1 = upstream binding factor-1; Htt = huntingtin; hipp = hippocampus; SOD1 = superoxide dismutase 1, EID1 = EP300-interacting inhibitor of differentiation 1; cKO = conditional knockout; HD = Huntington’s disease; PD Parkinson’s = disease; ALS = amyotrophic lateral sclerosis; AD = Alzheimer’s disease; LTM = long-term memory; LTP = long-term potentiation
HD
HD is a rare and fatal inheritable genetic disorder characterized by a loss of coordination (chorea), cognitive dysfunction, and psychiatric symptoms. At the cellular level, HD is characterized by intraneuronal inclusions, alterations in synaptic function and widespread neuronal death at the late stage of the disease, the striatum being the most affected region. Other brain regions, such as the cortex and the cerebellum, are also affected [79]. HD reflects an abnormally high number of unstable CAG repeats within the 5’-end of the coding region of the gene encoding the Huntingtin (Htt) protein. The number of repeats affects the severity and the onset of the disease, generally initiated at mid-life. The polyQ expansions alter the Htt protein functions and confer self-aggregation properties [80].
An initial study demonstrated that CBP is found in polyQ aggregates [81]. Indeed, CAG expansions bind and sequester transcriptional regulators, including CBP and PCAF in animal models [8, 9]. The resulting depletion of CBP from the nucleus induced a loss in its activity and a decrease in CBP-activated gene expression [9, 82, 83]. Interestingly, the soluble form of mutant Htt also promoted the degradation of CBP [84]. In addition, CBP sequestration into ubiquitin-positive nuclear inclusions and decreased CBP-mediated activation of gene expression [e.g., brain-derived neurotrophic factor (Bdnf)] were shown in a newly generated mouse model displaying HD-like-2, consistent with observations in HD patient brains [85]. In Drosophila models, reduced PCAF levels were associated with enhanced neurodegeneration. The CBP/p300 family also exhibited a strong influence over HD pathology, while the MYST family members had less of an effect [86].
HAT disruption in HD has several downstream consequences that might play a role in the pathophysiology of the disease. For instance, CBP alterations could directly affect striatal regulations, and thereby motor impairment, influencing the transcription dependent of its binding partner CREB, a key regulator of striatal vulnerability [87]. A decrease of histone H3ac, associated with decreased CBP/CREB-dependent gene expression, was observed in the hippocampus of the HdhQ7/Q111 transgenic mouse model, an event that might explain some cognitive deficits associated with HD [88]. A 30 % increase in Hdac1 expression was also observed in the cortices and striata of HD R6/2 mice, but is unlikely to be a cause of a detectable decrease in global histone acetylation [89]. Alterations in the acetylation of CBP-target nonhistone proteins, such as p53 [9, 90, 91] or the upstream binding factor-1 [92] also play a role in the progression of the disease. The Htt protein is subjected to acetylation that targets mutant Htt to autophagosomes for degradation [91]. The turnover of mutant Htt is thus regulated through the balance between CBP and HDAC1, and the loss of CBP in HD might explain the accumulation of mutant Htt [91]. Moreover, the acetylation of tubulin (likely through cytoplasmic acetyltransferases) regulates anterograde and retrograde axonal transport through signaling the anchoring of molecular motors, such as Kinesin-1 to microtubules [93]. As a decrease of acetylated-tubulin has been observed in postmortem brain samples [94], one could predict that a disruption of intracellular trafficking could occur in HD patients, as observed in HD mice neuronal culture and leading to the alteration to the alteration of vesicular BDNF transport [95].
At the level of bulk chromatin, a genome-wide deacetylation of H3 histone (H3K9K14ac) was observed in the striatum of the R6/2 mouse model, but no association with a specific HAT was observed [96]. However, the authors observed that an H3ac association at specific gene loci strongly correlated with expressed genes in both wild-type and transgenic striata, suggesting that these changes in histone H3ac, although genome-wide, were not sufficient to trigger the transcriptional dysfunctions associated with HD [96].
Several studies of HD using in vitro and in vivo models have confirmed CBP involvement in HD physiopathology; thus, the use of a molecule to activate HAT function would be of prime interest. The partial rescue of neuronal loss was described after CBP overexpression, whereas the genetic depletion of CBP worsened the phenotype of HD mice [83, 97, 98]. However, the overexpression of PCAF was not sufficient to ameliorate the HD phenotype in transgenic flies, suggesting that therapeutic strategies aimed at increasing PCAF protein levels are likely ineffective in ameliorating HD pathology; notably, increasing the HAT activity of PCAF has not been tested, and other trials targeting PCAF activity are needed [86]. Increasing the acetylation of histones and non-histone proteins via the use of HDAC inhibitors also counteracts neurodegeneration [8, 88, 99–101]. Intriguingly, McFarlan et al. [96] also demonstrated that HDAC inhibition re-established altered transcriptional levels in the striatum of the R6/2 mouse model, but only slightly improved H3ac binding to specific promoter, emphasizing the idea that the overall chromatin environment surrounding a gene would be more adapted to therapeutic targeting than simply increasing the acetylation of the histones.
PD
PD is a late-onset neurodegenerative disorder characterized by progressive motor dysfunctions (loss of muscle rigidity, tremor, and bradykinesia), the selective loss of nigrostriatal dopaminergic neurons and the presence of intraneuronal protein inclusions, called Lewy bodies (LBs), in the patient’s brain. PD has been associated with both environmental and genetic factors, with most of the cases (90 %) being sporadic. However, gene mutations or variations in the number of transcript copies lead to the appearance of hereditary PD forms. These genes include PARK2 (implicated in the targeting to proteasomal complex), PINK1 (involved in mitochondrial response to stress), and α-synuclein (which forms, when misfolded, the large intracellular aggregates termed LBs) [102, 103].
Several leads for PD pathophysiology have recently emerged. First, according to the range of mutated genes described in this disorder, defective proteasomal machinery or mitochondrial dysfunctions could intervene in the pathological mechanism. More recently, the involvement of deficient epigenetic regulations has also been described, despite the fact that no particular HAT has been implicated in PD. Indeed, in Drosophila, toxic nuclear aggregates of α-synuclein interact with histone H3 and promote its deacetylation, possibly through a histone-“masking” mechanism [104]. Consistent with this result, several studies targeting HDAC members through sodium butyrate or valproic acid have shown an increase in histone acetylation, associated with prosurvival and anti-inflammatory effects, and decreased neurotoxicity markers [105–110].
Surprisingly, other studies have associated increased HAT activity with PD. The exposure to Dieldrin, a neurotoxic pesticide associated with PD etiopathogenesis, was shown to induce histone H3 and H4 hyperacetylation via the time-dependent CBP accumulation in dopaminergic neurons [111]. Alternatively, wild-type α-synuclein reduces p300 expression level and HAT activity. As a consequence, the reduced acetylation of the p65 subunit of NFκB was observed, eventually leading to a decrease in the transactivation of the pro-apoptotic gene PKCδ. Native α-synuclein exhibits a neuroprotective effect through the inhibition of p300 HAT activity [112]; however, the interaction between mutant α-synuclein and p300 remains elusive.
ALS
ALS is a fatal adult-onset neuromuscular disorder, affecting upper and lower motor neurons, and leading to progressive muscle wasting, paralysis, and, eventually, death [113]. Motor neuron degeneration is associated primarily with the pathological aggregation of ubiquitin, fused in sarcoma protein, and the transactive response DNA binding protein (TAR) DNA-binding protein of 43-kDa in the cytoplasm of motor neuron cell bodies [114, 115]. In addition, ALS has also been associated with an important impairment of energy metabolism [116]. Although most ALS cases occur sporadically, 10 % of all ALS cases are inherited, among which 10–20 % are caused by mutations in the gene encoding superoxide dismutase-1 (SOD1). These mutations confer an adverse pro-apoptotic activity to SOD1, which is associated with oxidative damage and mitochondrial function disturbance, leading to increased neuronal vulnerability. Most of the SOD1 mutant proteins are prone to aggregation, and cytoplasmic inclusions have been detected in human patients and model systems [113, 117]. Even if the premature death of motor neurons is determinant in the onset of this disorder, the molecular mechanism of neuronal degeneration are multi-factorial, and the cause of ALS pathogenicity remains a matter of debate [118, 119].
The study of a symptomatic ALS mice model (SOD1 G86R) identified the loss of CBP and decreased histone H3ac in the lumbar spinal cord cholinergic motor neurons [76]. A gene expression profile analysis performed on laser microdissected degenerating motor neurons from sporadic ALS patients revealed the down-regulation of the cbp gene [120]. A comparison of HDAC expression revealed a reduction of HDAC 11 mRNA and increased HDAC 2 levels in postmortem ALS brain and spinal cord specimens [121]. Mouse models bearing a SOD1 mutation have been used to test therapeutic strategies with HDACi. Some studies have described an amelioration of motor performance and/or an increased survival of motor neurons in the G93A transgenic ALS mouse model, with phenylbutyrate [122] or valproate [123]. Sodium phenylbutyrate combined with an antioxidant [124] or riluzole [125] showed a beneficial effect. However, in the G86R mouse model, valproate restored histone acetylation in the spinal cord and CBP levels in motor neurons, but failed to improve mice survival, as a major detrimental effect on the neuromuscular junction was observed [126].
Altered microtubule-dependent trafficking associated with the damaged transport of mitochondria was identified in ALS [127]. Microtubule trafficking is affected by the acetylation of α-tubulin through the HAT ELP3 [128], and allelic variants of ELP3 were associated with sporadic cases of ALS [129]. Recent evidence obtained from the SOD1 G93A mouse model showed that the deletion of HDAC6, a HDAC that deacetylates α-tubulin, did not affect disease onset, but significantly extended mice survival. This protective effect was associated with an increased level of α-tubulin acetylation that maintains motor axon integrity [130]. Recently, a controversial study showed that HDAC6 knockdown increased the formation of large aggresome-like inclusions of mutant SOD1. This phenotype was associated with an increase of α-tubulin acetylation and retrograde transport of mutated proteins. Mutant SOD1 oligomers and small aggregates sequester and inactivate HDAC6, favoring tubulin acetylation and leading to the formation of large pathologic inclusions [131]. Thus, if ELP3 represents a strong candidate to design new activator-based therapies, then much effort must be made to understand the precise outcomes of tubulin acetylation. Moreover, ELP3 has been recently described to acetylate Bruchpilot, a large cytoskeletal-like protein detected in the presynaptic active zones in Drosophila, thereby regulating synaptic vesicle capture and neurotransmitter release efficiency. ELP3 loss-of-function or decreased expression, as in ALS, could thus result in the alteration of active zones function and morphology [132].
The site-specific acetylation of p53 leads to selective response to various signals [133]. The abnormal regulation of p53 has been observed in ALS patients [134], and cellular and animal ALS models [135, 136]. Interestingly, p53 acetylation at K320 through p300/CBP and PCAF serves as a prosurvival signal, and promotes neurite outgrowth and neuronal maturation [137]. Activating HAT activity in ALS motor neurons might mediate p53 K320 acetylation and facilitate targeting toward pro-survival rather than pro-apoptotic functions. However, an increase of p53 acetylation on K382, a known substrate for p300/CBP and HDAC SIRT1, induced apoptotic functions [138]. Thus, the CBP loss [76, 126] and SIRT1 overexpression [139] observed in motor neurons might induce a neuroprotective effect.
Overall, it is difficult to predict the expected effect of acetylation modulators in complex neurodegenerative diseases that affect several non-neuronal cell types [119, 140]. In particular, the effect of HAT activation in the neuromuscular junction, which is tightly regulated through chromatin acetylation [141, 142], has not been documented; however, studies have shown the deleterious effect of valproate [126].
AD and Related Diseases
AD is an age-related neurodegenerative disorder characterized by a progressive loss of memory and a deterioration of cognitive functions [143]. From the histopathologic point of view, neurofibrillary tangles (aggregates of cytoskeletal hyperphosphorylated tau protein), extracellular amyloid plaques [formed by the pathologic proteolytic processing of amyloid β precursor protein (APP)], and massive neuronal death are observed in several brain regions (cortex, hippocampus, and amygdala) [144, 145]. One distinctive hallmark of AD is the progressive and irreversible loss of cholinergic neurons in the basal forebrain [146]. Most AD cases are late in onset and are likely influenced by both genetic and environmental factors. Familial (FAD) forms represent 0.1 % of AD cases and have an autosomal dominant transmission mode associated with mutations in three genes: the APP gene, and the presinilins 1 and 2. In addition, one genetic risk factor, the ε4 allele of apolipoprotein, has been firmly implicated in AD, but the identification of many new risk genes for late-onset disease are still being discovered [147].
Recent literature shows that epigenetic regulations could stand as a valuable therapeutic strategy for AD [148]. Particularly, several studies report acetylation dysregulations associated with pathological markers of this disease. We have previously shown that the activation of the APP-dependent signaling targeted CBP to a caspase-6 degradation in primary cortical neuron cultures [76]. Notably, caspase-6 activity was highlighted in neurofibrillary tangles of AD brain patients [149–151]. Thus, the caspase 6-dependent degradation of CBP in AD, leading to an acetyltransferase loss in diseased neurons, has been hypothesized, suggesting HAT activation as an interesting strategy in AD. In addition, the genes encoding presenilins PS1 and PS2 have also been associated with CBP dysfunctions in specific cortical regions; the deletion of these two genes leads to a reduction of CBP protein and mRNA levels in both the cytoplasm and nucleus of cortical cells, whereas the total and phosphorylated forms of CREB remain unchanged. As a consequence, the genes regulated through the CREB/CBP complex are partially repressed [152]. This study suggests a positive regulation of CBP expression through presenilins. Interestingly, these conditional PS1−/−/PS2−/− mutants presented a decreased long-term potentiation (LTP), associated with long-term memory (LTM) deficits and hyperphosphorylated tau [152]. Thus, the effect of PS absence clearly alters the CBP-dependent pathway, and the effect of FAD-associated PS mutations remains unknown. Two studies conducted in cellular systems further reported that the overexpression of wild-type PS1 stimulated the transcriptional activity ability of CBP and of p300, while FAD-associated PS1 mutations did not produce this effect [153, 154]. However, Marambaud et al. [155] described a molecular pathway linking FAD-associated PS1 mutations with increased CBP function in cellular models. Briefly, the authors proposed that FAD mutations inhibit the production of an intracellular peptide N-Cad/CTF2 originally cleaved through wild-type PS1, thereby preventing CBP degradation and resulting in up-regulated CREB-mediated transcription. Importantly, whether this mechanism exists in vivo remains unknown.
A convincing experiment reporting the beneficial effect of CBP overexpression in an AD pathological model was recently published, showing that the in vivo hippocampal administration of viral particles carrying CBP restored the learning and memory deficits of AD triple transgenic mice [156]. Thus, by directly modulating CBP levels, the authors re-established CREB activity and BDNF expression in the hippocampus. Therefore, increasing CBP activity might be an interesting therapeutic tool in this case. In addition, an endogenous inhibitor of p300/CBP (EP300-interacting inhibitor of differentiation 1 or EID1) was shown to translocate from the cytoplasm to the nucleus in the hippocampus of the brain of an AD patient [157]. Interestingly, the authors showed that the neuron-specific expression of human EID1 gene in a mouse model reduced hippocampal LTP and impaired spatial memory. This phenotype was associated with the hypo-acetylation of histone H3 and p53 [157]. This study also confirmed an association between CBP activity deregulation and human pathology.
The acetylation modulation could also affect the pathological marks of AD. A recent study has shown an interesting action of PCAF in an AD non-transgenic mice model, as intracerebroventricular injections of Aβ peptides failed to induce toxicity in PCAF KO mice; indeed, these peptides that had a deleterious effect in wild-type mice and did not worsen PCAF KO mice memory deficits [158]. Consistent with these observations, neprelysin, a peptidase promoting Aβ degradation, presented increased expression in PCAF KO, rendering these mice insensitive to amyloid toxicity [158]. Moreover, restoring brain levels of CBP, while improving memory function, did not modify Aβ levels, plaques, or Tau immunoreactivity in a model exhibiting both amyloid plaques and neurofibrillary tangles [156]. Restoring memory functions with the HDAC inhibitor 4-phenylbutyrate increased dendritic spine density in CA1 and the clearance of intraneuronal Aβ accumulation [159, 160], even when a similar treatment did not affect Aβ levels and plaques [161]. Further, HDAC inhibition promotes decreased tau phosphorylation through, at least, down-regulating GSK3β activity [161]. Nonetheless, the inhibition of p300 and likely CBP has been associated with decreased Tau acetylation and the elimination of hyperphosphorylated Tau species [162]. In addition, an aggregation of acetylated-tau was found in pathological brains of AD patients and, hence, was suggested to be a new AD pathological hallmark [163]. These investigators further demonstrated in cellular systems that tau itself had a MYST-like acetyltransferase activity that could account for its self-acetylation [164]. Therefore, activating these HATs could result in increased Tau deposits. Thus, it is essential to determine whether the effects of HAT activation affect the mechanisms underlying AD pathology development.
Few studies have examined the role of acetylation in memory disorders other than AD, and no studies have shown a role for HAT in these diseases. One exception is frontotemporal dementia, a neurologic disease involving behavioral and language problems, leading to memory deficits. Some familial cases reflect progranulin haploinsufficiency [165]. A recent study showed that the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) increased progranulin mRNA and protein expression in haploinsufficiency progranulin human cells [166], suggesting that increasing acetylation should be beneficial. Notably, increased HDAC6 levels have been reported in the temporal cortex of patients with frontotemporal lobar degeneration with TAR DNA-binding protein of 43-kDa inclusions [167], suggesting that hypoacetylation could be involved in the pathophysiology. HDAC6 concentration had also been shown in LBs in PD and dementia with LBs [168]. However, LBs might represent a cytoprotective response to sequester toxic proteins [169]; thus, the effect of HDAC6 accumulation in α-synuclein aggresomes remains unknown. Overall, these studies suggest an important role for nuclear and cytoplasmic acetylation regulation in these diseases.
HAT in Synaptic Plasticity and Memory
During the last decade, several studies described the involvement of epigenetic regulations in cognitive functions. These mechanisms facilitate the adaptation of neuronal gene expression patterns according to experience, broadening the functions of neuronal networks. Indeed, transcriptional regulations are required in LTM formation in vivo [170, 171], as in ex vivo models of synaptic plasticity (LTP) [172, 173], and are highly dependent of CREB and NFκB activities [174, 175]. Among these regulations, histone acetylation plays an important and well-characterized role, whereby both HATs and HDACs are key regulators of cognitive processes. Moreover, emerging evidence has also implicated non-histone protein acetylation in cognitive functions. Although we present seminal studies supporting the involvement of acetylation mechanisms in memory and plasticity, we will focus on newly published findings, as extensive and complete literature is available on this topic [11, 176–178].
Histone Acetylation in Synaptic Plasticity and Memory
The induction of H3 acetylation has been previously reported in hippocampal CA1 formation at 1 h after contextual fear conditioning (CxFC; a commonly used task for hippocampus-dependent associative learning) [179]. This rise in histone acetylation was associated with an activation of both N-methyl-D-aspartate receptors and the extracellular signal-regulated kinase (ERK) signaling pathway, which are upstream factors for the stabilization and the formation of LTM. In 2010, Peleg et al. [15] revealed that CxFC causes an increase of several histones residues (H3K9K14, H4K5K8K12), as observed 1 h after exposure to the trial, in 3-month-old wild-type mice. Genome-wide analyses revealed that H4K12 acetylation was associated with transcriptional modifications after learning, and the differential expression of 2,229 genes was been identified through chromatin immunoprecipitation sequencing. The expression of the formin 2 gene (an actin nucleator, important for synaptic plasticity and memory) was increased in a H4K12 acetylation-dependent manner [15]. The same year, we [180] also demonstrated that hippocampal-dependent spatial memory consolidation in the Morris Water Maze (MWM) increased H3K9K14 and H4K12 histone residues in the H2B N-terminus region. Interestingly, these hyperacetylations were associated with the increased expression and activity of several HATs, among which the proximal promoter of CBP was enriched in the acetylated forms of histones. The up-regulation of several memory-related genes (cFos and Bdnf-IV) was also identified in associated with increased CBP activity [180]. In addition, the BDNF neurotrophin plays an important role in synaptic plasticity and behavioral adaptations. Interestingly, H3K9K14 acetylation was increased after BDNF treatment, another HDAC activity-dependent mechanism [181]. Indeed, the use of trichostatin A (TSA) and SAHA on hippocampal slices treated with BDNF abolished the histone acetylation induction and BDNF-positive effect on dendritic spines. A recent study from our laboratory further assessed acetylation responses during early stages of spatial and fear memory learning before trace consolidation. In both cases, H2B N-terminal (K5K12K15K20), H3K9K14, and H4K12 acetylation was increased in the learning paradigm, whereas H3K9K14 acetylation was already activated in response to the experimental context (swimming, exploration…) [182]. Thus, it is likely that specific acetylation marks on H2B and H4 histones play a role during memory formation, while the acetylation of H3K9K14 could be more easily modified in response to the environment (i.e., processing of the context). It is suggested that the rapid acetylation of the chromatin at H3K9K14 occurs within 1 h of the processing of the context whether a memory is formed or not, which could relax the chromatin to facilitate gene promoter access or tag chromatin for further acetylation on H2B- and H4-specific marks. Notably, chromatin structure modifications, such as the acetylation of H3 and H4 at specific residues, also occur in response to an enrichment of the environment [14]. Thus, increasing HAT activity could thus be an interesting tool to restore hippocampus-dependent memory formation.
Several studies, inhibiting or activating HDACs, have also shown their implications in memory processes. Thus, HDAC2 overexpression has been associated with an impairment of hippocampus-dependent memory formation (cued, and CxFC and MWM) and working memory in mice, while HDAC2 KO presented enhanced memory formation. HDAC1 overexpression had no effect on these tasks [183]. Transgenic mice with focal homozygous deletions in Hdac3 in the CA1 region of the dorsal hippocampus demonstrated enhanced LTM for object location together with increased H4K8 acetylation and the transcription of both Nr4a2 and c-fos [184]. More recently, HDAC1 activity has been associated with fear memory extinction for the inhibition of excess fear. The overexpression of HDAC1 in the adult mouse hippocampus increased this specific form of learning [185]. Furthermore, the loss of HDAC4 (in 2-month-old mice) and HDAC5 (in 10- but not 2-month-old mice) impaired memory functions, with an alteration in the CxFC and the MWM tasks in both cases [186, 187].
Changes in histone acetylation have also been observed in other brain regions, such as the lateral amygdala. In rats, an amygdala-dependent auditory fear conditioning training stimulates H3 acetylation in an ERK signaling-dependent manner. Consistent with this result, the treatment of amygdala slices with TSA causes an enhancement of LTP [188]. Cued fear conditioning was also impaired after HDAC5 knockdown (in 10-month-old mice) or HDAC2 overexpression (in 4-month-old mice) [183, 186], confirming a link between histone acetylation and amygdala proper functioning.
Non-histone Protein Acetylation in Synaptic Plasticity and Memory
In addition to histones, the acetylation of non-histone proteins has also been associated with the processes of synaptic plasticity and memory formation. In 2007, Zhang and collaborators identified an F-actin binding protein, cortactin, as a new target for acetylation. Cortactin acetylation is regulated through different factors (PCAF, p300, HDAC6, and SIRT1), leading to the reduction of cortactin binding to F-actin and cell mobility [189, 190]. Recently, Cortactin acetylation was also shown to affect dendritic spine morphogenesis, promoting the dendritic clustering of PSD95 and Shank1, two post-synaptic proteins, in excitatory hippocampal synapses. Cortactin acetylation can be promoted through BDNF and glutamate stimulation [191].
Nuclear factor kappa B (NFκB) is another non-histone substrate for acetylation; its function in synaptic plasticity and several types of LTM has previously been established and reviewed elsewhere [192, 193]. Interestingly, in cultured cells, it was shown that CBP and p300 acetylate the NFκB p65 subunit in a stimulus-dependent manner, which, subsequently, could be deacetylated through HDAC3. p65 acetylation regulates NFκB nuclear function, and acetylation increases its DNA binding affinity (K221) and transcriptional activity (K301) [194]. In contrast, IκBα binds and sequesters the deacetylated form, switching NFκB function off [195]. Consistent with these in cellulo studies, visual fear conditioning induces NFκB activity in the rat amygdala through increased CBP activity and p65-CBP interactions, associated with an enhancement of p65 acetylation and DNA binding [196]. More recently, a function for NFκB acetylation has been described in depression. Treatment of the depressed Flinders Sensitive Line rats with L-acetylcarnitine (LAC; an endogenous acetylating agent) had a rapid (3 days) and long-lasting (2 weeks after drug withdrawal) antidepressant action. LAC rescued mGlu2 receptor mRNA and protein expression in the hippocampus of these rats via increased p65 K310 acetylation and the downstream activation of NFκB. Moreover, LAC re-establishes H3 K27 acetylation of both grm2 (encoding the mGlu2 protein) and bdnf promoters, particularly in the depressed rats’ prefrontal cortex [197]. Notably, epigenetic modifications are consistently implicated in the physiopathology of major depressive diseases and psychiatric-related disorders, such addiction, schizophrenia, or vulnerability to stress, as reviewed in [11]. The use of an HAT activator could thus represent an interesting therapeutic option in such disorders, particularly in depressive and schizophrenic behaviors, where an impairment of acetylation was reported.
Transgenic HAT Animal Models
The involvement of lysine acetylation in memory and synaptic plasticity has led to the generation of several transgenic models to study the functions and consequences of the loss of HATs in these processes.
To gain insights into the functions of CBPs, several mouse strains were generated, including hemizygous, conditional KO, transgenic and region-specific (viral induction)-depleted mice. These models are reviewed in [198], and have been analyzed at the molecular level in different learning and memory tasks. Studies examining the effect of a partial or total loss of CBP function show the involvement of its HAT activity in LTP and LTM, and suggest that CBP is not required for the formation of short-term memory (STM) [47, 199–202]. Nevertheless, another study using a conditional model with complete CBP inactivation in excitatory neurons of the forebrain showed an impairment of both memory types [203]. However, more recent studies confirm CBP involvement in LTM. The use of a conditional KO CBP model in forebrain neurons identified an alteration of LTM in an object recognition task, whereas the performances of CBP in the MWM or in a CxFC task were not altered [204]. However, CBP loss in the dorsal CA1 hippocampus, through adenovirus injections, induced alterations of histones H2B, H3, and H4 acetylations, and a loss of CBP-dependent transcriptional programs [205]. These mice exhibited deficits in LTP and in the formation of LTM during object recognition and CxFC tasks, confirming that CBP-dependent chromatin modifications are involved in the consolidation of certain forms of memory. Thus, CBP HAT function is clearly involved in the normal adult CNS functioning, particularly in synaptic plasticity mechanisms associated with learning and memory.
p300 conditional transgenic mice, expressing an inhibitory truncated form of p300, have LTM deficits in object recognition and CxFC tasks. These mice lack activation domains and the p300 C-terminal domain containing the HAT function, causing an overall decrease in acetylation of H3K9K14 residues in the forebrain [206]. Moreover, p300 conditional ablation in specific forebrain subregions, during the postnatal period, leads to long-term recognition and contextual fear memory deficits [207].
PCAF KO mice exhibit STM impairments at 2 months of age, while no LTM deficits were detected [208]. These alterations are associated with a strong response to stress (corticosterone increase). At 6 months, these mice show altered LTM, as shown with the MWM task, and this phenotypes remain, even after 12 months. This transgenic model is characterized by a complete PCAF inactivation, resulting in the altered expression or activation of different factors involved in STM processes (cFos and ERK1).
Potential use of HAT Activator as a Therapeutic Option
The impairment (e.g., deregulation, degradation, sequestration, etc.) of HAT(s) has been described in several neurodegenerative diseases (‘HAT Impairment in Neurodegenerative Disorders’ section) and neurodevelopmental disorders. Thus, the specific activation of HAT activity could be useful in pathological situations where a decrease in the levels of these enzymes has been described. One example is the case of the Rubinstein–Taybi syndrome that presents a haploinsufficiency in CBP and, in some cases, p300 [45, 209–211]. A therapeutic strategy based on CBP/p300 reactivation could be useful, even in the adult, for re-establishing at least some cognitive functions [198, 212], particularly for ALS, in which motor neurons present lower quantities of CBP owing to degradation [76]. In addition, HDACi are poorly efficient in this context, as these molecules were deleterious for the neuromuscular junction, despite a significant improvement of motor neuron survival [126]. A HAT activator could also be relevant for its use in AD and related diseases, as this molecule could improve both neuronal survival, neurogenesis, and memory-related alterations (see below). However, the efficacy of HAT activators in pathological animal models remains undocumented.
HAT Activation vs HDAC Inhibition as a Therapeutic Strategy
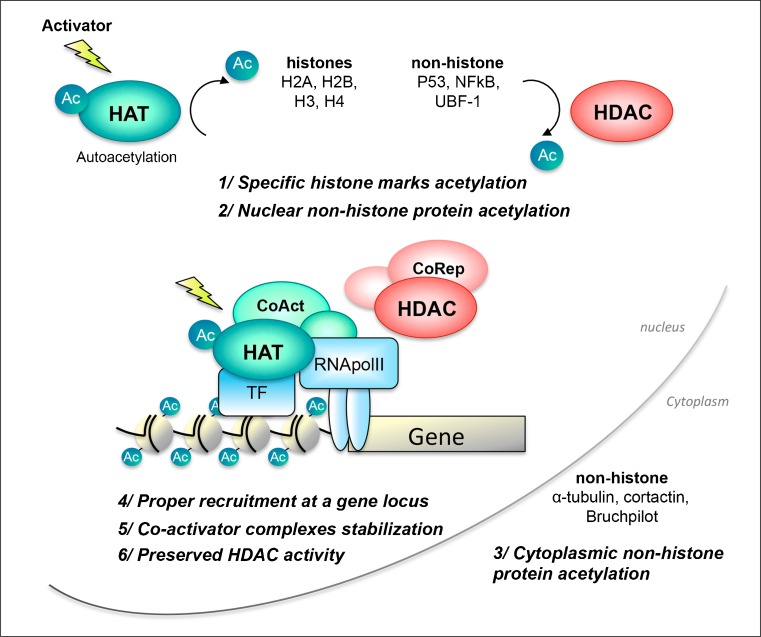
How HDACi efficiently affects pathological mechanisms is not well understood [5]. Notably, amongst the 14 HDAC isoforms described so far, class I HDAC2 and HDAC3 (but not HDAC1) have been associated with the promoters of genes implicated in synaptic plasticity [183, 213], potentially explaining some of the HDACi beneficial effects on memory formation. HDAC2 was further increased in AD postmortem brains [214]. Interestingly, at least one wild-type allele of cbp was required to mediate the effects of HDAC inhibition on memory functions [215]. A supportive study demonstrated that HDACi were inefficient in a complete knock down of cbp [203]. Thus, simply inducing a histone hyperacetylated state with HDACi does not adequately replace HAT activation, at least in certain functions (i.e., memory formation), likely reflecting the other potential roles of HAT (e.g., CBP). The indirect (HDAC inhibitor-mediated) activation of p300/CBP HAT auto-acetylation, which stimulates HAT activity [36, 216], could also contribute to the beneficial effects of HDACi. Thus, it seems essential to envisage direct stimulation of the HAT function, as a new therapeutic tool in neurodegenerative diseases; by directly targeting the remaining HAT(s), one could re-activate the deficient enzymatic function, as well as all the other related functions that are not typically considered using an HDACi strategy. HATs are recruited at promoter loci through, in part, lysine acetylation recognition of conserved bromodomains [20]; together with the proper bound coactivator complexes, HATs acetylate nucleosomes at specific promoter sites (see 4, 5/ in Fig. 3). Following an HDACi treatment, the global levels of acetylation are indeed increased, but this activation will not necessarily occur at specific defective HAT-targeted sites.
Fig. 3.
Histone acetyltransferase (HAT) activation as a therapeutic strategy. Illustration of the beneficial effects of the use of a HAT activator at the cellular level (see ‘HAT Activation vs HDAC Inhibition as a Therapeutic Strategy’). The activation of the HAT preserves the “acetylation code” of nuclear and cytoplasmic proteins (1–3), along with a proper positioning at gene loci (4) and recruitment of coactivators (5). Importantly, this strategy will preserve the histone deacetylase (HDAC) activity surrounding gene loci (6), otherwise impaired when following an HDAC inhibition strategy. Therefore, as both HAT and HDAC activities are present, a correct acetylation turn-over is possible, allowing transcriptional proceedings, see [217]. Ac = Acetyl; NFκB = nuclear factor kappa B; UBF-1 = upstream binding factor-1; CoAct = coactivator; CoRep = co-repressor; TF = transcription factor; RNApolII = RNA polymerase II
In addition, Hazzalin and Mahadevan [217] showed that the turnover of histone acetylation is important to produce gene induction, as observed for the fos and jun genes. Indeed, HDACi rapidly enhanced histone acetylation at these genes, but inhibited their transcription; these results are in contrast to the predominant view that increased histone acetylation is characteristic of enhanced transcription [217]. Thus, on certain genes, HDACi directly inhibits gene induction, reminiscent of the low percentage of genes that are actually activated after HDACi treatment [218]. This result also suggests that both HAT and HDAC complexes have to be present and active to the specific acetylation mark to activate transcription, suggesting that a strategy aimed at activating the HAT function might be more efficient than inhibiting the HDAC to re-establish proper transcription in a given pathological context (see 6/ in Fig. 3).
Last, the effects of HDAC inhibition and HAT activation on nonhistone proteins (e.g., p53, NFkB, cortactin, tubulin, etc.), i.e., those not directly associated with transcription, should be further documented in pathological conditions.
In vivo Effect of HAT Activators on Brain Functions
Only a few HAT activators have been produced, and most of these molecules are cell impermeant [69, 70]. So far, only two HAT activator molecules, a PCAF activator, SPV106 [219], and a CBP/P300 activator, CSP-TTK21 [72], have been shown to cross the BBB, consistent with the use of these compounds to study brain functions in vivo. Both of these compounds were active after systemic administration in adult mice (Fig. 4).
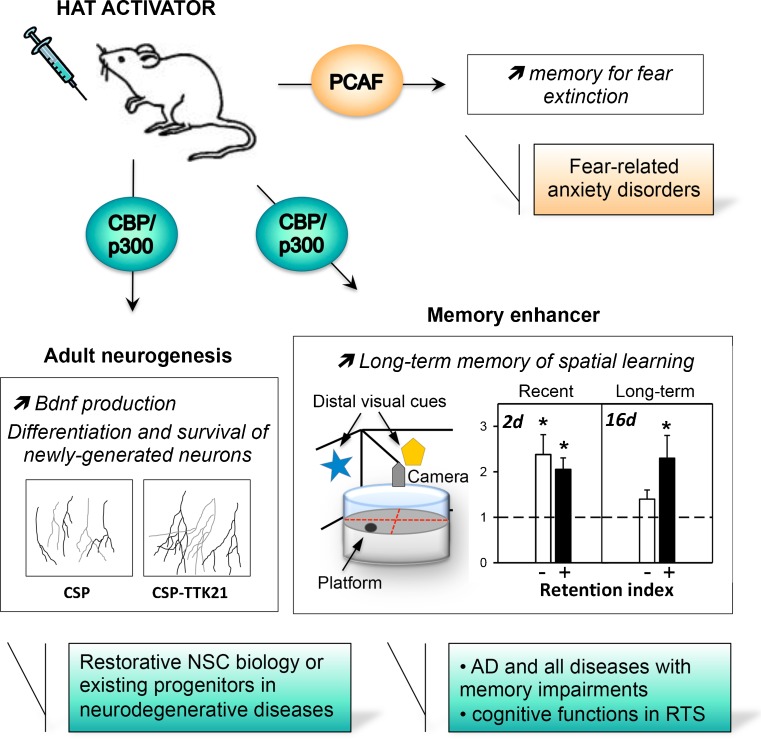
Fig. 4.
Effects of histone acetyltransferase (HAT) activator molecules on brain functions and their potential therapeutic use in diseases. In rodent models, the described effects of different HAT activators [SPV106 for p300/cyclic adenomonophosphate response element-binding (CREB)-associated factor (PCAF) [219] or carbon nanosphere (CSP)-TTK21 for CREB-binding protein (CBP)/p300 [72] are depicted]. The drawings represent the effect of CSP-TTK21 on newly generated (doublecortin-positive) neurons with increased dendritic branching measured in the hippocampus of adult mice 3 days after CSP-TTK21 injection. For spatial memory, CSP (−)- or CSP-TTK21 (+)-injected mice were tested in the Morris water maze and retention was evaluated 2 days (recent, 2d) or 16 days (long-term, 16d) after a weak protocol of acquisition. The retention index indicates significant retention of the platform location in the CSP-TTK21 injected group at long delays without effect on recent memory (*p < 0.05). For further details see [72]. The potential applications in diseases are summarized in the boxes (see text). NSC = neural stem cells; AD = Alzheimer’s disease; RTS = Rubinstein Taybi syndrome
Wei et al. [219] used SPV106 to demonstrate a role for PCAF in fear extinction. Fear conditioning and extinction are two distinct forms of learning that engage different molecular pathways. Fear extinction involves the gradual reduction in the fear response through the repeated presentation of a non-reinforced conditioned stimulus, which generates a new memory that competes with the original fear memory trace. As hippocampal acetylation or HATs such as CBP/P300 are essential to form contextual fear, fear extinction also involves epigenetic mechanisms [185, 220–222]. Notably, as discussed above, HDAC1 regulates extinction learning via a mechanism involving H3K9 deacetylation and the subsequent trimethylation of target genes [185]. Interestingly, HDAC1 was up-regulated in postmortem brain samples from schizophrenic patients [223, 224]. Using a pharmacological approach with the PCAF activator SPV106 and the PCAF inhibitor (H3-CoA-20-Tat), these authors demonstrated that PCAF activity facilitated the formation of fear extinction memory, but was not essential for fear acquisition [219]. These results suggest that PCAF regulation is an attractive target for the treatment of fear-related anxiety disorders.
Based on the small molecule HAT activator CTPB [69], we have synthesized several derivatives, from which TTK21 [N-(4-chloro-3-trifluoromethyl-phenyl)-2-n-propoxy-benzamide] was selected using a low-throughput enzyme assay, as this molecule efficiently activated the HAT activity of the CBP and p300 HATs [72]. However, similar to the parent compound CTPB, TTK21 was poorly permeable to living cells, but when conjugated to a glucose-derived CSP, CSP-TTK21 was able to cross membranes and induce histone acetylation. Moreover, after systemic injection, we detected acetylated histones in the frontal cortex and dorsal hippocampus of adult C57BL6/J mice, showing that the new CSP-TTK21 compound passes the BBB and reaches different parts of the brain. The effect of this HAT activator was also tested on 2 important hippocampal functions: adult neurogenesis and spatial memory formation. Remarkably, CSP-TTK21 treatment promoted the formation of long and highly-branched doublecortin-positive neurons in the subgranular zone of the dentate gyrus, suggesting that CBP/p300 activation favors the maturation and differentiation of adult neuronal progenitors. At the molecular level, we observed that CSP-TTK21 induced mRNA levels of the neuroD1 differentiation marker and BDNF, a neurotrophin required for the terminal differentiation of newly generated neurons. The concomitant enrichment of acetylated-histone was measured on the proximal promoter of these two genes. Finally, we observed that CBP/p300 activation during spatial training, while not improving the retention of recent memory (2 days), resulted in a significant extension of memory duration over 16 days. Thus, by inducing adult neurogenesis and LTM formation, our data show that the direct stimulation of CBP/P300 HAT function could have important effects in terms of therapeutic options for brain diseases (Fig. 4).
Concluding Remarks
Taken together, the results obtained in vivo show that new HAT activator molecules represent an important scientific advance and provide new therapeutic options for brain diseases. Indeed, molecules that favor and promote the in vivo maturation and differentiation of newly generated neurons in the adult present an obvious advantage in several neurodegenerative diseases. For instance, the transplantation of neural stem cells (NSC) rescued dysfunctional neurons in 2 studies associated with PD [225] and spinal cord injury [226]. Beneficial results were also observed in a mutant SOD mouse model of ALS [227]. Thus, the transplantation of undifferentiated NSCs achieves functional effects in animal models of neurologic disorders [228, 229], and it is likely that restorative NSC biology could be assisted by HAT activator molecules, thereby promoting neurotrophin production and the maturation of progenitors on site ([72]; Figs. 2 and 4). Notably, impaired neurogenesis, and specifically the survival and differentiation of neuronal progenitors, have been reported in mouse models of AD [230, 231]. Thus, the activation of differentiation programs (neuroD1, bdnf) in pathological brains might be a promising strategy in AD.
Moreover, the positive effect of environmental enrichment (EE) is associated with increased histone-tail acetylation in young rodents [14, 232, 233]. EE promotes various plasticity mechanisms in the hippocampus, including bdnf gene up-regulation, enhanced dendritic branching, and the stimulation of adult neurogenesis [234–237], reminiscent of our observations that CSP-TTK21 activates CBP/p300 activity. Furthermore, CBP-deficient mice present no response to the beneficial effect of EE on the induction and enhancement of spatial navigation capabilities during neurogenesis, highlighting the contribution of CBP to EE effects [233]. However, EE efficiently restored brain functions in animal models of neurodegenerative diseases [14, 238]. Together, these data suggest that therapeutic approaches with HAT activator molecules could present regulations similar to those induced through EE. This approach might combat pathological signaling or improve successful aging, as the aging brain retains considerable functional plasticity.
Thus, HAT activator molecules provide a promising and exciting area of research, but improvement of drug design, specificity, and targeting to the brain is still required. Notably, HAT activator molecules need further testing in animal models of neurologic pathologies.
Electronic supplementary material
(PDF 1224 kb)
Acknowledgments
This work was supported by funding from the Centre National de la Recherche Scientifique (CNRS), the University of Strasbourg, the Department of Biotechnology, the Government of India (Grant/ DBT/ CSH/ GIA/ 1752 to TKK), the Jawaharlal Nehru Centre for Advanced Scientific Research (JNCASR), Agence Nationale de la Recherche (ANR-12-MALZ-0002-01 to ALB), the Indo-French Centre for the Promotion of Advanced Research (IFCPAR/CEFIPRA No.4803-3 to T.K. Kundu and A.L. Boutillier), and Alsace Alzheimer 67 association (to A.L. Boutillier and F. Blanc). T.K. Kundu is a recipient of the Sir JC Bose national fellowship from the Department of Science and Technology through the government of India. Raphaelle Cassel received a doctoral fellowship from the French government. A. Schneider was supported through the Agence Nationale pour la Recherche (ANR-12-MALZ-0002). Frédéric Blanc reports receiving personal fees from Eisai, Piramal, Biogen Novartis, Janssen, Lundbeck, Pfizer, and Biogen, and non-financial support from Novartis, Lundbeck, Bayer Schering, Merck Serono, TEVA Neurosciences, and Pfizer, outside of the submitted work. Amrutha Swaminathan reports receiving grants from the Council for Scientific and Industrial Research, outside of the submitted work. Anne L. Boutillier, S. Chatterjee, and T.K. Kundu have a patent HAT Activator CSP-TTK21 pending. Olivier Bousiges, R. Cassel, A. Schneider, and B.R. Selvi. have nothing to disclose.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Disclosures
None.
References
- 1.Izzo A, Schneider R. Chatting histone modifications in mammals. Brief Funct Genomics. 2010;9:429–443. doi: 10.1093/bfgp/elq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allis CD, Berger SL, Cote J, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 4.McBrian MA, Behbahan IS, Ferrari R, et al. Histone acetylation regulates intracellular pH. Mol Cell. 2013;49:310–321. doi: 10.1016/j.molcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selvi BR, Cassel JC, Kundu TK, Boutillier AL. Tuning acetylation levels with HAT activators: therapeutic strategy in neurodegenerative diseases. Biochim Biophys Acta. 2010;1799:840–853. doi: 10.1016/j.bbagrm.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Steffan JS, Bodai L, Pallos J, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 9.Steffan JS, Kazantsev A, Spasic-Boskovic O, et al. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci U S A 2000;97:6763–6768. [DOI] [PMC free article] [PubMed]
- 10.Minamiyama M, Katsuno M, Adachi H, et al. Sodium butyrate ameliorates phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2004;13:1183–1192. doi: 10.1093/hmg/ddh131. [DOI] [PubMed] [Google Scholar]
- 11.Graff J, Kim D, Dobbin MM, Tsai LH. Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiol Rev. 2011;91:603–649. doi: 10.1152/physrev.00012.2010. [DOI] [PubMed] [Google Scholar]
- 12.Sleiman SF, Basso M, Mahishi L, et al. Putting the 'HAT' back on survival signalling: the promises and challenges of HDAC inhibition in the treatment of neurological conditions. Expert Opin Investig Drugs. 2009;18:573–584. doi: 10.1517/13543780902810345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dash PK, Orsi SA, Moore AN. Histone deactylase inhibition combined with behavioral therapy enhances learning and memory following traumatic brain injury. Neuroscience. 2009;163:1–8. doi: 10.1016/j.neuroscience.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 15.Peleg S, Sananbenesi F, Zovoilis A, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 16.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 17.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 18.Zhao S, Xu W, Jiang W, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundby A, Lage K, Weinert BT, et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012;2:419–431. doi: 10.1016/j.celrep.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 21.You L, Nie J, Sun WJ, Zheng ZQ, Yang XJ. Lysine acetylation: enzymes, bromodomains and links to different diseases. Essays Biochem. 2012;52:1–12. doi: 10.1042/bse0520001. [DOI] [PubMed] [Google Scholar]
- 22.Livengood JA, Scoggin KE, Van Orden K, et al. p53 Transcriptional activity is mediated through the SRC1-interacting domain of CBP/p300. J Biol Chem. 2002;277:9054–9061. doi: 10.1074/jbc.M108870200. [DOI] [PubMed] [Google Scholar]
- 23.Sanjuan R, Marin I. Tracing the origin of the compensasome: evolutionary history of DEAH helicase and MYST acetyltransferase gene families. Mol Biol Evol. 2001;18:330–343. doi: 10.1093/oxfordjournals.molbev.a003809. [DOI] [PubMed] [Google Scholar]
- 24.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lassar AB, Martin PL, Roeder RG. Transcription of class III genes: formation of preinitiation complexes. Science. 1983;222:740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- 26.Kundu TK, Wang Z, Roeder RG. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol Cell Biol. 1999;19:1605–1615. doi: 10.1128/mcb.19.2.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadoul K, Wang J, Diagouraga B, Khochbin S. The tale of protein lysine acetylation in the cytoplasm. J Biomed Biotechnol. 2011;2011:970382. doi: 10.1155/2011/970382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornberg RD, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Develop. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 29.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Develop. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 30.Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iizuka M, Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem. 1999;274:23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- 32.Magnaghi-Jaulin L, Ait-Si-Ali S, Harel-Bellan A. Histone acetylation and the control of the cell cycle. Prog Cell Cycle Res. 2000;4:41–47. doi: 10.1007/978-1-4615-4253-7_4. [DOI] [PubMed] [Google Scholar]
- 33.Smith ER, Eisen A, Gu W, et al. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci U S A. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatterjee S, Senapati P, Kundu TK. Post-translational modifications of lysine in DNA-damage repair. Essays Biochem. 2012;52:93–111. doi: 10.1042/bse0520093. [DOI] [PubMed] [Google Scholar]
- 35.Blanco JC, Minucci S, Lu J, et al. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Develop. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson PR, Wang D, Wang L, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 37.Black JC, Mosley A, Kitada T, Washburn M, Carey M. The SIRT2 deacetylase regulates autoacetylation of p300. Mol Cell. 2008;32:449–455. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gregoire S, Xiao L, Nie J, et al. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol Cell Biol. 2007;27:1280–1295. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Chen J. SIRT1 regulates autoacetylation and histone acetyltransferase activity of TIP60. J Biol Chem. 2010;285:11458–11464. doi: 10.1074/jbc.M109.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bengtson CP, Bading H. Nuclear calcium signaling. Adv Exp Med Biol. 2012;970:377–405. doi: 10.1007/978-3-7091-0932-8_17. [DOI] [PubMed] [Google Scholar]
- 41.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 42.Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002;21:5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol. 2002;12:2090–2097. doi: 10.1016/s0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- 44.Soliman ML, Rosenberger TA. Acetate supplementation increases brain histone acetylation and inhibits histone deacetylase activity and expression. Mol Cell Biochem. 2011;352:173–180. doi: 10.1007/s11010-011-0751-3. [DOI] [PubMed] [Google Scholar]
- 45.Kalkhoven E, Roelfsema JH, Teunissen H, et al. Loss of CBP acetyltransferase activity by PHD finger mutations in Rubinstein-Taybi syndrome. Hum Mol Genet. 2003;12:441–450. doi: 10.1093/hmg/ddg039. [DOI] [PubMed] [Google Scholar]
- 46.Kung AL, Rebel VI, Bronson RT, et al. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Develop. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 47.Oike Y, Hata A, Mamiya T, et al. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: implications for a dominant-negative mechanism. Hum Mol Genet. 1999;8:387–396. doi: 10.1093/hmg/8.3.387. [DOI] [PubMed] [Google Scholar]
- 48.Shikama N, Lutz W, Kretzschmar R, et al. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J. 2003;22:5175–5185. doi: 10.1093/emboj/cdg502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao TP, Oh SP, Fuchs M, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka Y, Naruse I, Hongo T, et al. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech Develop. 2000;95:133–145. doi: 10.1016/s0925-4773(00)00360-9. [DOI] [PubMed] [Google Scholar]
- 51.Nakashima K, Yanagisawa M, Arakawa H, et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- 52.Lee S, Lee B, Lee JW, Lee SK. Retinoid signaling and neurogenin2 function are coupled for the specification of spinal motor neurons through a chromatin modifier CBP. Neuron. 2009;62:641–654. doi: 10.1016/j.neuron.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Y, Nadal-Vicens M, Misono S, et al. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Weaver IC, Gauthier-Fisher A, et al. CBP histone acetyltransferase activity regulates embryonic neural differentiation in the normal and Rubinstein-Taybi syndrome brain. Develop Cell. 2010;18:114–125. doi: 10.1016/j.devcel.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 55.Bu P, Evrard YA, Lozano G, Dent SY. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol. 2007;27:3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin W, Zhang Z, Srajer G, et al. Proper expression of the Gcn5 histone acetyltransferase is required for neural tube closure in mouse embryos. Dev Dyn. 2008;237:928–940. doi: 10.1002/dvdy.21479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genetics. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- 58.Yamauchi T, Yamauchi J, Kuwata T, et al. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci U S A. 2000;97:11303–11306. doi: 10.1073/pnas.97.21.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen YC, Gatchel JR, Lewis RW, et al. Gcn5 loss-of-function accelerates cerebellar and retinal degeneration in a SCA7 mouse model. Hum Mol Genetics. 2012;21:394–405. doi: 10.1093/hmg/ddr474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez-Cerdeno V, Lemen JM, Chan V, et al. N-Myc and GCN5 regulate significantly overlapping transcriptional programs in neural stem cells. PloS One. 2012;7:e39456. doi: 10.1371/journal.pone.0039456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gehrking KM, Andresen JM, Duvick L, Lough J, Zoghbi HY, Orr HT. Partial loss of Tip60 slows mid-stage neurodegeneration in a spinocerebellar ataxia type 1 (SCA1) mouse model. Hum Mol Genetics. 2011;20:2204–2212. doi: 10.1093/hmg/ddr108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye F, Chen Y, Hoang T, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lyssiotis CA, Walker J, Wu C, Kondo T, Schultz PG, Wu X. Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proc Natl Acad Sci U S A 2007;104:14982–14987. [DOI] [PMC free article] [PubMed]
- 64.Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lau OD, Kundu TK, Soccio RE, et al. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol Cell. 2000;5:589–595. doi: 10.1016/s1097-2765(00)80452-9. [DOI] [PubMed] [Google Scholar]
- 66.Collins HM, Abdelghany MK, Messmer M, et al. Differential effects of garcinol and curcumin on histone and p53 modifications in tumour cells. BMC Cancer. 2013;13:37. doi: 10.1186/1471-2407-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Selvi BR, Chatterjee S, Modak R, Eswaramoorthy M, Kundu TK. Histone acetylation as a therapeutic target. Subcell Biochem. 2012;61:567–596. doi: 10.1007/978-94-007-4525-4_25. [DOI] [PubMed] [Google Scholar]
- 68.Swaminathan V, Reddy BA, Ruthrotha Selvi B, Sukanya MS, Kundu TK. Small molecule modulators in epigenetics: implications in gene expression and therapeutics. Subcell Biochem. 2007;41:397–428. [PubMed] [Google Scholar]
- 69.Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. Small molecule modulators of histone acetyltransferase p300. J Biol Chem. 2003;278:19134–19140. doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- 70.Souto JA, Benedetti R, Otto K, et al. New anacardic acid-inspired benzamides: histone lysine acetyltransferase activators. ChemMedChem. 2010;5:1530–1540. doi: 10.1002/cmdc.201000158. [DOI] [PubMed] [Google Scholar]
- 71.Mantelingu K, Kishore AH, Balasubramanyam K, et al. Activation of p300 histone acetyltransferase by small molecules altering enzyme structure: probed by surface-enhanced Raman spectroscopy. J Phys Chem B. 2007;111:4527–4534. doi: 10.1021/jp067655s. [DOI] [PubMed] [Google Scholar]
- 72.Chatterjee S, Mizar P, Cassel R, et al. A novel activator of CBP/p300 acetyltransferases promotes neurogenesis and extends memory duration in adult mice. J Neurosci. 2013;33:10698–10712. doi: 10.1523/JNEUROSCI.5772-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dal Piaz F, Tosco A, Eletto D, et al. The identification of a novel natural activator of p300 histone acetyltranferase provides new insights into the modulation mechanism of this enzyme. Chembiochem. 2010;11:818–827. doi: 10.1002/cbic.200900721. [DOI] [PubMed] [Google Scholar]
- 74.Milite C, Castellano S, Benedetti R, et al. Modulation of the activity of histone acetyltransferases by long chain alkylidenemalonates (LoCAMs) Bioorg Med Chem. 2011;19:3690–3701. doi: 10.1016/j.bmc.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 75.Sbardella G, Castellano S, Vicidomini C, et al. Identification of long chain alkylidenemalonates as novel small molecule modulators of histone acetyltransferases. Bioorg Med Chem Lett. 2008;18:2788–2792. doi: 10.1016/j.bmcl.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 76.Rouaux C, Jokic N, Mbebi C, Boutillier S, Loeffler JP, Boutillier AL. Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J. 2003;22:6537–6549. doi: 10.1093/emboj/cdg615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rouaux C, Loeffler JP, Boutillier AL. Targeting CREB-binding protein (CBP) loss of function as a therapeutic strategy in neurological disorders. Biochem Pharmacol. 2004;68:1157–1164. doi: 10.1016/j.bcp.2004.05.035. [DOI] [PubMed] [Google Scholar]
- 78.Saha RN, Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13:539–550. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kumar P, Kalonia H, Kumar A. Huntington's disease: pathogenesis to animal models. Pharmacol Rep. 2010;62:1–14. doi: 10.1016/s1734-1140(10)70238-3. [DOI] [PubMed] [Google Scholar]
- 80.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 81.Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc Natl Acad Sci U S A. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cong SY, Pepers BA, Evert BO, et al. Mutant huntingtin represses CBP, but not p300, by binding and protein degradation. Mol Cell Neurosci. 2005;30:560–571. [PubMed] [Google Scholar]
- 83.Jiang H, Poirier MA, Liang Y, et al. Depletion of CBP is directly linked with cellular toxicity caused by mutant huntingtin. Neurobiol Dis. 2006;23:543–551. doi: 10.1016/j.nbd.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 84.Choi YJ, Kim SI, Lee JW, et al. Suppression of aggregate formation of mutant huntingtin potentiates CREB-binding protein sequestration and apoptotic cell death. Mol Cell Neurosci. 2012;49:127–137. doi: 10.1016/j.mcn.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 85.Wilburn B, Rudnicki DD, Zhao J, et al. An antisense CAG repeat transcript at JPH3 locus mediates expanded polyglutamine protein toxicity in Huntington's disease-like 2 mice. Neuron. 2011;70:427–440. doi: 10.1016/j.neuron.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bodai L, Pallos J, Thompson LM, Marsh JL. Pcaf modulates polyglutamine pathology in a Drosophila model of Huntington's disease. Neurodegener Dis. 2012;9:104–106. doi: 10.1159/000330505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choi YS, Lee B, Cho HY, et al. CREB is a key regulator of striatal vulnerability in chemical and genetic models of Huntington's disease. Neurobiol Dis. 2009;36:259–268. doi: 10.1016/j.nbd.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Giralt A, Puigdellivol M, Carreton O, et al. Long-term memory deficits in Huntington's disease are associated with reduced CBP histone acetylase activity. Hum Mol Genet. 2012;21:1203–1216. doi: 10.1093/hmg/ddr552. [DOI] [PubMed] [Google Scholar]
- 89.Quinti L, Chopra V, Rotili D, et al. Evaluation of histone deacetylases as drug targets in Huntington's disease models. Study of HDACs in brain tissues from R6/2 and CAG140 knock-in HD mouse models and human patients and in a neuronal HD cell model. PLoS Curr 2010;2. [DOI] [PMC free article] [PubMed]
- 90.Bae BI, Xu H, Igarashi S, et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron. 2005;47:29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 91.Jeong H, Then F, Melia TJ, Jr, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee J, Hwang YJ, Boo JH, et al. Dysregulation of upstream binding factor-1 acetylation at K352 is linked to impaired ribosomal DNA transcription in Huntington's disease. Cell Death Differ. 2011;18:1726–1735. doi: 10.1038/cdd.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reed NA, Cai D, Blasius TL, et al. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 94.Nguyen T, Mehta NR, Conant K, et al. Axonal protective effects of the myelin-associated glycoprotein. J Neurosci. 2009;29:630–637. doi: 10.1523/JNEUROSCI.5204-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gauthier LR, Charrin BC, Borrell-Pages M, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 96.McFarland KN, Das S, Sun TT, et al. Genome-wide histone acetylation is altered in a transgenic mouse model of Huntington's disease. PloS One. 2012;7:e41423. doi: 10.1371/journal.pone.0041423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klevytska AM, Tebbenkamp AT, Savonenko AV, Borchelt DR. Partial depletion of CREB-binding protein reduces life expectancy in a mouse model of Huntington disease. J Neuropathol Exp Neurol. 2010;69:396–404. doi: 10.1097/NEN.0b013e3181d6c436. [DOI] [PubMed] [Google Scholar]
- 98.Taylor JP, Taye AA, Campbell C, Kazemi-Esfarjani P, Fischbeck KH, Min KT. Aberrant histone acetylation, altered transcription, and retinal degeneration in a Drosophila model of polyglutamine disease are rescued by CREB-binding protein. Genes Develop. 2003;17:1463–1468. doi: 10.1101/gad.1087503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dompierre JP, Godin JD, Charrin BC, et al. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington's disease by increasing tubulin acetylation. J Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ferrante RJ, Kubilus JK, Lee J, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington's disease mice. J Neurosci. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gardian G, Browne SE, Choi DK, et al. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington's disease. J Biol Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- 102.Goedert M, Spillantini MG, Del Tredici K, Braak H. 100 years of Lewy pathology. Nat Rev Neurol. 2013;9:13–24. doi: 10.1038/nrneurol.2012.242. [DOI] [PubMed] [Google Scholar]
- 103.Yasuda T, Mochizuki H. The regulatory role of alpha-synuclein and parkin in neuronal cell apoptosis; possible implications for the pathogenesis of Parkinson's disease. Apoptosis. 2010;15:1312–1321. doi: 10.1007/s10495-010-0486-8. [DOI] [PubMed] [Google Scholar]
- 104.Kontopoulos E, Parvin JD, Feany MB. Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet. 2006;15:3012–3023. doi: 10.1093/hmg/ddl243. [DOI] [PubMed] [Google Scholar]
- 105.Chen PS, Peng GS, Li G, et al. Valproate protects dopaminergic neurons in midbrain neuron/glia cultures by stimulating the release of neurotrophic factors from astrocytes. Mol Psychiatry. 2006;11:1116–1125. doi: 10.1038/sj.mp.4001893. [DOI] [PubMed] [Google Scholar]
- 106.Chen PS, Wang CC, Bortner CD, et al. Valproic acid and other histone deacetylase inhibitors induce microglial apoptosis and attenuate lipopolysaccharide-induced dopaminergic neurotoxicity. Neuroscience. 2007;149:203–212. doi: 10.1016/j.neuroscience.2007.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gardian G, Yang L, Cleren C, Calingasan NY, Klivenyi P, Beal MF. Neuroprotective effects of phenylbutyrate against MPTP neurotoxicity. Neuromol Med. 2004;5:235–241. doi: 10.1385/NMM:5:3:235. [DOI] [PubMed] [Google Scholar]
- 108.Gottlicher M, Minucci S, Zhu P, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Monti B, Gatta V, Piretti F, Raffaelli SS, Virgili M, Contestabile A. Valproic acid is neuroprotective in the rotenone rat model of Parkinson's disease: involvement of alpha-synuclein. Neurotox Res. 2010;17:130–141. doi: 10.1007/s12640-009-9090-5. [DOI] [PubMed] [Google Scholar]
- 110.Outeiro TF, Kontopoulos E, Altmann SM, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 111.Song C, Kanthasamy A, Anantharam V, Sun F, Kanthasamy AG. Environmental neurotoxic pesticide increases histone acetylation to promote apoptosis in dopaminergic neuronal cells: relevance to epigenetic mechanisms of neurodegeneration. Mol Pharmacol. 2010;77:621–632. doi: 10.1124/mol.109.062174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jin H, Kanthasamy A, Ghosh A, Yang Y, Anantharam V, Kanthasamy AG. alpha-Synuclein negatively regulates protein kinase Cdelta expression to suppress apoptosis in dopaminergic neurons by reducing p300 histone acetyltransferase activity. J Neurosci. 2011;31:2035–2051. doi: 10.1523/JNEUROSCI.5634-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Turner MR, Hardiman O, Benatar M, et al. Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurol. 2013;12:310–322. doi: 10.1016/S1474-4422(13)70036-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mackenzie IR, Bigio EH, Ince PG, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61:427–434. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 115.Deng HX, Zhai H, Bigio EH, et al. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann Neurol. 2010;67:739–748. doi: 10.1002/ana.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dupuis L, Pradat PF, Ludolph AC, Loeffler JP. Energy metabolism in amyotrophic lateral sclerosis. Lancet Neurol. 2011;10:75–82. doi: 10.1016/S1474-4422(10)70224-6. [DOI] [PubMed] [Google Scholar]
- 117.Gibson SB, Bromberg MB. Amyotrophic lateral sclerosis: drug therapy from the bench to the bedside. Semin Neurol. 2012;32:173–178. doi: 10.1055/s-0032-1329193. [DOI] [PubMed] [Google Scholar]
- 118.Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dupuis L, Loeffler JP. Neuromuscular junction destruction during amyotrophic lateral sclerosis: insights from transgenic models. Curr Opin Pharmacol. 2009;9:341–346. doi: 10.1016/j.coph.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 120.Jiang YM, Yamamoto M, Kobayashi Y, et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2005;57:236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- 121.Janssen C, Schmalbach S, Boeselt S, Sarlette A, Dengler R, Petri S. Differential histone deacetylase mRNA expression patterns in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2010;69:573–581. doi: 10.1097/NEN.0b013e3181ddd404. [DOI] [PubMed] [Google Scholar]
- 122.Ryu H, Smith K, Camelo SI, et al. Sodium phenylbutyrate prolongs survival and regulates expression of anti-apoptotic genes in transgenic amyotrophic lateral sclerosis mice. J Neurochem. 2005;93:1087–1098. doi: 10.1111/j.1471-4159.2005.03077.x. [DOI] [PubMed] [Google Scholar]
- 123.Sugai F, Yamamoto Y, Miyaguchi K, et al. Benefit of valproic acid in suppressing disease progression of ALS model mice. Eur J Neurosci. 2004;20:3179–3183. doi: 10.1111/j.1460-9568.2004.03765.x. [DOI] [PubMed] [Google Scholar]
- 124.Petri S, Kiaei M, Kipiani K, et al. Additive neuroprotective effects of a histone deacetylase inhibitor and a catalytic antioxidant in a transgenic mouse model of amyotrophic lateral sclerosis. Neurobiol Dis. 2006;22:40–49. doi: 10.1016/j.nbd.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 125.Del Signore SJ, Amante DJ, Kim J, et al. Combined riluzole and sodium phenylbutyrate therapy in transgenic amyotrophic lateral sclerosis mice. Amyotroph Lateral Scler. 2009;10:85–94. doi: 10.1080/17482960802226148. [DOI] [PubMed] [Google Scholar]
- 126.Rouaux C, Panteleeva I, Rene F, et al. Sodium valproate exerts neuroprotective effects in vivo through CREB-binding protein-dependent mechanisms but does not improve survival in an amyotrophic lateral sclerosis mouse model. J Neurosci. 2007;27:5535–5545. doi: 10.1523/JNEUROSCI.1139-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.De Vos KJ, Chapman AL, Tennant ME, et al. Familial amyotrophic lateral sclerosis-linked SOD1 mutants perturb fast axonal transport to reduce axonal mitochondria content. Hum Mol Genet. 2007;16:2720–2728. doi: 10.1093/hmg/ddm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Creppe C, Malinouskaya L, Volvert ML, et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 129.Simpson CL, Lemmens R, Miskiewicz K, et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum Mol Genet. 2009;18:472–481. doi: 10.1093/hmg/ddn375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Taes I, Timmers M, Hersmus N, et al. Hdac6 deletion delays disease progression in the SOD1G93A mouse model of ALS. Hum Mol Genet. 2013;22:1783–1790. doi: 10.1093/hmg/ddt028. [DOI] [PubMed] [Google Scholar]
- 131.Gal J, Chen J, Barnett KR, Yang L, Brumley E, Zhu H. HDAC6 regulates mutant SOD1 aggregation through two SMIR motifs and tubulin acetylation. J Biol Chem. 2013;288:15035–15045. doi: 10.1074/jbc.M112.431957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Miskiewicz K, Jose LE, Bento-Abreu A, et al. ELP3 controls active zone morphology by acetylating the ELKS family member Bruchpilot. Neuron. 2011;72:776–788. doi: 10.1016/j.neuron.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 133.Roy S, Tenniswood M. Site-specific acetylation of p53 directs selective transcription complex assembly. J Biol Chem. 2007;282:4765–4771. doi: 10.1074/jbc.M609588200. [DOI] [PubMed] [Google Scholar]
- 134.Martin LJ, Liu Z. DNA damage profiling in motor neurons: a single-cell analysis by comet assay. Neurochem Res. 2002;27:1093–1104. doi: 10.1023/a:1020961006216. [DOI] [PubMed] [Google Scholar]
- 135.Barbosa LF, Cerqueira FM, Macedo AF, et al. Increased SOD1 association with chromatin, DNA damage, p53 activation, and apoptosis in a cellular model of SOD1-linked ALS. Biochim Biophys Acta. 1802;2010:462–471. doi: 10.1016/j.bbadis.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 136.de Aguilar JL G, Gordon JW, Rene F, et al. Alteration of the Bcl-x/Bax ratio in a transgenic mouse model of amyotrophic lateral sclerosis: evidence for the implication of the p53 signaling pathway. Neurobiol Dis. 2000;7:406–415. doi: 10.1006/nbdi.2000.0295. [DOI] [PubMed] [Google Scholar]
- 137.Di Giovanni S, Knights CD, Rao M, et al. The tumor suppressor protein p53 is required for neurite outgrowth and axon regeneration. EMBO J. 2006;25:4084–4096. doi: 10.1038/sj.emboj.7601292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pearson M, Carbone R, Sebastiani C, et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 139.Kim D, Nguyen MD, Dobbin MM, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 141.Mejat A, Ramond F, Bassel-Duby R, Khochbin S, Olson EN, Schaeffer L. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat Neurosci. 2005;8:313–321. doi: 10.1038/nn1408. [DOI] [PubMed] [Google Scholar]
- 142.Ravel-Chapuis A, Vandromme M, Thomas JL, Schaeffer L. Postsynaptic chromatin is under neural control at the neuromuscular junction. EMBO J. 2007;26:1117–1128. doi: 10.1038/sj.emboj.7601572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Selkoe DJ. Preventing Alzheimer's disease. Science. 2012;337:1488–1492. doi: 10.1126/science.1228541. [DOI] [PubMed] [Google Scholar]
- 144.Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 145.Iqbal K, Liu F, Gong CX, Alonso Adel C, Grundke-Iqbal I. Mechanisms of tau-induced neurodegeneration. Acta Neuropathol. 2009;118:53–69. doi: 10.1007/s00401-009-0486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Davies P, Maloney AJ. Selective loss of central cholinergic neurons in Alzheimer's disease. Lancet. 1976;2:1403. doi: 10.1016/s0140-6736(76)91936-x. [DOI] [PubMed] [Google Scholar]
- 147.Bettens K, Sleegers K, Van Broeckhoven C. Genetic insights in Alzheimer's disease. Lancet Neurol. 2013;12:92–104. doi: 10.1016/S1474-4422(12)70259-4. [DOI] [PubMed] [Google Scholar]
- 148.Adwan L, Zawia NH. Epigenetics: A novel therapeutic approach for the treatment of Alzheimer's disease. Pharmacol Therap. 2013;139:41–50. doi: 10.1016/j.pharmthera.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Albrecht S, Bourdeau M, Bennett D, Mufson EJ, Bhattacharjee M, LeBlanc AC. Activation of caspase-6 in aging and mild cognitive impairment. Am J Pathol. 2007;170:1200–1209. doi: 10.2353/ajpath.2007.060974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Guo H, Albrecht S, Bourdeau M, Petzke T, Bergeron C, LeBlanc AC. Active caspase-6 and caspase-6-cleaved tau in neuropil threads, neuritic plaques, and neurofibrillary tangles of Alzheimer's disease. Am J Pathol. 2004;165:523–531. doi: 10.1016/S0002-9440(10)63317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Guo H, Petrin D, Zhang Y, Bergeron C, Goodyer CG, LeBlanc AC. Caspase-1 activation of caspase-6 in human apoptotic neurons. Cell Death Differ. 2006;13:285–292. doi: 10.1038/sj.cdd.4401753. [DOI] [PubMed] [Google Scholar]
- 152.Saura CA, Choi SY, Beglopoulos V, et al. Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron. 2004;42:23–36. doi: 10.1016/s0896-6273(04)00182-5. [DOI] [PubMed] [Google Scholar]
- 153.Francis YI, Diss JK, Kariti M, Stephanou A, Latchman DS. p300 activation by Presenilin 1 but not by its M146L mutant. Neurosci Lett. 2007;413:137–140. doi: 10.1016/j.neulet.2006.11.036. [DOI] [PubMed] [Google Scholar]
- 154.Francis YI, Stephanou A, Latchman DS. CREB-binding protein activation by presenilin 1 but not by its M146L mutant. Neuroreport. 2006;17:917–921. doi: 10.1097/01.wnr.0000220137.06542.a0. [DOI] [PubMed] [Google Scholar]
- 155.Marambaud P, Wen PH, Dutt A, et al. A CBP binding transcriptional repressor produced by the PS1/epsilon-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell. 2003;114:635–645. doi: 10.1016/j.cell.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 156.Caccamo A, Maldonado MA, Bokov AF, Majumder S, Oddo S. CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2010;107:22687–22692. doi: 10.1073/pnas.1012851108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu R, Lei JX, Luo C, et al. Increased EID1 nuclear translocation impairs synaptic plasticity and memory function associated with pathogenesis of Alzheimer's disease. Neurobiol Dis. 2012;45:902–912. doi: 10.1016/j.nbd.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Duclot F, Meffre J, Jacquet C, Gongora C, Maurice T. Mice knock out for the histone acetyltransferase p300/CREB binding protein-associated factor develop a resistance to amyloid toxicity. Neuroscience. 2010;167:850–863. doi: 10.1016/j.neuroscience.2010.02.055. [DOI] [PubMed] [Google Scholar]
- 159.Ricobaraza A, Cuadrado-Tejedor M, Garcia-Osta A. Long-term phenylbutyrate administration prevents memory deficits in Tg2576 mice by decreasing Abeta. Front Biosci (Elite Ed) 2011;3:1375–1384. doi: 10.2741/e340. [DOI] [PubMed] [Google Scholar]
- 160.Ricobaraza A, Cuadrado-Tejedor M, Marco S, Perez-Otano I, Garcia-Osta A. Phenylbutyrate rescues dendritic spine loss associated with memory deficits in a mouse model of Alzheimer disease. Hippocampus. 2012;22:1040–1050. doi: 10.1002/hipo.20883. [DOI] [PubMed] [Google Scholar]
- 161.Ricobaraza A, Cuadrado-Tejedor M, Perez-Mediavilla A, Frechilla D, Del Rio J, Garcia-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer's disease mouse model. Neuropsychopharmacology. 2009;34:1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- 162.Min SW, Cho SH, Zhou Y, et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Irwin DJ, Cohen TJ, Grossman M, et al. Acetylated tau, a novel pathological signature in Alzheimer's disease and other tauopathies. Brain. 2012;135:807–818. doi: 10.1093/brain/aws013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cohen TJ, Friedmann D, Hwang AW, Marmorstein R, Lee VM. The microtubule-associated tau protein has intrinsic acetyltransferase activity. Nat Struct Mol Biol. 2013;20:756–762. doi: 10.1038/nsmb.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mackenzie IR, Baker M, Pickering-Brown S, et al. The neuropathology of frontotemporal lobar degeneration caused by mutations in the progranulin gene. Brain. 2006;129:3081–3090. doi: 10.1093/brain/awl271. [DOI] [PubMed] [Google Scholar]
- 166.Cenik B, Sephton CF, Dewey CM, et al. Suberoylanilide hydroxamic acid (vorinostat) up-regulates progranulin transcription: rational therapeutic approach to frontotemporal dementia. J Biol Chem. 2011;286:16101–16108. doi: 10.1074/jbc.M110.193433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Odagiri S, Tanji K, Mori F, et al. Brain expression level and activity of HDAC6 protein in neurodegenerative dementia. Biochem Biophys Res Commun. 2013;430:394–399. doi: 10.1016/j.bbrc.2012.11.034. [DOI] [PubMed] [Google Scholar]
- 168.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 169.Miki Y, Mori F, Tanji K, Kakita A, Takahashi H, Wakabayashi K. Accumulation of histone deacetylase 6, an aggresome-related protein, is specific to Lewy bodies and glial cytoplasmic inclusions. Neuropathology. 2011;31:561–568. doi: 10.1111/j.1440-1789.2011.01200.x. [DOI] [PubMed] [Google Scholar]
- 170.Agranoff BW. Memory and protein synthesis. Sci Am. 1967;216:115–122. doi: 10.1038/scientificamerican0667-115. [DOI] [PubMed] [Google Scholar]
- 171.Flood JF, Rosenzweig MR, Bennett EL, Orme AE. The influence of duration of protein synthesis inhibition on memory. Physiol Behav. 1973;10:555–562. doi: 10.1016/0031-9384(73)90221-7. [DOI] [PubMed] [Google Scholar]
- 172.Nguyen PV, Abel T, Kandel ER. Requirement of a critical period of transcription for induction of a late phase of LTP. Science. 1994;265:1104–1107. doi: 10.1126/science.8066450. [DOI] [PubMed] [Google Scholar]
- 173.Stanton PK, Sarvey JM. Blockade of long-term potentiation in rat hippocampal CA1 region by inhibitors of protein synthesis. J Neurosci. 1984;4:3080–3088. doi: 10.1523/JNEUROSCI.04-12-03080.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gutierrez H, Davies AM. Regulation of neural process growth, elaboration and structural plasticity by NF-kappaB. Trends Neurosci. 2011;34:316–325. doi: 10.1016/j.tins.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sakamoto K, Karelina K, Obrietan K. CREB: a multifaceted regulator of neuronal plasticity and protection. J Neurochem. 2011;116:1–9. doi: 10.1111/j.1471-4159.2010.07080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Graff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14:97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 177.Peixoto L, Abel T. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology. 2013;38:62–76. doi: 10.1038/npp.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zocchi L, Sassone-Corsi P. Joining the dots: from chromatin remodeling to neuronal plasticity. Curr Opin Neurobiol. 2010;20:432–440. doi: 10.1016/j.conb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 180.Bousiges O, Vasconcelos AP, Neidl R, et al. Spatial memory consolidation is associated with induction of several lysine-acetyltransferase (histone acetyltransferase) expression levels and H2B/H4 acetylation-dependent transcriptional events in the rat hippocampus. Neuropsychopharmacology. 2010;35:2521–2537. doi: 10.1038/npp.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Calfa G, Chapleau CA, Campbell S, et al. HDAC activity is required for BDNF to increase quantal neurotransmitter release and dendritic spine density in CA1 pyramidal neurons. Hippocampus. 2012;22:1493–1500. doi: 10.1002/hipo.20990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bousiges O, Neidl R, Majchrzak M, et al. Detection of Histone acetylation levels in the dorsal hippocampus reveals early tagging on specific residues of H2B and H4 histones in response to learning. PloS One. 2013;8:e57816. doi: 10.1371/journal.pone.0057816. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 183.Guan JS, Haggarty SJ, Giacometti E, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.McQuown SC, Barrett RM, Matheos DP, et al. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bahari-Javan S, Maddalena A, Kerimoglu C, et al. HDAC1 regulates fear extinction in mice. J Neurosci. 2012;32:5062–5073. doi: 10.1523/JNEUROSCI.0079-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Agis-Balboa RC, Pavelka Z, Kerimoglu C, Fischer A. Loss of HDAC5 impairs memory function: implications for Alzheimer's disease. J Alzheimers Dis. 2013;33:35–44. doi: 10.3233/JAD-2012-121009. [DOI] [PubMed] [Google Scholar]
- 187.Kim MS, Akhtar MW, Adachi M, et al. An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J Neurosci. 2012;32:10879–10886. doi: 10.1523/JNEUROSCI.2089-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Monsey MS, Ota KT, Akingbade IF, Hong ES, Schafe GE. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PloS One. 2011;6:e19958. doi: 10.1371/journal.pone.0019958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang X, Yuan Z, Zhang Y, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27:197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhang Y, Zhang M, Dong H, et al. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene. 2009;28:445–460. doi: 10.1038/onc.2008.388. [DOI] [PubMed] [Google Scholar]
- 191.Catarino T, Ribeiro L, Santos SD, Carvalho AL. Regulation of synapse composition by protein acetylation: the role of acetylated cortactin. J Cell Sc. 2013;126:149–162. doi: 10.1242/jcs.110742. [DOI] [PubMed] [Google Scholar]
- 192.Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mattson MP, Meffert MK. Roles for NF-kappaB in nerve cell survival, plasticity, and disease. Cell Death Differ. 2006;13:852–860. doi: 10.1038/sj.cdd.4401837. [DOI] [PubMed] [Google Scholar]
- 194.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 196.Yeh SH, Lin CH, Gean PW. Acetylation of nuclear factor-kappaB in rat amygdala improves long-term but not short-term retention of fear memory. Mol Pharmacol. 2004;65:1286–1292. doi: 10.1124/mol.65.5.1286. [DOI] [PubMed] [Google Scholar]
- 197.Nasca C, Xenos D, Barone Y, et al. L-acetylcarnitine causes rapid antidepressant effects through the epigenetic induction of mGlu2 receptors. Proc Natl Acad Sci U S A. 2013;110:4804–4809. doi: 10.1073/pnas.1216100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Valor LM, Viosca J, Lopez-Atalaya JP, Barco A. Lysine acetyltransferases CBP and p300 as therapeutic targets in cognitive and neurodegenerative disorders. Curr Pharm Design 2013 Feb 19 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 199.Alarcon JM, Malleret G, Touzani K, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 200.Bourtchouladze R, Lidge R, Catapano R, et al. A mouse model of Rubinstein-Taybi syndrome: defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proc Natl Acad Sci U S A. 2003;100:10518–10522. doi: 10.1073/pnas.1834280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wood MA, Kaplan MP, Park A, et al. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chen G, Zou X, Watanabe H, van Deursen JM, Shen J. CREB binding protein is required for both short-term and long-term memory formation. J Neurosci. 2010;30:13066–13077. doi: 10.1523/JNEUROSCI.2378-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Valor LM, Pulopulos MM, Jimenez-Minchan M, Olivares R, Lutz B, Barco A. Ablation of CBP in forebrain principal neurons causes modest memory and transcriptional defects and a dramatic reduction of histone acetylation but does not affect cell viability. J Neurosci. 2011;31:1652–1663. doi: 10.1523/JNEUROSCI.4737-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Barrett RM, Malvaez M, Kramar E, et al. Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology. 2011;36:1545–1556. doi: 10.1038/npp.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Oliveira AM, Wood MA, McDonough CB, Abel T. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learning Memory. 2007;14:564–572. doi: 10.1101/lm.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Oliveira AM, Estevez MA, Hawk JD, Grimes S, Brindle PK, Abel T. Subregion-specific p300 conditional knock-out mice exhibit long-term memory impairments. Learn Mem. 2011;18:161–169. doi: 10.1101/lm.1939811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Maurice T, Duclot F, Meunier J, et al. Altered memory capacities and response to stress in p300/CBP-associated factor (PCAF) histone acetylase knockout mice. Neuropsychopharmacology. 2008;33:1584–1602. doi: 10.1038/sj.npp.1301551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bartholdi D, Roelfsema JH, Papadia F, et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: delineation of the phenotype of the first patients carrying mutations in EP300. J Med Genet. 2007;44:327–333. doi: 10.1136/jmg.2006.046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Roelfsema JH, White SJ, Ariyurek Y, et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet. 2005;76:572–580. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Viosca J, Lopez-Atalaya JP, Olivares R, Eckner R, Barco A. Syndromic features and mild cognitive impairment in mice with genetic reduction on p300 activity: Differential contribution of p300 and CBP to Rubinstein-Taybi syndrome etiology. Neurobiol Dis. 2010;37:186–194. doi: 10.1016/j.nbd.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 212.Hallam TM, Bourtchouladze R. Rubinstein-Taybi syndrome: molecular findings and therapeutic approaches to improve cognitive dysfunction. Cell Mol Life Sci. 2006;63:1725–1735. doi: 10.1007/s00018-005-5555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.McQuown SC, Wood MA. HDAC3 and the molecular brake pad hypothesis. Neurobiol Learn Mem. 2011;96:27–34. doi: 10.1016/j.nlm.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Graff J, Rei D, Guan JS, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Vecsey CG, Hawk JD, Lattal KM, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Dancy BM, Crump NT, Peterson DJ, et al. Live-cell studies of p300/CBP histone acetyltransferase activity and inhibition. Chembiochem. 2012;13:2113–2121. doi: 10.1002/cbic.201200381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hazzalin CA, Mahadevan LC. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol. 2005;3:e393. doi: 10.1371/journal.pbio.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 219.Wei W, Coelho CM, Li X, et al. p300/CBP-associated factor selectively regulates the extinction of conditioned fear. J Neurosci. 2012;32:11930–11941. doi: 10.1523/JNEUROSCI.0178-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14:268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lattal KM, Barrett RM, Wood MA. Systemic or intrahippocampal delivery of histone deacetylase inhibitors facilitates fear extinction. Behav Neurosci. 2007;121:1125–1131. doi: 10.1037/0735-7044.121.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Stafford JM, Raybuck JD, Ryabinin AE, Lattal KM. Increasing histone acetylation in the hippocampus-infralimbic network enhances fear extinction. Biol Psychiatry. 2012;72:25–33. doi: 10.1016/j.biopsych.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Benes FM. Searching for unique endophenotypes for schizophrenia and bipolar disorder within neural circuits and their molecular regulatory mechanisms. Schizophr Bull. 2007;33:932–936. doi: 10.1093/schbul/sbm064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sharma RP, Grayson DR, Gavin DP. Histone deactylase 1 expression is increased in the prefrontal cortex of schizophrenia subjects: analysis of the National Brain Databank microarray collection. Schizophr Res. 2008;98:111–117. doi: 10.1016/j.schres.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ourednik J, Ourednik V, Lynch WP, Schachner M, Snyder EY. Neural stem cells display an inherent mechanism for rescuing dysfunctional neurons. Nat Biotechnol. 2002;20:1103–1110. doi: 10.1038/nbt750. [DOI] [PubMed] [Google Scholar]
- 226.Teng YD, Lavik EB, Qu X, et al. Functional recovery following traumatic spinal cord injury mediated by a unique polymer scaffold seeded with neural stem cells. Proc Natl Acad Sci U S A. 2002;99:3024–3029. doi: 10.1073/pnas.052678899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Clement AM, Nguyen MD, Roberts EA, et al. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302:113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- 228.Carletti B, Piemonte F, Rossi F. Neuroprotection: the emerging concept of restorative neural stem cell biology for the treatment of neurodegenerative diseases. Curr Neuropharmacol. 2011;9:313–317. doi: 10.2174/157015911795596603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Rossi F, Cattaneo E. Opinion: neural stem cell therapy for neurological diseases: dreams and reality. Nature reviews Neurosci. 2002;3:401–409. doi: 10.1038/nrn809. [DOI] [PubMed] [Google Scholar]
- 230.Faure A, Verret L, Bozon B, et al. Impaired neurogenesis, neuronal loss, and brain functional deficits in the APPxPS1-Ki mouse model of Alzheimer's disease. Neurobiol Aging. 2011;32:407–418. doi: 10.1016/j.neurobiolaging.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 231.Verret L, Jankowsky JL, Xu GM, Borchelt DR, Rampon C. Alzheimer's-type amyloidosis in transgenic mice impairs survival of newborn neurons derived from adult hippocampal neurogenesis. J Neurosci. 2007;27:6771–6780. doi: 10.1523/JNEUROSCI.5564-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Branchi I, Karpova NN, D'Andrea I, Castren E, Alleva E. Epigenetic modifications induced by early enrichment are associated with changes in timing of induction of BDNF expression. Neurosci Lett. 2011;495:168–172. doi: 10.1016/j.neulet.2011.03.038. [DOI] [PubMed] [Google Scholar]
- 233.Lopez-Atalaya JP, Ciccarelli A, Viosca J, et al. CBP is required for environmental enrichment-induced neurogenesis and cognitive enhancement. EMBO J. 2011;30:4287–4298. doi: 10.1038/emboj.2011.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Nithianantharajah J, Hannan AJ. Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat Rev Neurosci. 2006;7:697–709. doi: 10.1038/nrn1970. [DOI] [PubMed] [Google Scholar]
- 235.O'Callaghan RM, Griffin EW, Kelly AM. Long-term treadmill exposure protects against age-related neurodegenerative change in the rat hippocampus. Hippocampus. 2009;19:1019–1029. doi: 10.1002/hipo.20591. [DOI] [PubMed] [Google Scholar]
- 236.Rossi C, Angelucci A, Costantin L, et al. Brain-derived neurotrophic factor (BDNF) is required for the enhancement of hippocampal neurogenesis following environmental enrichment. Eur J Neurosci. 2006;24:1850–1856. doi: 10.1111/j.1460-9568.2006.05059.x. [DOI] [PubMed] [Google Scholar]
- 237.Simpson J, Kelly JP. The impact of environmental enrichment in laboratory rats--behavioural and neurochemical aspects. Behav Brain Res 2011;222:246–264. [DOI] [PubMed]
- 238.Nithianantharajah J, Hannan AJ. Mechanisms mediating brain and cognitive reserve: experience-dependent neuroprotection and functional compensation in animal models of neurodegenerative diseases. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:331–339. doi: 10.1016/j.pnpbp.2010.10.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1224 kb)