Abstract
Antimicrobial peptides/proteins (AMPs) are a group of immune proteins that exhibit strong antibiotic properties against numerous infectious bacterial strains. They are evolutionarily conserved and present in every kingdom and phylum, ranging from prokaryotes to humans. We analyzed the transciptome from the larvae of Asian corn borer, Ostrinia furnacalis (Guenée), and identified several putative AMP transcripts, OfgLys5, OfgLys6, OfgLys10, OfgAtt, and OfgIID. OfgLys5, OfgLys6, and OfgLys10 are all highly homologous with c-type lysozymes, and OfgAtt shows significant identities with Lepidoptera attacin. The amino acid sequence of OfgLys5 and OfgLys6 possessed all conserved features critical for fundamental structure and function of c-type lysozyme, including the two catalytic sites, Glu32 and Asp50. OfgAtt is a typical glycine-rich protein. The antimicrobial activity of O. furnacalis hemolymph increased significantly after injection with Escherichia coli, Micrococcus luteus, or Beauveria bassiana. OfgAtt, IDD, and Lys6 are expressed at low level prior to the challenge, but strongly induced against Gram-positive and negative bacteria, and fungi. Under the same inducement conditions, the transcripts of these three genes elevated most when fifth instar larvae were injected. Therefore, O. furnacalis larvae are induced to produce antimicrobial materials in the hemolymph after the infection, and increase of lysozyme and attacin may contribute to the antimicrobial activity.
Keywords: Ostrinia furnacalis Guenée, antimicrobial peptide, Lysozyme, Attacin
Introduction
Insects lack an adaptive immune system, and have to rely solely on innate immune system to defend against microbial infections 1. Innate immunity of insects is divided into two major reaction types: cellular immunity and humoral immunity 1-3. Humoral immune system relies mainly on antimicrobial peptides (AMPs), which are directly active against the invasive microorganisms. Insect AMPs are synthesized mainly in the fat body (equivalent of liver in mammals) and secreted into the hemolymph to combat a variety of pathogenic microorganisms 4.
Since the first AMP was purified from the hemolymph of Hyalophora cecropia in 1980 5, a large number of AMPs have been identified from every kingdom and phylum, ranging from prokaryotes to humans. These molecules share common features such as relatively low molecular weight, positive charge, high heat stability, good water solubility, and broad-spectrum antimicrobial activity 2,6,7. Based on their amino acid composition and antimicrobial activities, AMPs are generally classified into five groups: cecropin, insect defensins, lysozymes, proline-rich proteins, and glycine-rich proteins such as attacins 8,9. Thereinto, lysozyme (EC 3.2.1.17) function as an antibacterial protein by catalyzing the hydrolysis of β-1,4-glycosidic bond between N-acetylglucosamine and N-acetylmuramic acids of peptidoglycans in bacterial cell walls 10. Several types of lysozymes have been described, i.e., the c (chicken), g (goose), phage, bacteria, plant, and i (invertebrate) types 11. C- and i-type lysozymes have been reported in several insect orders, including Diptera, Lepidoptera, Orthoptera, Isoptera, and Hemiptera 12-14. For example, thirteen, eight, and six c-type lysozyme genes are identified in the genome of Drosophila melanogaster 15, Anopheles gambiae 16, and Aedes agypti 17, respectively. Five of eighteen lysozyme genes in D. melanogaster encode i-type lysozymes 15. cDNA of over fifteen c-type lyszoyme have been sequenced from Bombyx mori 18, Galleria mellonella 19, Hyphantria cunea 20, Manduca sexta 14, Ostrinia furnacalis 21, Trichoplusia ni 22 etc.
Among the above five major groups of AMPs, attacins are rich in glycine residues. They are thought to increase the permeability of the outer membrane of Gram-negative bacteria by interacting with lipopolysaccharides and inhibit the synthesis of outer membrane protein at the transcriptional level 23. Families of Attacin-like peptides have been described in the lepidopteron species B. mori 24, H. cunea 25, H.ni 22, M. sexta 26, and Spodoptera exigua 27, as well as in the dipteran species D. melanogaster 28, Glossina morsitans 23 etc. Mature attacin peptides are typically ~ 190 amino acids in length and adopt a “random coil” structure in solution 23,29. This loose, flexible structure allows relatively free amino acid substitutions, explaining the low level of amino acid identity among the attacin homologs in distant taxa 23.
Another common characteristic for AMPs is the high inducibility by injury and microorganism infection. The signaling mechanisms that elicit expression of AMPs after inducement are best understood in D. melanogaster. There are at least two major signal transduction pathways, Imd and Toll, which regulate the induced expression of AMP genes 30,31. The Imd pathway mainly regulates the response to Gram-negative bacterial infection and some Gram-positive bacterial infections, and the Toll pathway accounts primarily for the response to infections by fungi and other Gram-positive bacteria 32,33. Upon infection, the signaling pathways are rapidly activated to induce the synthesis of large amount of AMPs, finally resulting in strong antimicrobial activities of hemolymph to kill the invasive microorganisms. For example, in M. sexta, the transcriptional level of AMP increased around 200-fold after the injection of Micrococcus luteus 34.
Lepidopteran insect Asian corn borer, Ostrinia furnacalis (Guenée), is an important insect pest in Asia and causes serious damage on corn, sorghum, millet and cotton 35. Deep understanding of the innate immunity, especially AMP production, in O. furnacalis can afford basis for biologically controlling this insect pest. Here we report the identification of AMP transcripts from the transcriptome of O. furnacalis, and the expression profile analysis of these genes. We identified five putative full-lengthed AMPs, OfgLys5, OfgLys6, OfgLys10, OfgAtt, and OfgIID, and discovered that antimicrobial activity of O. furnacalis hemolymph significantly increased upon microbial challenge, and described that OfgLys6, OfgAtt, and OfgIID were greatly induced after infection, especially in fifth instar larvae.
Material and Methods
Insects rearing
Asian corn borer (O. furnacalis (Guenée)) was kindly gifted by Dr. Kanglai He from the Institute of Plant Protection, Chinese Academy of Agricultural Sciences. O. furnacalis larvae were reared on an artificial diet at 28 ℃ under a relative humidity of 70-90% and a photoperiod of 16 h light and 8 h darkness 36. Insects at different larval stages were collected for further experiments.
Identification of AMP transcripts in O. furnacalis transcriptome
The transcriptome from the fifth larvae of O. furnacalis was sequenced by Illumina technology at the Beijing Isntitute of Genomics (data not published). To find AMP genes, a local BLASTN search was performed using sequences of known AMP from D. melanogaster and M. sexta as queries (http://cegg.unige.ch/insecta/immunodb/) 14. The obtained sequencing components with the P-value lower than 0.01 were retrieved and assembled into contigs in CAP3 (http://pbil.univ-lyon1.fr/cap3.php). The resulting similar sequences were examined manually.
Sequence analysis
The deduced amino acid sequences of potential O. furnacalis AMPs were aligned using CLUSTAL W. Phylogenetic trees were constructed by the neighbor-joining method with a Poisson correction model, using MEGA version 4.0 37. For the neighbor-joining method, gaps were treated as characters, and statistical analysis was performed by the bootstrap method, using 1000 repetitions.
Induction of antimicrobial activity of O. furnacalis hemolymph
To determine the optimized inducement conditions for the production of AMP in O. furnacalis larvae and further study the expression profiles of AMP transcripts under these conditions, fifth instar day 0 larvae were injected with formalin-killed Escherichia coli DH5α, Micrococcus luteus, Beauveria bassiana, or sterile PBS solution as a control (3 μl/larva).
In particular, for E. coli injection, E. coli DH5α cultured freshly was treated with formalin and diluted with 0.85% saline into three concentration series: 2×105, 1×106, and 2×106 cells/μl. Fifth instar day 0 larvae from the same batch were divided into four groups, and injected with different concentration of E. coli and sterile PBS, respectively. At 20 h after injection, hemolymph was collected as described previously 35, and 3 larvae from each group were collected and stored at -80ºC for further RNA extraction as the following descriptions. For M. luteus treatment, dried M. luteus powder was dissolved in sterile 0.85% saline and diluted into three concentration series: 3, 15, and 30 μg/μl. The O. furnacalis larvae were injected with differently diluted M. luteus, and samples were collected as described for E. coli injection. For the induction by B. bassiana, B. bassiana was firstly inoculated on Potato Dextrose Agar plates and incubated at 26°C for 7-10 days. The produced conidia were then scraped and diluted with sterile water containing 0.1% Tween-80 into three concentration series: 1×104, 1×105, and 2×105 conidia/μl. The O. furnacalis larvae were injected with differently diluted B. bassiana. After 10 h, sampled were collected for further experiments. For each treatment, totally three independent biological replicates were encompassed.
Antimicrobial activity assay
For antimicrobial activity assay, hemolymph from around 150 larvae in each group in each treatment were pooled and boiled for 10 min to remove most high molecular weight proteins and then centrifuged at 12000 rpm for 10 min. The supernatant was used for the antimicrobial activity assay with agar well diffusion method described by Hultimark using E. coli DH5α 38. For each heat-treated hemolymph sample, duplicate 50-μl aliquots were assayed. The antimicrobial activity (U) is defined as the square of the difference between the radius of inhibition zone and the radius of loading well.
Reverse transcriptase (RT)-PCR analysis of AMP mRNA levels under different inducement conditions
To check the expression profiles of O. furnacalis AMP under different inducement conditions mentioned above, total RNA samples were prepared using TRizol Reagent (TIANGEN) from individual larva. Equal amount of total mRNA samples from 3 larvae were combined for the cDNA synthesis. First-strand cDNA was synthesized from an oligo(dT) primer following the instructions for QuantScriptRT Kit (TIANGEN). One μl of 1:5 diluted cDNA was used as template for RT-PCR experiments. O. furnacalis ribosomal protein L8 (rpL8) cDNA was used as an internal standard to adjust the template amounts in a preliminary PCR experiments. The primers for amplifying OfgLys6, OfgAtt, OfgIID, and rpL8 were: Lys6-RTf (5′- AATTAGCAGTCGTTGTAG-3′) and Lys6-RTr (5′-GAATAGTCCGTAGTCCTT-3′), Att-RTf (5′-CTGGATTACATGTTCAAGAA-3′) and Att-RTr (5′-TCTTTAACATAGAATTAGCG-3′), IID-RTf (5′-TGCTGACTTGAACGACGATA-3′) and IID-RTr (5′-GCTTAGGTATGGTTTCCACTTAG-3′), rpL8-RTf (5′-AAGCGAGGAACATCAGCC-3′) and rpL8-RTr (5′-GGTCTTGCCACCACGAAT-3′). The thermal cycling conditions were 94 ºC for 3min, then 25 cycles of 94 ºC for 30 s, 50 ºC for 30 s and 72 ºC for 1min followed by incubation at 72 ºC for 5 min. The PCR products were separated by electrophoresis on a 1.5% agarose gel.
Moreover, to investigate the expression difference of O. furnacalis AMP in different larval stages under the same inducement conditions, third-, fourth-, fifth-instar day 0 larvae were injected with 2×106 cells/μl of E. coli DH5α, or 30 μg/μl of M. luteus, or 2×105 conidia/μl of B. bassiana, respectively. Sterile PBS injection was used as a control (3μl/larva). Larvae were collected and RT-PCR analysis was performed as described above.
Results and Discussion
Identification of AMP transcripts in O. furnacalis transcriptome
A local BLASTN search of O. furnacalis transcriptome database identified 5 transcripts with deduced amino acid sequences similar to those of other insect AMPs. They include 3 lysozymes, 1 attacin, and 1 potential AMP with the high identity (67%) to M. sexta immune-induced protein-1 (IID-1)26. So far, little is known about IID except it was reported that it was highly immune induced 26 and putatively defensive against the pathogens 39. Based on the amino acid sequence similarities, these five transcripts were named as OfgLys5, Ofglys6, OfgLys10, OfgAtt, and OfgIID. Their corresponding nucleotide acid sequences have been submitted to GenBank with accession numbers KF146180, KF146181, KF146182, KF146178, and KF146179, respectively. With the exception of OfgLys10, all other AMP sequences were successfully cloned in full length and verified by DNA sequencing. Therefore, along with another lysozyme (OstrinLysC, EF12065) reported previously 21, totally 3 lysozyme transcripts have been isolated from O. furnacalis, with the fourth one (OfgLys10) to be cloned.
It is notable that a classical AMP gene, Cecropin, was not identified in this research. Cecropins are cationic α-helical AMPs, They were initially isolated from H. cecropia 5, and later discovered in lepidopteran and dipteran species 7,24. There are two possible reasons to explain why it failed to obtain cecropin homologues by searching O. furnacalis transcriptome. Firstly, Cecropin does exist in O. furnacalis, but was not sequenced with some unknown reasons. Secondly, O. furnacalis larvae utilize other AMPs to replace the roles of cecropins in defending against bacteria.
Sequence analysis of O. furnacalis lysozyme
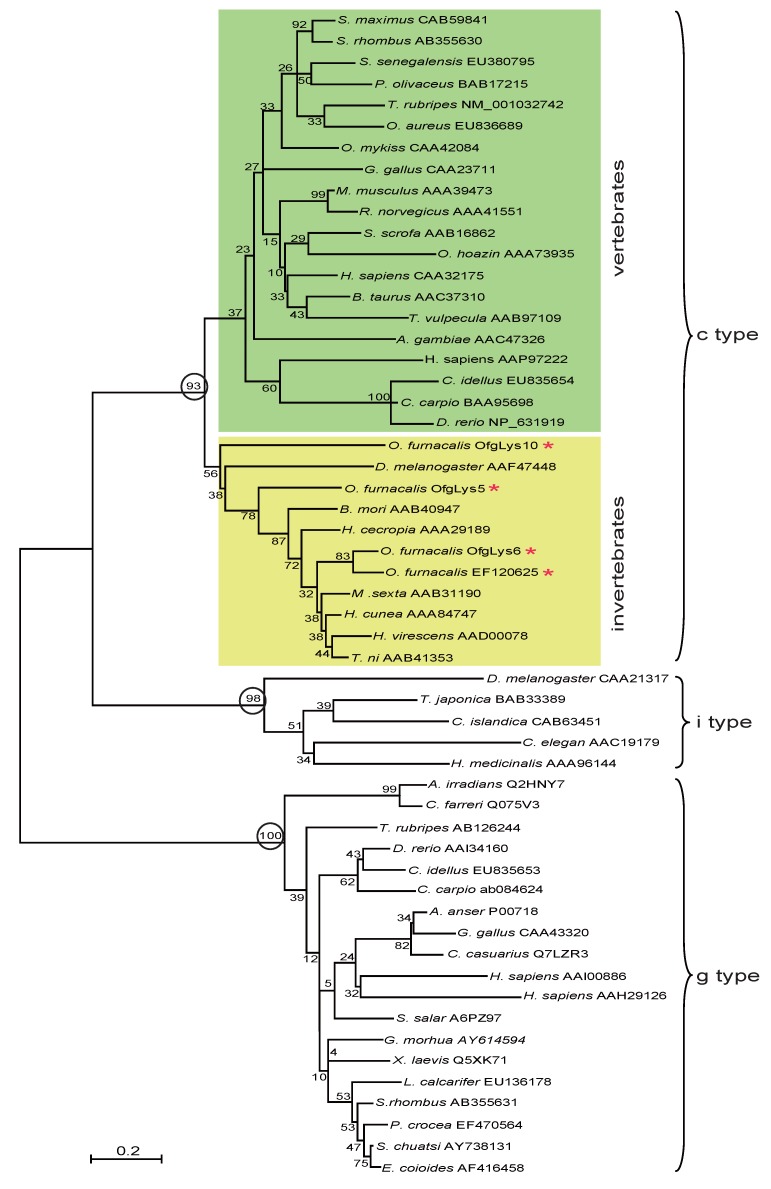
Lysozyme is an important molecule in the innate immune system, and widely distributed in the different organs or secretions of vertebrates, invertebrates, phages, fungi, bacteria, and plants. To investigate the relationship between O. furnacalis lysozymes and others, a phylogenetic tree was constructed based on the amino acid sequences of selected various types of lysozyme using the neighbor-joining method. As Fig.1 shows, the c-, g- and i-type lysozymes are each clustered together correctly. Among c-type cluster, the invertebrate and vertebrate formed a subgroup respectively. All O. furnacalis lysozyme involves in the invertebrate c-type lysozyme subgroup.
Figure 1.
Phylogenetic analysis of deduced amino acid sequences for various c-, g-, and i-type lysozymes. The tree was derived from sequence alignment using ClustalW. Numbers at the nodes indicates the bootstrap confidence values of 1000 replicates. The scale bar indicates the number of substitutions per site. The names of lysozyme genes used in the alignment and phylogenetic tree were shown as scientific name of species followed by GenBank accession number of this specific gene.
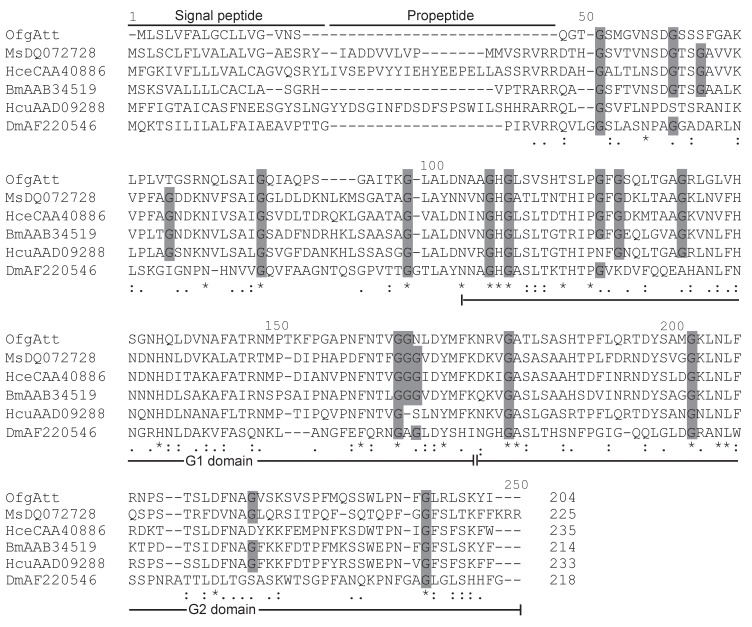
The amino acid sequences of mature O. furnacalis lysozymes excluding the signal peptide were aligned with c-type lysozymes, including six lepidopteran lysozymes, as well as lysozymes from the fruit fly, fish, chicken, mouse, pig, cattle, and human. Considering OfgLys10 was a truncated form, it was omitted from this alignment. As shown in Fig.2, both OfgLys5 and OfgLys6 possess all conserved residues which are fundamental for the three dimensional structure and the biological activity of the c-type lysozyme: 8 cysteine residues (Cys6, Cys27, Cys62, Cys72, Cys76, Cys90, Cys110, Cys120) and two catalytic sites of glutamic acid (Glu32) and aspartic acid (Asp50). Based on the structures determined in silkworm and chicken, there are two loops in the secondary structure of lysozymes (Fig.2). The size of loop-1 is identical in lepodopteran lysozymes and fruit fly lysozyme (4 residues) while it is longer in fish, chicken, mouse, pig, cattle, and human lysozymes (7 residues). The size of loop-2 is conserved among lepidopteran lysozymes (7 residues) while it is longer in other lysozymes, 8 residues in fish and 9 ones in chicken, mouse, pig, cattle, human, and even in fruit fly.
Figure 2.
Sequence alignment of O. furnacalis lysozymes with fruit fly, fish, chicken, mouse, pig, cattle, human and lepodipteran lysozymes. Gaps are indicated by “-”. Completely conserved amino acids are indicated by “*”, and conservative substitutions by “:” and “.” below the sequences. Eight absolutely conserved cysteines are shaded in light grey, and two catalytic sites are shaded in dark grey. Residues forming secondary structures of lysozymes in B. mori and chicken are presented in boxes. Two loops in the secondary structures are underlined.
Sequence analysis of O. furnacalis attacin
Attacins are glycine-rich immune proteins, and are synthesized as pre/pro-mature peptides. After removing the signal peptide, the resulting propeptides of attacins are further processed to be mature peptides by cleaving at the site Arg Ala/Val Arg Arg which is very similar to the potential furin processing site (RARR) 23. In this study, we isolated a new attacin transcript (OfgAtt), which is the most highly similar to M. sexta att-2 with the identity of 51% in putative amino acid sequences. We also performed alignment to compare the putative attacin protein sequences from corn borer with other related insects (Fig.3). Interestingly, O. furnacalis attacin lacks the propeptide. It seems that OfgAtt becomes mature immediately after cleaving the signal peptide at Ser17, without the necessary of further removing the propetide. In addition, it is known from the alignment that O. furnacalis attacin sequence conforms to the G-module structure and contains two G domains, namely a G1 domain from Asn68 to Lys136, and G2 domain from Asn137 to Arg204. The conservation of the G domains implies that they may correspond to functional units. Besides, the recurrent glycine residues seem highly conserved in aligned attacin protein sequences, consisting with the characteristic that attacins are glycine-rich proteins. However, among 18 conserved glycine residues, only half are present in all aligned insects, while the amino acid residues at the other 9 sites exhibit more or less varieties (Fig.3).
Figure 3.
Alignment between the deduced amino acid sequences of attacin from O. furnacalis (OfKF146178), M. sexta (MsDQ072728), H. secropia (HceCAA40886) B. mori (BmAAB34519), H. cunea (HcuAAD09288), and D. melanogaster (DmAF220546). Gaps are indicated by “-”. Completely conserved amino acids are indicated by “*”, and conservative substitutions by “:” and “.” below the sequences. Amino acid residues that have been conserved in all or most of analyzed species are shaded in dark grey. Signal peptide, propeptide, and G1 and G2 domain of mature peptide are shown.
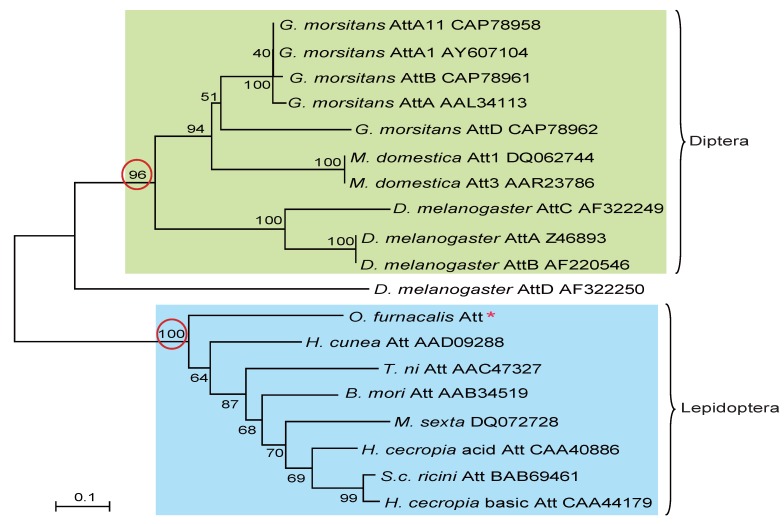
To understand the evolutionary relationships among attacins, phylogenetic comparative analysis was conducted based on the deduced amino acid sequences of attacin. As shown in Fig.4, attacins in lepidopteron and dipteran species formed two branches with high bootstrap value, 100 and 96 respectively. Even in the same clade, attacins analyzed from G. morsitans, M. domestica, D. melanogaster represent species-specific expansions of each gene family and their phylogenetic placement mirrors that of their host relationships. As expected, O. furnacalis attacin is included in the branch containing all analyzed lepidopteran attacins (Fig.4).
Figure 4.
Phylogenetic relationship of attacin among corn borer and other species. The phylogenetic tree was estimated by the neighbor-joining method in MEGA version 4.0 based on sequence alignment using ClustalW. Numbers at the nodes represent bootstrap proportions on 1000 replicates. The scale bar indicates the number of substitutions per site. The circled bootstrap value indicates that analyzed attacins forms into two separate branches. The names of attacins in the tree were shown as scientific name of species followed by GenBank accession number of this specific gene.
Increase of antimicrobial activity of hemolymph upon bacterial and fungal infection in O. furnacalis
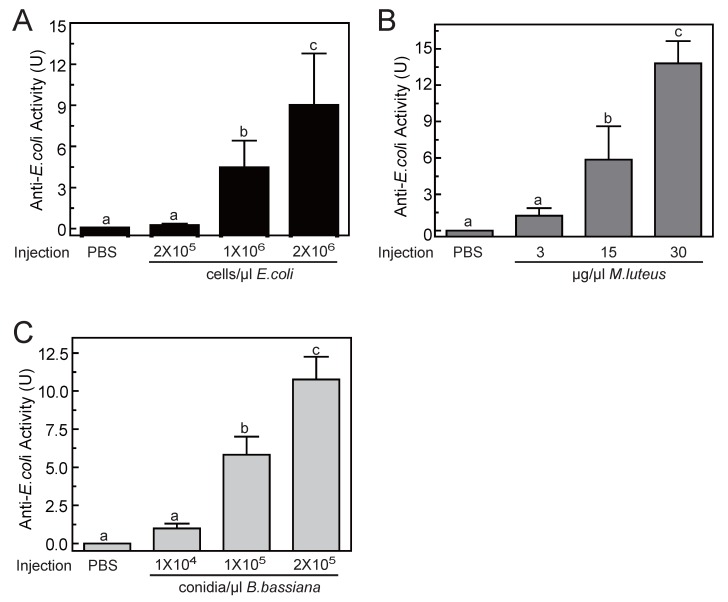
To determine the optimal inducement conditions for the AMP production in O. furnacalis larvae, we injected different concentrations of E. coli (Gram-negative bacteria), M. luteus (Gram-positive bacteria), and B. bassiana (fungi) into fifth instar larvae, and checked the antimicrobial activities of hemolymph using agar well diffusion method. As shown in Fig.5, among three selected concentrations, only the lowest injected concentration (2×105 cells/μl for E. coli, or 3 μg/μl for M. luteus, or 1×104 conidia/μl for B. bassiana) failed to induce detectable antimicrobial activity. The other two injections with higher dose both resulted in significant elevation in activity. This increase was even greater in larvae that were injected with most microbes. The antimicrobial activity of hemolymph increased up to 8.96±3.84, 13.80±1.84, 10.77±1.49 units, respectively, when the larvae was injected by 2×106 cells/μl of E. coli, or 30 μg/μl of M. luteus, or 2×105 conidia/μl of B. bassiana (Fig. 5). It suggested that bacterial or fungi infections both could induce the production of AMPs in O. furnacalis larvae, which may conferred the antimicrobial activity on the hemolymph to eliminate the invasive microbes. Normally, the larger amount of microorganisms, the more AMP synthesis they may induce. Taking induction effects and convenient operability together, the injection of M. luteus at the concentration of 30μg/μl might be a good choice for AMP production in future experiments.
Figure 5.
Antimicrobial activity assay of O. furnacalis hemolymph after the inducement by E. coli (A), M. luteus (B), and B. bassiana (C). The bars represent mean ± S.D. (n = 3). Bars labeled with different letters (a, b, and c) are significantly different (one-way ANOVA, followed by the Newman-Keuls test, P <0.05).
Expression profiles of OfgLys6, OfgAtt, and OfgIID
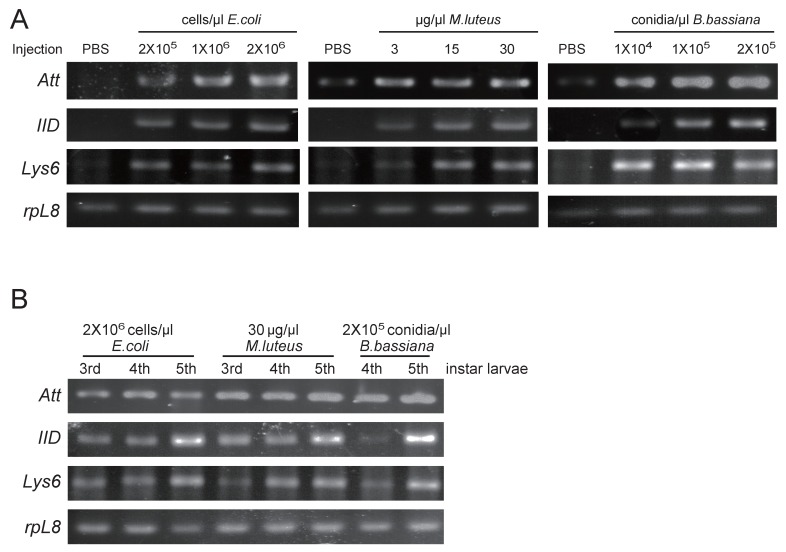
Among three identified O. furnacalis lysozymes, OfgLys10 is partial, and OfgLys6 is more similar to other known lysozymes. Therefore, we only focused on OfgLys6 in the expression profile assay. Along with the other two identified transcripts, we used RT-PCR methods to analyze the transcriptional expression of OfgLys6, OfgAtt, and OfgIID after immune challenge, as well as in different larval stages (Fig. 6). OfgLys6, OfgAtt, and OfgIID were all expressed at trace level in the control group with the injection of sterile PBS. When larvae were injected with E. coli, M. luteus, and B. bassiana, transcript abundance of these three genes increased strongly (Fig.6A). Generally speaking, the injection at the highest concentration led to the strongest induction of OfgLys6, OfgAtt, and OfgIID. However, the transcript level of OfgLys6 remained nearly unchanged in larvae that were challenged by different concentration of E. coli. Similar case occurred for OfyLys6 responding to the injection of B. bassiana.
Figure 6.
RT-PCR analysis of transcriptional levels of OfgAtt, IID, Lys6 in O. furnacalis larvae induced by different microbes (A) and in different stages (B). Ribosomal protein L8 (rpL8) was used as an internal standard to indicate a consistent total mRNA amount. The injection dose of E. coli, M. luteus, and B. bassiana, and larval stage are shown above each figure. More details about the treatments were seen under “Material and Methods”.
We also detected the transcript abundance of OfgLys6, OfgAtt, and OfgIID in the larvae in different stages of development. Considering it was difficult to detect the expression level of OfgLys6, OfgAtt, and OfgIID in the larvae without any immune inducement, we compared their transcript level in third-, fourth-, and fifth-instar larvae with immune injection instead of in the un-challenged larvae. As Fig.6B shows, after the injection of E.coli, M.luteus, or B. bassiana, transcripts of OfgLys6, OfgAtt, and OfgIID were detected in all injected larvae. This is consistent with the result in Fig.6A. Moreover, no obvious difference was observed for the transcript abundance of these three AMPs in third-instar larvae and in fourth-instar larvae. The transcripts of OfgLys6, OfgAtt, and OfgIID reached to the highest level in fifth instar stage. Taken together, such results suggested that expression of the OfgLys6, OfgAtt, and OfgIID is inducible in O. furnacalis larvae, and they are induced to the largest extent in fifth instar larvae.
Conclusions
Production of AMP is an important immune response against the infection of invading microorganisms because AMPs have broad-spectrum antimicrobial activities. Studies on identification and functioning mechanism of AMP have been extensively investigated in invertebrates and vertebrates. Here we identified 5 AMP transcripts from O. furnacalis in which OfgLys5 and OfgLys6 belong to c-type lysozymes, and OfgAtt is lepidopteran attacin rich in glycine residues. We also discovered that antimicrobial activity of O. furnacalis hemolymph increased significantly and expression levels of OfgLys6, OfgAtt, and OfgIID induced highly after the injection with bacteria or fungi. We hope that this work could provide the basis for further studying the induction mechanism of AMP in important agriculture pest, O. furnacalis, and even in other insects and higher animals. The acquired knowledge might contribute to biological control of this serious insect pest.
Acknowledgments
We thank Dr. Le Kang in Institute of Zoology, Chinese Academy for generously providing Micrococcus luteus strains. This work was supported by National Natural Science Foundation of China (31172090), Program for New Century Excellent Talents in University (NCET-11-0476), and the 973 program (2013CB127603).
References
- 1.Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunological reviews. 2004;198:97. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- 2.Iwanaga S, Lee BL. Recent advances in the innate immunity of invertebrate animals. J Biochem Mol Biol. 2005;38:128–150. doi: 10.5483/bmbrep.2005.38.2.128. [DOI] [PubMed] [Google Scholar]
- 3.Zou Z, Souza-Neto JA, Xi Z, Shin SW, Alvarez KS, Kokoza K, Dimopoulos G, Raikhel AS. Genome-wide transcriptome analysis of immunity related genes in the mosquito, Aedes aegypti. PLoS Pathogens. 2011;7:e1002394. doi: 10.1371/journal.ppat.1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nature immunology. 2002;3:121. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- 5.Hultmark D, Steiner H, Rasmuson T, Boman HG. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur J Biochem. 1980;106:7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x. [DOI] [PubMed] [Google Scholar]
- 6.Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 7.Bulet P, Stocklin R. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept Lett. 2005;12:3–11. doi: 10.2174/0929866053406011. [DOI] [PubMed] [Google Scholar]
- 8.Andreu D, Rivas L. Animal antimicrobial peptides: an overview. Biopolymers. 1998;47:415–433. doi: 10.1002/(SICI)1097-0282(1998)47:6<415::AID-BIP2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 9.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria. Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 10.Bachali S, Jager M, Hassanin A, Schoentgen F, Jolles P. et al. Phylogenetic analysis of invertebrate lysozymes and the evolution of lysozyme function. J Mol Evol. 2002;54:652–664. doi: 10.1007/s00239-001-0061-6. [DOI] [PubMed] [Google Scholar]
- 11.Jimenez-Cantizano RM, Infante C, Martin-Antonio B, Ponce M, Hachero I. et al. Molecular characterization, phylogeny, and expression of c-type and g-type lysozymes in brill (Scophthalmus rhombus) Fish Shellfish Immunol. 2008;25:57–65. doi: 10.1016/j.fsi.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Araujo CA, Waniek PJ, Stock P, Mayer C, Jansen AM. et al. Sequence characterization and expression patterns of defensin and lysozyme encoding genes from the gut of the reduviid bug Triatoma brasiliensis. Insect Biochem Mol Biol. 2006;36:547–560. doi: 10.1016/j.ibmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Fujita A, Minamoto T, Shimizu I, Abe T. Molecular cloning of lysozyme-encoding cDNAs expressed in the salivary gland of a wood-feeding termite, Reticulitermes speratus. Insect Biochem Mol Biol. 2002;32:1615–1624. doi: 10.1016/s0965-1748(02)00100-5. [DOI] [PubMed] [Google Scholar]
- 14.Gorman MJ, Kankanala P, Kanost MR. Bacterial challenge stimulates innate immune responses in extra-embryonic tissues of tobacco hornworm eggs. Insect Mol Biol. 2004;13:19–24. doi: 10.1111/j.1365-2583.2004.00454.x. [DOI] [PubMed] [Google Scholar]
- 15.Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS. et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Calvo E, Marinotti O, James AA, Paskewitz SM. Characterization of the c-type lysozyme gene family in Anopheles gambiae. Gene. 2005;360:131–139. doi: 10.1016/j.gene.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Ursic Bedoya RJ, Mitzey AM, Obraztsova M, Lowenberger C. Molecular cloning and transcriptional activation of lysozyme-encoding cDNAs in the mosquito Aedes aegypti. Insect Mol Biol. 2005;14:89–94. doi: 10.1111/j.1365-2583.2004.00534.x. [DOI] [PubMed] [Google Scholar]
- 18.Lee WJ, Brey PT. Isolation and characterization of the lysozyme-encoding gene from the silkworm Bombyx mori. Gene. 1995;161:199–203. doi: 10.1016/0378-1119(95)00199-g. [DOI] [PubMed] [Google Scholar]
- 19.Zdybicka-Barabas A, Staczek S, Mak P, Skrzypiec K, Mendyk E. et al. Synergistic action of Galleria mellonella apolipophorin III and lysozyme against Gram-negative bacteria. Biochim Biophys Acta. 2013;1828:1449–1456. doi: 10.1016/j.bbamem.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Park HY, Park SS, Shin SW, Park DS, Kim MG. et al. Protein purification and nucleotide sequence of a lysozyme from the bacteria-induced larvae of the fall webworm, Hyphantria cunea. Arch Insect Biochem Physiol. 1997;35:335–345. doi: 10.1002/(SICI)1520-6327(199705)35:3<335::AID-ARCH7>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 21.Wang WX, Wang YP, Deng XJ, Dang XL, Tian JH. et al. Molecular and functional characterization of a c-type lysozyme from the Asian corn borer, Ostrinia furnacalis. J Insect Sci. 2009;9:17. doi: 10.1673/031.009.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang D, Liu G, Gunne H, Steiner H. PCR differential display of immune gene expression in Trichoplusia ni. Insect Biochem Mol Biol. 1996;26:177–184. doi: 10.1016/0965-1748(95)00080-1. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Hu C, Wu Y, Stuart A, Amemiya C. et al. Characterization of the antimicrobial peptide attacin loci from Glossina morsitans. Insect Mol Biol. 2008;17:293–302. doi: 10.1111/j.1365-2583.2008.00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng T, Zhao P, Liu C, Xu P, Gao Z. et al. Structures, regulatory regions, and inductive expression patterns of antimicrobial peptide genes in the silkworm Bombyx mori. Genomics. 2006;87:356–365. doi: 10.1016/j.ygeno.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 25.Kwon YM, Kim HJ, Kim YI, Kang YJ, Lee IH. et al. Comparative analysis of two attacin genes from Hyphantria cunea. Comp Biochem Physiol B Biochem Mol Biol. 2008;151:213–220. doi: 10.1016/j.cbpb.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y, Johnson TJ, Myers AA, Kanost MR. Identification by subtractive suppression hybridization of bacteria-induced genes expressed in Manduca sexta fat body. Insect Biochem Mol Biol. 2003;33:541–559. doi: 10.1016/s0965-1748(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 27.Bang K, Park S, Yoo JY, Cho S. Characterization and expression of attacin, an antibacterial protein-encoding gene, from the beet armyworm, Spodoptera exigua (Hubner) (Insecta: Lepidoptera: Noctuidae) Mol Biol Rep. 2012;39:5151–5159. doi: 10.1007/s11033-011-1311-3. [DOI] [PubMed] [Google Scholar]
- 28.Hedengren M, Borge K, Hultmark D. Expression and evolution of the Drosophila attacin/diptericin gene family. Biochem Biophys Res Commun. 2000;279:574–581. doi: 10.1006/bbrc.2000.3988. [DOI] [PubMed] [Google Scholar]
- 29.Gunne H, Hellers M, Steiner H. Structure of preproattacin and its processing in insect cells infected with a recombinant baculovirus. Eur J Biochem. 1990;187:699–703. doi: 10.1111/j.1432-1033.1990.tb15356.x. [DOI] [PubMed] [Google Scholar]
- 30.Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 31.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 32.Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H. et al. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci U S A. 2002;99:13705–13710. doi: 10.1073/pnas.212301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanji T, Hu X, Weber AN, Ip YT. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Molecular and cellular biology. 2007;27:4578. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An C, Ishibashi J, Ragan EJ, Jiang H, Kanost MR. Functions of Manduca sexta hemolymph proteinases HP6 and HP8 in two innate immune pathways. J Biol Chem. 2009;284:19716–19726. doi: 10.1074/jbc.M109.007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afidchao M, Musters CJM, Snoo GR. Asian corn borer (ACB) and non-ACB pests in GM corn (Zea mays L.) in the Philippines. Pest Manag. Sci. 2013;69:792–801. doi: 10.1002/ps.3471. [DOI] [PubMed] [Google Scholar]
- 36.Qu M, Liu T, Yang J, Yang Q. The gene, expression pattern and subcellular localization of chitin synthase B from the insect Ostrinia furnacalis. Biochem Biophys Res Commun. 2001;404:302–307. doi: 10.1016/j.bbrc.2010.11.111. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in bioinformatics. 2004;5:150. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 38.Hultmark D, Engstrom A, Bennich H, Kapur R, Boman HG. Insect immunity: isolation and structure of cecropin D and four minor antibacterial components from Cecropia pupae. EurJBiochem. 1982;127:207. doi: 10.1111/j.1432-1033.1982.tb06857.x. [DOI] [PubMed] [Google Scholar]
- 39.Gandhe A, Arunkumar KP, John SH, Nagaraju J. Analysis of bacteria-challenged wild silkmoth, Antheraea mylitta (Lepidoptera) transcriptome reveals potential immune genes. BMC Genomics. 2006;7:184–193. doi: 10.1186/1471-2164-7-184. [DOI] [PMC free article] [PubMed] [Google Scholar]