Abstract
Electron microscopy (EM) has dominated high-resolution cellular imaging for over 50 years thanks to its ability to resolve on a nanometer-scale intracellular structures such as the microtubules of the mitotic spindle. It is advantageous to view the cell of interest prior to processing the sample for EM. Correlative light electron microscopy (CLEM) is a technique that allows one to visualize cells of interest by light microscopy (LM) before being transferred to EM for ultra-structural examination.
Here we describe how CLEM can be applied as an effective tool to study the spindle apparatus of mitotic cells. This approach allows transfected cells of interest, in desirable stages of mitosis, to be followed from LM through to EM. CLEM has often been considered as a technically challenging and laborious technique. In this chapter we provide step-by-step pictorial guides that allow successful CLEM to be achieved. In addition we explain how it is possible to vary the sectioning plane, allowing spindles and microtubules to be analyzed from different angles, and the outputs that can be obtained from these methods when applied to the study of kinetochore fiber (K-fiber) ultrastructure.
Keywords: correlative electron microscopy, kinetochore-fiber, mitosis, microtubule, mitotic spindle
I. Introduction
The mitotic spindle is a complex machine consisted of microtubules, motor proteins, and non-motor proteins which, together, generate the forces needed to separate the sister chromatids between the two daughter cells (Scholey et al., 2003). A better visualization of its ultrastructure is necessary to understand the mechanisms underlying its functions.
LM, and the discovery of the green fluorescent protein (GFP) led to a many important discoveries due to the possibility of tracking protein dynamics in live cells. However, LM has a relatively low resolution which does not allow one to visualize structures smaller than ~200 nm. This diffraction limit has been a major imaging weakness, and EM has been one of the few techniques to overcome it. Another disadvantage of LM is the restricted number of separate wavelength channels which can be used on a single sample without overlap, the rest of the cell remaining unobservable.
EM also possesses its share of drawbacks, other than the tricky and time-consuming nature of sample preparation. Only static samples can be observed, making the analysis of dynamic changes impossible. Also, routine EM does not allow one to easily locate cells of interest, such as cells expressing a fluorescent protein or in a particular stage of the cell cycle. It is possible to overcome these limitations by combining the ease and dynamic nature of LM with the sub-nanometer resolving power of EM in the form of correlative light-electron microscopy (CLEM).
CLEM techniques are useful for studying the mitotic spindle. The complexity of spindle microtubules means that they cannot be viewed individually by LM. Also, mitosis is a very dynamic process; each of its stages lasts less than 30 minutes, so pinpointing the exact stage of the cell cycle for a particular cell is critical before engaging in time-costly EM sample preparation. This is why the ability to observe and select cells of interest using LM prior to EM sample processing is a great advantage; allowing both the stage of mitosis to be chosen carefully, and to ensure that the cell is adequately expressing a fluorescent protein of interest.
Studies using EM to research mitotic spindles have yielded outstanding data, such as the quantification of microtubule polarity by Euteneuer and McIntosh (1981), the study of microtubule spacing, position, displacement and length (McDonald et al., 1992), or the more recent whole-cell reconstruction by electron tomography to study cytoskeletal elements (Hoog et al., 2007).
Here, we describe our own application of CLEM to study the ultrastructure of the mitotic spindle, particularly K-fibers (Booth et al., 2011; Cheeseman et al., 2011). We describe both longitudinal and orthogonal sectioning relative to the spindle axis (Figure 1), which reveal different information about spindle architecture (Figure 1D), and how we can quantify such results. Longitudinal sectioning has allowed us to quantify microtubule cross-linkers between K-fiber microtubules, whereas sample-tilting of orthogonally-sectioned K-fibers allowed the quantification of the number of microtubules forming the fiber. Subsequent analysis of the spacing of these microtubules allows us to measure their density and distribution.
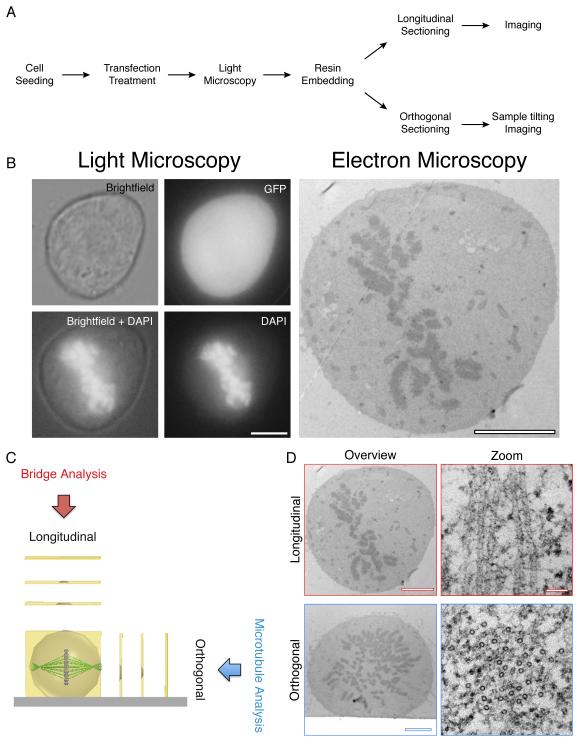
Figure 1. CLEM performed on mitotic cells.
(A) A workflow to achieve CLEM using longitudinal or orthogonal sectioning. (B) A transfected mitotic HeLa observed by LM (Brightfield, GFP and DAPI) and by electron microscopy. Scale bar 5 μm. (C) Schematic of Longitudinal and orthogonal EM sectioning, and examples of output analysis. (D) Representative electron micrographs of cells sectioned longitudinally (above) and orthogonally (below) with high magnification of microtubules (right). Scale bar 4 μm (overview) and 50 nm (zoom).
II. Materials
35 mm glass-bottomed dishes with etched coordinates (MatTek Corporation, P35G- 2-14-C-grid) – referred to here as CLEM dishes
0.1M Phosphate buffer (PB): mix 0.2M Na2HPO4 with 0.2M NaH2PO4 and dilute to 0.1M. Solution should be at pH 7.4.
Fix solution (EM grade fixatives: 3% w/v glutaraldehyde (Agar Scientific R1020), 0.5% w/v paraformaldehyde (Agar Scientific R1026) in 0.05M PB)
Wash solution (0.05M PB, 0.1M sucrose)
DNA stain solution (0.1% w/v Hoechst-33342 in wash solution, or other similar DNA dye)
1% osmium tetroxide (Agar Scientific R1015) in water
0.5% w/v uranyl acetate (Agar Scientific R1260A) in 30% ethanol
Molecular grade 100% ethanol (Sigma-Aldrich 270741-1L)
EPON resin (Agar Scientific R1031, made up using the supplier’s specifications for a ‘medium’ block. Make sure resin mix is fully homogenized, and containing as few bubbles as possible. 200 ml of resin can be made up at a time, aliquoted into small glass vials, and kept frozen at −20°C)
Gelatin capsules (Size 0, Agar Scientific G292-10)
Copper mesh sample grids, coated with formvar (Agar Scientific R1202). We routinely use 200 hexagonal mesh grids (TAAB GG017/C), but 100 mesh or slot grids can also be used. Beware, as the larger the gaps between the copper bars, the more easily the sample will distort and can tear.
High precision tweezers. We prefer self-closing tweezers as they facilitate the handling of sample grids.
5% w/v uranyl acetate (Agar Scientific R1260A) in 50% ethanol
Reynold’s lead citrate solution (see Reynolds, 1963)
III. Methods
A. Cell Transfection and Observation
Cells are seeded into CLEM dishes that contain a coordinate-engraved glass coverslip, providing a pattern to be left in the base of the resin, once embedded. The coordinates are essential for the LM to EM transfer as they allow cells of interest to be tracked throughout the entire CLEM process.
Seeding the appropriate amount of cells into the dishes is important: too many will make locating the cell of interest among many other unwanted cells difficult once the sample is embedded in resin; it will also make reading the coordinates under the light microscope difficult. However, seeding too few cells reduces the chances of finding a suitable cell of interest. We therefore seed cells at 5% density, or 40 000 HeLa cells per 35 mm dish in preparation for imaging and resin-embedding the sample the following day. If the cells require transfection for over 24 hours, we usually transfect in separate plates (such as 6-well plates), and reseed them into the CLEM dishes at the appropriate time to attain the required density. Aim for a cell density of 10-15% on the day of processing for CLEM.
B. Fixation and Sample Preparation
a. Fixative solution osmolarity
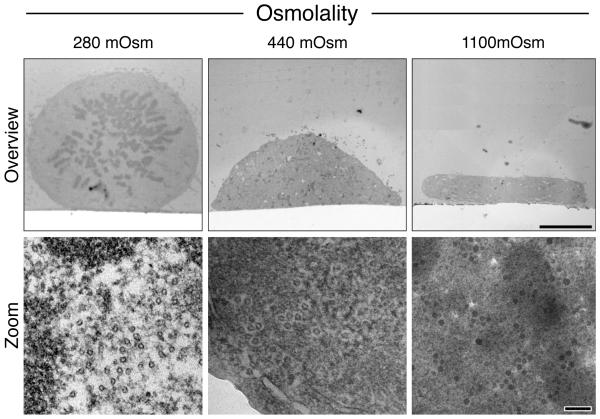
The physiological osmolality of mammalian tissue is ~290 mOsm, depending on species, tissue type and hydration status (Loqman et al., 2010; Mathieu et al., 1978). Fixative solutions should mimic physiological osmolality, providing an iso-osmotic equilibrium between intracellular and extracellular fluids. Figure 2 shows examples of orthogonally sectioned cells fixed with solutions of varying osmotic strengths. At 440 mOsm and 1100 mOsm, a large amount of cell shrinkage can be observed, with poor spindle apparatus preservation and unusually dense cytosol. Therefore, we routinely use a fixing solution of ~280 mOsm, consisting of 3% glutaraldehyde, 0.5% paraformaldehyde in 0.05M PB.
Figure 2. Optimization of mitotic spindle and cell structure preservation.
Orthogonal sections of cells fixed with 280, 440 or 1100 mOsm. Representative high magnification electron micrographs of the cytosol in each condition are shown below. Scale bars 5 μm (overview) and 100 nm (bottom).
b. Light microscopy
On the day of sample processing, cells of interest can be identified under the light microscope with a 20× air objective. The low magnification allows images to be acquired containing a large field of view, useful for cell re-location during later processing. Some cells expressing fluorescent proteins require pre-fixation imaging as their fluorescence is obscured by the auto-fluorescence created by glutaraldehyde. Once the cell of interest has been located and imaged, add fixative solution for 1 hour. It should be noted that microtubules are sensitive to temperature changes (Engelborghs et al., 1976), we therefore recommend that imaging of unfixed cells is carried out using an appropriate live imaging chamber at 37 °C.
After fixation, replace the fixative solution with 1-2 ml wash solution with 0.1% Hoechst-33342 (or similar DNA dye) incubate for ~20 minutes, rinse three times with wash solution (leaving on 1 ml of the final wash) and return the dish to the microscope. This second round of imaging is an opportunity to acquire high magnification images of cells of interest, using 60× or 100× oil-immersion objectives (Figure 1B, left). Take fluorescent and white light images of the cell, and also images of the same field of view focused on the coordinates. These will serve as references later to pinpoint the cell in the resin block and determine the orientation of the spindle axis. It helps at this stage to carefully wipe off any immersion oil with ethanol, and mark the approximate location of the cell with a fine marker pen on the underside of the dish.
c. Resin embedding
Cells become round during mitosis and are therefore less adherent to their substrate. This means that during all steps up to resin embedding, dishes must be handled with extreme care so as to not detach or change the orientation of the cell.
Next, replace the wash solution with a few drops of 1% osmium tetroxide on the coverslip for 1 hour. Remove the osmium, and gently rinse the cells twice for 30 minutes with double-distilled water. Remove the water and replace with 30% ethanol for 30 minutes, then replace with a small amount of 0.5% uranyl acetate in 30% ethanol for 1 hour. Next, the cells need to be dehydrated using a gradient of sequential solutions containing increasing amounts of ethanol. Replace stepwise with each of the following solutions: 30%, 50%, 60%, 70%, 80% and 90% ethanol, then twice with 100% ethanol, incubating at each step for 10 minutes.
The cells can now be infiltrated with resin. Mix a 1:2 ratio of resin:ethanol solution, making sure that it is fully homogenized using a 3 ml plastic Pasteur pipette. Remove the ethanol from the dish, and lightly cover the bottom of the dish with the resin infiltration mix for 20 minutes. Remove, and replace with a 1:1 ratio of resin:ethanol solution for 20 minutes. Remove the mix, and replace with a ~2 mm layer of 100% resin covering the bottom of the dish. If the sample is to be sectioned orthogonally, the dish can be placed in a 60°C oven for 48-72 hours. For longitudinal sectioning, fill either half of an embedding capsule with 100% resin, and gently place it open-side down onto the cell (Figure 3A), which you should be able locate using the pen mark placed earlier. The dish can then be placed in the oven.
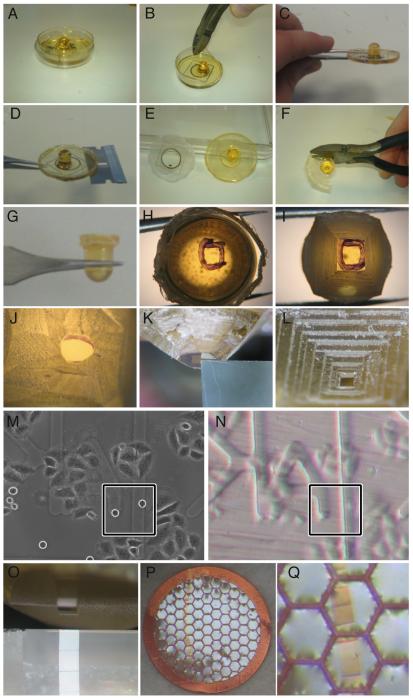
Figure 3. Pictorial guide to CLEM processing for sectioning longitudinally to the spindle axis.
Following polymerization, resin was separated from the CLEM dish. Unwanted plastic was removed from the edges of the dish using pliers (A-C) allowing a razor to be inserted between the resin and the dish base (D). Following the separation of resin and dish (E) excess resin was removed using pliers (F) until just the capsule remained (G). The cell of interest was marked (H) with the aid of LM images (M) and resin coordinates (N). Unwanted resin was removed using a junior hacksaw (I) and a razor (J). Resin was trimmed using a microtome and a glass knife (K) until a neat block was generated at the top of a pyramid (L). Blocks were sectioned using a diamond knife (O) and ribbons collected using 100 mesh copper grids (P & Q), coated with formvar.
We recommend the use of EPON resin as other resin types (for example, Agar Scientific Low Viscosity Resin) that we have tested react and bind the CLEM dish, making the separation steps (below) much more difficult.
C. Longitudinal Sectioning
Longitudinal sectioning is the conventional EM method for viewing cells. Sections parallel to the plane of the coverslip are taken from the base of the cell moving progressively upwards (see Figure 1C). This plane of sectioning allows extended lengths of microtubules to be observed, and is therefore particularly useful for analyzing microtubule attachment to the kinetochore, or quantifying microtubule crosslinkers (Figure 1D).
Once the resin has fully polymerized, the dishes can be removed from the oven. Figure 3 contains a pictorial guide of the steps required to separate the resin and dish up to the sectioning. Using pliers, start by cutting off the edges of the dish entirely (Figure 3 A-C), so that the seam between resin and plastic is accessible all the way around the dish. Very carefully insert a razor blade between the plastic and resin (Figure 3 D), slowly forcing the razor towards the centre of the dish all the way around the resin, to separate them (Figure 3 E). This must be performed with extreme care, as too much leverage by the razor will shatter the glass. This shattering usually renders the sample unusable, as removing all the glass fragments from the resin is very difficult without damaging the sample, and any microscopic shards of glass remaining will damage the diamond knife during sectioning. Dipping the resin and dish into liquid nitrogen for 1-2 seconds can help separate them, as the difference in thermal expansion between resin and plastic will eventually detach them. Other protocols use 40% hydrofluoric acid to dissolve the glass, bypassing this tricky step (Polishchuk et al., 2012).
Once detached, excess resin can be trimmed away until only the capsule remains (Figure 3 F, G). The coordinates imprinted on the underside of the capsule can be observed using a tissue dissection microscope; draw around the coordinate containing your cell of interest using a thin marker pen, the LM images serving as reference (Figure 3 H). Using a microtome chuck and bench-top vice to firmly hold the resin block in place, remove excess resin around the coordinate using a junior hacksaw (Figure 3 I), making sure that it never scratches the coordinate surface. This risk can be minimized by trimming away the resin using razor blades (Figure 3 J).
The remainder of the resin trimming and sectioning is performed using an ultramicrotome with glass knives (Figure 3 K) and a diamond knife (Figure 3 O), respectively. It is possible to make out the cell of interest and the etched coordinate using microtome binoculars (Figure 3 M, N). We routinely trim a square block face, up to ~50 μm from the cell edge (Figure 3 L), but a wider space can be left according to one’s experience. The larger the block face created during sectioning, the more difficult it will be to locate the cell in the sections under the EM. A square block face is optimal, as this helps acquire serial sections during sectioning. Sections 80 nm in thickness are taken using the diamond knife, and collected using the copper grids coated with formvar (Figure 3 P, Q). To handle the grids, high-precision tweezers should be used at all times, carefully gripping the grid by its outer edge only, so as to not tear or damage the formvar or sample sections.
To attain optimal contrast under the microscope, we post-stain the sections by placing each grid section-side-down onto a drop of 5% uranyl acetate in 50% ethanol for 7-8 min, gently rinse in distilled water for 1 min, and place face-down on a drip of Reynold’s lead citrate solution for 7-8 min. The grid is then rinsed in water again for 1 min. Both solutions should be centrifuged in 1.5 ml Eppendorf tubes at 8 000 g for 5 min before use to remove unwanted precipitate. Grids should be dried face-up for at least 2 h on clean filter paper before imaging, and kept in a clean, dust-free environment (such as a Petri dish or grid storage box).
D. Orthogonal Sectioning
Orthogonal sectioning involves taking sections that are perpendicular to the spindle axis. In mitotic cells, this is useful to view and quantify most K-fiber microtubules within a single section. Quantifying K-fiber microtubules is possible using longitudinal serial sections (McEwen et al., 1997), however we avoided this method because: (1) serial sections are particularly difficult to acquire, and (2) spatial distribution analysis cannot be performed, as the compression forces exerted on each section of a serial reconstruction by the knife will likely deform the sample more than a single orthogonal cross-section through a K-fiber.
Figure 4 contains a pictorial guide of the steps required to prepare the sample for orthogonal sectioning. The samples to be sectioned orthogonally should consist of a flat layer of resin (without the resin capsule; Figure 4 A). Once removed from the oven, separate the dish from the resin using the same method as for the longitudinal samples (see above) and with the same amount of care (Figure 4 B, C, D). Using a tissue dissection microscope, find the coordinate containing your cell of interest and circle it using a marker pen (Figure 4 H). Using the coordinate grid and the LM images taken previously as references (Figure 4 E, F, G), determine the position and direction of the spindle axis, and draw an elongated rectangle around the cell of interest (Figure 4 H, I, J). Cut out the rectangle using the hacksaw (Figure 4 K, L), paying attention not to touch or damage the surface of the marked coordinate. You should end up with a strip of resin (Figure 4 M). Carefully remove excess off one end, so the cell is near the tip, and insert it into a microtome chuck (Figure 4 N, O).
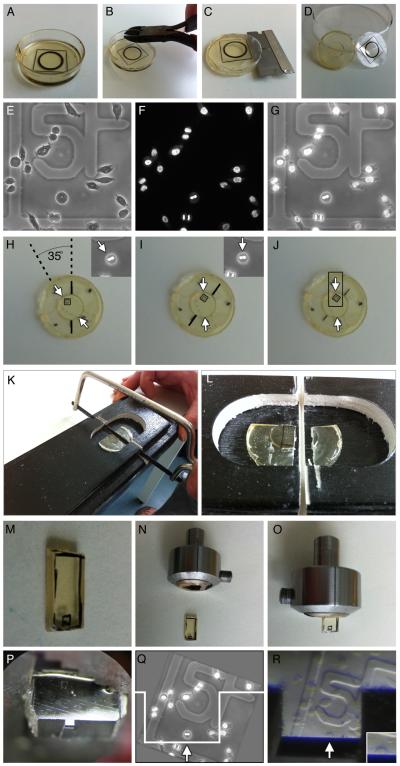
Figure 4. A pictorial guide to orthogonal CLEM processing.
Following polymerization, resin was separated from the CLEM dish (A-D). Unwanted plastic was removed from the edges of the CLEM dish using pliers (B) allowing a razor to be inserted between the resin and the dish base (C). Following separation (D) the position of the spindle was estimated using the reference LM images (E-G). These images allowed the re-orientation of the resin (H & I) so that an appropriate block could be marked (J) before excision using a junior hacksaw and a mitre block (K & L). The excised block (M) was inserted into a microtome chuck (N & O) and fine trimmed using a glass knife (P) before serial sections were taken of the cell, in the desired orientation (Q & R).
Using an ultramicrotome, you should be able to see the coordinates, and can trim excess resin from the tip using glass knives until you approach the cell of interest (Figure 4 P, Q, R). Trim away resin from either side of the cell to a depth of ~100 μm, leaving a 50-100 μm buffer zone around the cell. Finally, trim away excess resin from the “upper” side of the strip, which is the block face positioned reverse-parallel to the one imprinted with coordinates. The thickness should be similar to the width either side of the cell edge, so that the block face is square shaped. The cell can now be sectioned using a diamond knife; we routinely take 80-100 nm slices. Sections should be collected and treated as described for longitudinal sectioning (see above), along with the same post-staining method.
E. Imaging and Sample Tilting
In longitudinal sections, K-fibers can be identified as bundles of microtubules in parallel conformation terminating at the kinetochore. During image capture, we typically take 4 μm by 3 μm images at 60 000× magnification. This allows us to distinguish adjacent microtubules and the material that crosslinks them with enough resolution to measure the length of each element. One particular type of analysis that we have performed on such images is the quantification of microtubule crosslinker frequency (Booth et al., 2011; Cheeseman et al., 2011), but the qualitative assessment of microtubule attachment to kinetochores and of the overall organization of the fiber is also possible.
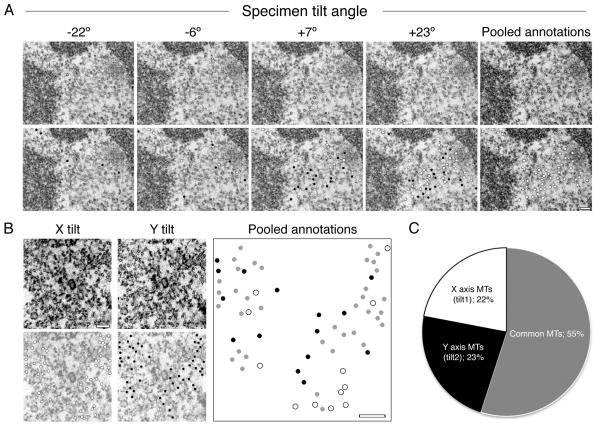
To image orthogonal sections under the microscope, full analysis of the K-fiber requires sample tilting. This is because microtubules are most easily recognizable when they are perpendicular to the imaging plane, as they appear as characteristic electron dense rings. Not all microtubules will be at the correct angle, which increases the risk of quantification error. We can minimize this error by imaging the sample at various tilt angles (Figure 5 A). Our optimization shows that a single axis tilt of ± 45° (90° in total) is sufficient to reveal 80% of microtubules in a K-fiber (Figure 5 B, C). A dual tilt along perpendicular axes is necessary to obtain 100% coverage. We perform image acquisition for a typical mammalian K-fiber at 60 000× to 90 000× magnification.
Figure 5. Sample tilting is necessary to achieve full coverage of microtubules in orthogonal sections.
(A) Example electron micrographs taken from a −45° to +45° tilt series of a K-fiber. Observable microtubules are marked with dots (bottom row). Black dots represent microtubules that are unique to that tilt frame. White dots represent the accumulating microtubules identified in previous tilt frames. The total number of microtubule annotations were pooled together onto one frame (far right) giving a fair overview of the whole K-fiber. Scale bar 100 nm. (B & C) A dual tilt series of one K-fiber was carried out. (B) Representative electron micrographs taken from the central region of both X and Y tilts (A-top). All microtubules observed in each tilt series were annotated (bottom; X axis in white, Y axis in black). The sum of microtubules from both tilts were pooled together on to a single blank image, any microtubules that were common to both tilts were marked grey. Scale bar, 100 nm. (C) A pie chart showing the percentage of total microtubules that were unique to each tilt and also the common ones.
Additionally, once the image tilt series (.raw file) is acquired, it can be assembled into a tomogram using IMOD software’s Etomo package (Boulder Laboratory for 3-Dimensional Electron Microscopy). The final tomogram is a stack of images detailing the sample section in 3 dimensions, and also removes some background compared to an unaltered electron micrograph.
We use ImageJ/Fiji software and IMOD’s Neighbor Density Analysis (NDA) package to analyze the spatial distribution of microtubules within a fiber (McDonald et al., 1992). The output is a probability distribution graph, indicating the distance from any given microtubule at which one is most likely to find another microtubule. Other examples of types of spatial analyses which can be performed are (1) nearest neighbor analysis, which calculates the average distance between a microtubule and its nearest neighbor in the fiber, (2) the angular distribution of neighboring microtubules surrounding any given microtubule (also performed using the IMOD NDA package) (Ding et al., 1993).
IV. Discussion
CLEM remains among the most powerful imaging techniques available. The ability to view a live cell, in any state, or undergoing any particular or rare event, and to take an EM snapshot to be viewed at ~100 000× magnification remains an outstanding tool to study cellular processes. However, this technique is often overlooked as it is considered too time-consuming and technically challenging.
Our CLEM protocol has been optimized for the study of mitotic spindles, where a great amount of attention has been placed on the preservation of the ultrastructure of microtubules and other spindle components. Our osmolality tests have shown that fixative solution osmolality must be as close to physiological conditions as possible. But further improvements could potentially be achieved during sample dehydration steps, at which point cell shrinkage can occur, as well as partial cytosolic washout.
Our protocol uses chemical fixation which is sub-optimal for microtubule preservation. High-pressure freezing is an alternative fixation method, which uses the combination of ultra-low temperature to snap freeze the cell while applying pressure to inhibit the formation of ice crystals which would rupture and damage cellular structures. This fixation has been shown to substantially improve preservation of cellular architecture and organelle appearance (Wolf et al., 1981). Some studies have used CLEM with high-pressure freezing to study mitotic or meiotic events with remarkable success (Pelletier et al., 2006). However, the implementation of this method with CLEM considerably increases the difficulty of the overall protocol, particularly when studying mitotic spindles. The size of the sample that can be frozen is very small and as microtubules are particularly sensitive to temperature variation, a fast transfer from the light microscope/incubator to the high-pressure freezer is needed.
A useful addition to our CLEM protocol would be the ability to readily view proteins of interest under both light and electron microscopes. There has been recent focus on developing hybrid genetic tags that are both fluorescent and can be converted into an electron-dense signal to serve this purpose such as MiniSOG (Shu et al., 2011) and GFP-APEX (Martell et al., 2012). However, these tags have yet to be used to study the mitotic spindle.
Although our experimental purposes have only required standard epifluorescence micrographs before switching to EM, confocal microscopy could easily be implemented instead. This would allow the above protocol to be expanded, by combining confocal Z stacks and serial EM section imaging to create correlated 3D reconstructions in both light (confocal) and electron micrographs or electron tomograms. However, obtaining serial sections remains a challenge even for experienced electron microscopists. Nonetheless, there are currently several labs attempting the EM reconstruction of entire mitotic spindles, and whole-cell tomographic reconstruction has been achieved to study cytoskeletal structures (Hoog and Antony, 2007; Hoog et al., 2007), indicating the feasibility of this approach. Moreover, the recent effort by EM equipment suppliers to develop dual-beam EM microscopes which are able to both section and image the sample in an automated fashion could revolutionize this field. These machines, which are able to repeatedly remove 5 nm layers of sample and image the back-scattering of electrons using high-resolution scanning EM, bypass all the major difficulties involved with EM. So far, the resolution of this equipment is sufficient to comfortably reconstruct synaptic vesicles and other organelles (Knott et al., 2011), but it is not yet enough to view cytoskeletal elements such as microtubules in high detail.
Overall, CLEM is a powerful imaging method, able to give unrivalled cellular structural detail, which we have applied to the study of kinetochore-fiber ultrastructure. We believe that the further integration of such tools as hybrid tags and dual-beam microscopes with CLEM will unlock a vast potential for the field of electron microscopy, which will maintain a firm place in research, regardless of the development of other super-resolution imaging systems.
Acknowledgments
We would like to thank Alison Beckett for technical help and discussions. DGB and LPC were supported by Prize Studentships from The Wellcome Trust. IAP is a Royal Society University Research Fellow. SJR is a Senior Cancer Research Fellow for Cancer Research UK.
References
- Booth DG, Hood FE, Prior IA, Royle SJ. A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. Embo Journal. 2011;30:906–919. doi: 10.1038/emboj.2011.15. DOI: 10.1038/emboj.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman LP, Booth DG, Hood FE, Prior IA, Royle SJ. Aurora A kinase activity is required for localization of TACC3/ch-TOG/clathrin inter-microtubule bridges. Communicative & integrative biology. 2011;4:409–12. doi: 10.4161/cib.4.4.15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–51. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelborghs Y, Heremans KAH, Demaeyer LCM, Hoebeke J. Effect of temperature and pressure on polymerization equilibrium of neuronal microtubules. Nature. 1976;259:686–689. doi: 10.1038/259686a0. DOI: 10.1038/259686a0. [DOI] [PubMed] [Google Scholar]
- Euteneuer U, McIntosh JR. Structural polarity of kinetochore microtubules in Ptk1-cells. Journal of Cell Biology. 1981;89:338–345. doi: 10.1083/jcb.89.2.338. DOI: 10.1083/jcb.89.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoog JL, Antony C. Whole-cell investigation of microtubule cytoskeleton architecture by electron tomography. In: McIntosh JR, editor. Cellular Electron Microscopy. 2007. pp. 145–167. [DOI] [PubMed] [Google Scholar]
- Hoog JL, Schwartz C, Noon AT, O’Toole ET, Mastronarde DN, McIntosh JR, Antony C. Organization of interphase microtubules in fission yeast analyzed by electron tomography. Developmental Cell. 2007;12:349–361. doi: 10.1016/j.devcel.2007.01.020. DOI: 10.1016/j.devcel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Knott G, Rosset S, Cantoni M. Focussed ion beam milling and scanning electron microscopy of brain tissue. Journal of visualized experiments: JoVE. 2011:e2588. doi: 10.3791/2588. DOI: 10.3791/2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqman MY, Bush PG, Farquharson C, Hall AC. A cell shrinkage artefact in growth plate chondrocytes with common fixative solutions: importance of fixative osmolarity for maintaining morphology. European Cells & Materials. 2010;19:214–227. doi: 10.22203/ecm.v019a21. [DOI] [PubMed] [Google Scholar]
- Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–8. doi: 10.1038/nbt.2375. DOI: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Claassen H, Weibel ER. Differential effect of glutaraldehyde and buffer osmolarity on cell dimensions - study on lung-tissue. Journal of Ultrastructure Research. 1978;63:20–34. doi: 10.1016/s0022-5320(78)80041-0. DOI: 10.1016/s0022-5320(78)80041-0. [DOI] [PubMed] [Google Scholar]
- McDonald KL, O’Toole ET, Mastronarde DN, McIntosh JR. Kinetochore microtubules in Ptk cells. Journal of Cell Biology. 1992;118:369–383. doi: 10.1083/jcb.118.2.369. DOI: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. Journal of Cell Biology. 1997;137:1567–1580. doi: 10.1083/jcb.137.7.1567. DOI: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Mueller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. DOI: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Polishchuk RS, Polishchuk EV, Luini A. Visualizing live dynamics and ultrastructure of intracellular organelles with preembedding correlative light-electron microscopy. Methods in cell biology. 2012;111:21–35. doi: 10.1016/B978-0-12-416026-2.00002-9. [DOI] [PubMed] [Google Scholar]
- Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. The Journal of cell biology. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM, Brust-Mascher I, Mogilner A. Cell division. Nature. 2003;422:746–752. doi: 10.1038/nature01599. [DOI] [PubMed] [Google Scholar]
- Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, Tsien RY. A Genetically Encoded Tag for Correlated Light and Electron Microscopy of Intact Cells, Tissues, and Organisms. Plos Biology. 2011;9 doi: 10.1371/journal.pbio.1001041. DOI: e1001041 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf KV, Stockem W, Wohlfarth-Bottermann KE, Moor H. Cytoplasmic actomyosin fibrils after preservation with high-pressure freezing. Cell and Tissue Research. 1981;217:479–495. doi: 10.1007/BF00219359. [DOI] [PubMed] [Google Scholar]