Abstract
The success of tyrosine kinase inhibitors (TKIs) in select patients with non–small-cell lung cancer (NSCLC) has transformed management of the disease, placing new emphasis on understanding the molecular characteristics of tumor specimens. It is now recognized that genetic alterations in the epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) define two unique subtypes of NSCLC that are highly responsive to genotype-directed TKIs. Despite this initial sensitivity, however, the long-term effectiveness of such therapies is universally limited by the development of resistance. Identifying the mechanisms underlying this resistance is an area of intense, ongoing investigation. In this review, we provide an overview of recent experience in the field, focusing on results from preclinical resistance models and studies of patient-derived, TKI-resistant tumor specimens. Although diverse TKI resistance mechanisms have been identified within EGFR-mutant and ALK-positive patients, we highlight common principles of resistance shared between these groups. These include the development of secondary mutations in the kinase target, gene amplification of the primary oncogene, and upregulation of bypass signaling tracts. In EGFR-mutant and ALK-positive patients alike, acquired resistance may also be a dynamic and multifactorial process that may necessitate the use of treatment combinations. We believe that insights into the mechanisms of TKI resistance in patients with EGFR mutations or ALK rearrangements may inform the development of novel treatment strategies in NSCLC, which may also be generalizable to other kinase-driven malignancies.
INTRODUCTION
Advances in molecular biology have highlighted the genomic complexity of cancer cells. Within this diverse genetic landscape, however, certain cancers are dependent on single oncogenic pathways for survival. This state of “oncogene addiction” commonly involves aberrant kinase activation, providing a therapeutic basis for agents directed against the corresponding dysregulated kinases.1 The success of the tyrosine kinase inhibitor (TKI) imatinib in chronic myeloid leukemia (CML) originally validated this treatment paradigm.2 Subsequently, this approach has been translated to other oncogene-driven malignancies, including gastrointestinal stromal tumors (GIST), BRAF-mutant melanoma, and epidermal growth factor receptor (EGFR) –mutant non–small-cell lung cancer (NSCLC).3–7 Although selective kinase inhibitors produce high objective response rates (ORRs) in these molecularly defined populations, the long-term impact of such therapies are limited to variable degrees by the development of resistance.8–10
NSCLC provides an instructive conceptual framework for examining kinase-directed therapies and the mechanisms that ultimately limit their effectiveness. In the last decade, NSCLC management has evolved toward stratification of patients based on genetic alterations within “driver” oncogenes, such as EGFR and anaplastic lymphoma kinase (ALK). Somatic mutations in EGFR are identified in 10% to 30% of patients with NSCLC.6,7,11 Common EGFR alterations include the L858R point mutation and exon 19 deletions.12 These mutations result in enhanced EGFR signaling and confer sensitivity to the EGFR TKIs gefitinib and erlotinib.6,7,11 In first-line treatment, EGFR inhibitors produce ORRs nearing 75% in patients with typical EGFR mutations.12 Randomized trials have also demonstrated improved progression-free survival (PFS) for EGFR-mutant patients receiving TKIs compared with chemotherapy.13–15
Like EGFR mutations, ALK rearrangements define a unique molecular subset of NSCLC. Most ALK rearrangements arise from chromosomal inversions that generate novel ALK fusion transcripts, commonly involving echinoderm microtubule-associated protein-like 4 (EML4) as the 5′ fusion partner (EML4-ALK).16,17 Identified in 4% to 6% of patients with NSCLC,16,18–20 ALK rearrangements are associated with unique clinicopathologic features and sensitivity to the ALK TKI crizotinib.20 Initial clinical studies of crizotinib demonstrated ORRs of 60% and a median PFS of 8 to 10 months.21–23 Given its high response rate, the US Food and Drug Administration (FDA) granted accelerated approval of crizotinib in 2011.
Despite the success of genotype-directed therapies in EGFR-mutant and ALK-positive patients, resistance inevitably develops. Indeed, the median PFS after treatment with EGFR or ALK inhibitors in target populations is generally less than 1 year.13–15,21–23 Thereafter, standard management usually consists of cytotoxic chemotherapy. It is therefore critical to develop new insights into the mechanisms of TKI resistance to develop more effective treatment strategies.
DEFINITIONS OF RESISTANCE
Resistance to targeted therapies is generally classified as either primary (ie, intrinsic) or secondary (ie, acquired). Primary resistance describes a de novo lack of treatment response, whereas acquired resistance denotes disease progression after an initial response. Like CML and GIST,24,25 criteria for acquired resistance were recently proposed for EGFR-mutant NSCLC (Table 1).26 Although similar criteria have not been established for ALK-positive NSCLC, our definition will largely mirror that of EGFR for this review.
Table 1.
Criteria for Acquired Resistance to EGFR Tyrosine Kinase Inhibitors
| 1. Patient has received prior therapy with an EGFR TKI (monotherapy). |
| 2. Tumor genotyping confirms the presence of a typical EGFR mutation that is associated with sensitivity to EGFR TKIs. Examples include exon 19 deletions, L858R, and G719X. |
| OR |
| Patient achieves either a documented partial or complete response OR prolonged stable disease (≥ 6 months) based on RECIST or WHO criteria. |
| 3. Disease progression occurs despite uninterrupted exposure to an EGFR TKI within 30 days. |
| 4. Patient has not received additional systemic therapy since discontinuation of EGFR TKIs. |
Adapted from Jackman et al.26
Abbreviations: EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
PRIMARY RESISTANCE
EGFR
Although ORRs to EGFR TKIs are high among EGFR-mutant patients, some patients exhibit intrinsic resistance. The mechanistic basis for these observations is largely unknown. Primary resistance may be due in part to differential TKI sensitivities across various EGFR mutations. “Classic” EGFR mutations, namely exon 19 deletions and L858R, are associated with marked sensitivity to TKIs.27 Conversely, exon 20 insertions or duplications (∼4% of EGFR mutations) seem to be resistant to EGFR inhibitors despite in vitro evidence suggesting that these alterations result in aberrant kinase activation.28–30
Intrinsic resistance to EGFR inhibitors may also be due to secondary genetic alterations that co-occur with sensitizing EGFR mutations. For instance, a T790M mutation within EGFR has been occasionally identified as a minor clone within treatment-naive tumor specimens containing classic EGFR mutations.31–33 Similarly, MET amplification has been reported in EGFR-mutant tumors before TKI exposure.33–35 As is discussed later, MET amplification and T790M are common mechanisms of acquired resistance. When present de novo, it has been suggested that these genetic alterations may also promote intrinsic resistance if present at sufficiently high allelic frequencies. Alternatively, selective pressure from TKIs may permit cells containing T790M or MET amplification to emerge as dominant clones early during therapy.
ALK
A small number of ALK-positive patients experience disease progression immediately after starting crizotinib. Recent preclinical data suggest that differences in specific ALK fusion gene products may partially account for heterogeneous treatment responses.36 A number of different 5′ ALK fusion partners have been identified.37 Additionally, multiple different EML4-ALK variants exist, all of which preserve the ALK kinase domain but differ with respect to the EML4 breakpoint. In one cell line model, differences in crizotinib sensitivity were observed between different EML4-ALK fusion variants and ALK fusion partners.36 Despite these in vitro observations, subgroup analysis from a phase I trial of crizotinib showed no correlation between EML4-ALK variant type and response.21
Another explanation for primary resistance to crizotinib may be false-positive genotyping. ALK rearrangements may be detected by various techniques, but only ALK fluorescence in situ hybridization (FISH) testing is currently approved by the FDA.38 This assay is technically challenging because EML4 and ALK both map to chromosome 2 and are normally separated by only ∼12 megabases.16,38 False-positive results may occur as a result of sectioning artifact, poor nucleus morphology, aberrant probe hybridization, or misinterpretation at pathologic review.39 It is therefore possible that rare cases of “primary resistance” to crizotinib may be due to technical factors rather than intrinsic biology. Lastly, ALK FISH may identify true-positive ALK translocations, but these may not generate functional rearrangements in all patients.
Heterogeneity of TKI Response
ALK and EGFR TKIs can produce wide spectrums of response, even among those with identical genetic alterations. One intriguing explanation for this heterogeneity involves differences within the cellular apoptotic machinery. In particular, recent data have suggested that the pro-apoptotic protein BIM is a biomarker and mediator of TKI-induced apoptosis in several oncogene-driven malignancies.40–46 In EGFR-mutant cell lines, BIM is upregulated in response to EGFR TKIs, and BIM levels correlate with the degree of apoptotic response.42–45 Likewise, inhibition of BIM expression promotes intrinsic resistance to EGFR TKIs. Consistent with these preclinical findings, low pretreatment BIM RNA levels from EGFR-mutant patients were associated with decreased tumor shrinkage and a shorter PFS after treatment with EGFR TKIs.45 The reasons for these differences in baseline BIM levels remain unclear. One recent report suggests that a genetic polymorphism in BIM results in alternative splicing and altered BIM function, which may contribute to intrinsic resistance in some patients.47
ACQUIRED RESISTANCE
Mechanisms of acquired resistance in oncogene-driven malignancies are broadly divided into two categories. The first involves development of additional genetic alterations in the primary oncogene, which facilitates continued downstream signaling. This commonly arises through secondary mutations in the kinase target or through gene amplification of the kinase itself (Table 2).8,62–64 Alternatively, resistance can develop independent of genetic changes in the target. This occurs through activation of downstream signaling pathways, changes in tumor histology, or alterations in drug metabolism.8,27,62,65,66
Table 2.
Major Mechanisms of Acquired Resistance Identified in Clinical Specimens
| Mechanism | Estimated Frequency (%) | References |
|---|---|---|
| EGFR TKI resistance | ||
| Genetic alterations in EGFR | ||
| T790M mutations | 50 | 48–51 |
| D761Y, T854A, and L747S mutations | < 5 | 42, 52, 53 |
| EGFR amplification | 8 | 50 |
| Bypass signaling tracts | ||
| MET amplification | 5–22 | 35, 50, 51 |
| HER2 amplification | 12 | 54 |
| PIK3CA mutations | 5 | 50 |
| BRAF mutations | 1 | 55 |
| CRKL amplification | 9 | 56 |
| HGF overexpression | 1 of 2 cases | 57 |
| Phenotypic alterations | ||
| Transformation to small-cell lung cancer | 3–14 | 50, 51 |
| ALK TKI resistance | ||
| Genetic alterations in ALK | ||
| ALK secondary mutations (eg, L1196M) | 22-36 | 58–61 |
| ALK gene amplification | 7-18 | 60, 61 |
| Bypass signaling tracts | ||
| EGFR activation | 44 | 60 |
| KIT gene amplification | 15 | 60 |
Abbreviations: EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor; HER2, human epidermal growth factor receptor 2; HGF, hepatocyte growth factor; ALK, anaplastic lymphoma kinase.
Secondary Mutations
Experience with imatinib resistance in CML has informed approaches to acquired resistance in other malignancies. Secondary mutations in the ABL kinase domain are common causes of TKI resistance in CML.67,68 Among the more than 50 secondary mutations identified to date, the most common involves a threonine-to-isoleucine substitution at position 315 (T315I) of ABL, the so-called gatekeeper residue.8 This mutation reduces imatinib binding but preserves ABL kinase activity.67 In EGFR-mutant NSCLC, the earliest reports of TKI resistance identified an analogous secondary mutation in exon 20 of EGFR, leading to a threonine-to-methionine substitution within the gatekeeper residue at position 790 (T790M).48,49 Secondary T790M mutations have since been found in approximately 50% of TKI-resistant, EGFR-mutant patients, establishing this alteration as the dominant resistance mechanism in the clinic.50,51
Although other gatekeeper mutations sterically impede TKI binding, T790M causes resistance predominantly through changes in adenosine triphosphate (ATP) affinity.69 EGFR-mutant tumors are generally sensitive to competitive inhibitors because such mutations reduce the receptor's affinity for ATP. The addition of T790M, however, restores the ATP affinity of the kinase back to wild-type levels, re-establishing ATP as the favored substrate rather than the TKI. When coexpressed with classic EGFR sensitizing mutations, T790M confers resistance in vitro and in transgenic mice.48,49,70 Furthermore, biochemical studies demonstrate that such dual mutations result in enhanced kinase activity and oncogenicity.71,72 Despite these findings, preclinical and retrospective clinical studies suggest that T790M may actually confer a growth disadvantage relative to TKI-sensitive parental cells.73,74
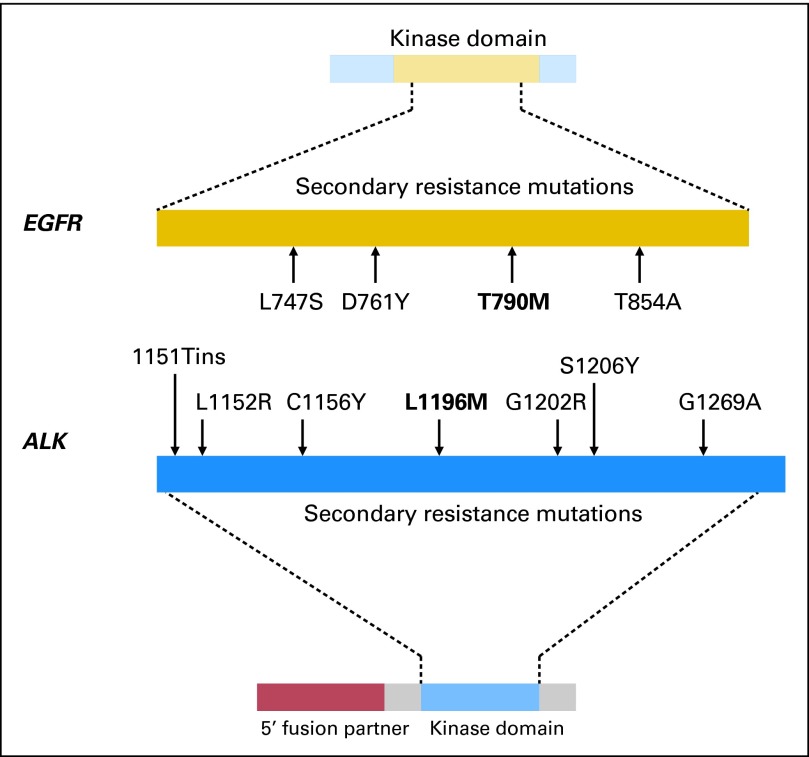
T790M mutations have also been rarely detected within treatment-naive specimens.31–33 In most patients, however, it is unclear whether these mutations arise de novo during therapy or whether EGFR-directed therapies select for preexisting clones. In addition to T790M, three additional secondary EGFR mutations have been associated with TKI resistance: D761Y, T854A, and L747S (Fig 1).42,52,53 The structural basis for how these mutations confer resistance is unknown. Collectively, non-T790M secondary mutations are relatively uncommon and, in vitro, confer less pronounced resistance.
Fig 1.
Comparison of the number and distribution of secondary resistance mutations in the tyrosine kinase domains of the epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK). In EGFR-positive patients with acquired tyrosine kinase inhibitor (TKI) resistance, four different second-site mutations in EGFR have been identified in clinical specimens. The gatekeeper mutation T790M (bold) is the most common, present in approximately 50% of patients at the time of resistance. The remaining EGFR secondary mutations are present at low frequencies. In contrast, seven different secondary mutations have been identified in ALK-positive patients at the time of TKI resistance, including the L1196M gatekeeper mutation (bold). Despite this wider distribution of secondary mutations within the ALK tyrosine kinase domain, such mutations are found in only approximately 30% of patients.
Secondary mutations in the ALK kinase domain have been identified in approximately 30% of ALK-positive patients with crizotinib resistance.58–61,75 The earliest description involved an ALK-positive patient who developed disease progression after receiving crizotinib for 5 months.58 Analysis of pleural fluid from this patient revealed two nonoverlapping mutations, L1196M and C1156Y, within the ALK kinase domain. Each independently conferred crizotinib resistance in vitro. The L1196M substitution is notable because it involves the ALK gatekeeper residue, analogous to T790M in EGFR. The L1196M mutation, which replaces a leucine moiety with a bulkier methionine residue, likely causes resistance by steric interference with crizotinib binding.
Since the initial case report of crizotinib resistance, additional second-site ALK mutations have been identified in patient-derived NSCLC specimens (1151Tins, L1152R, G1202R, S1206Y, and G1269A).59–61 Mutations in a majority of these residues were also found in accelerated in vitro mutagenesis screening.76 A separate ALK secondary mutation, F1174L, has also been reported in a crizotinib-resistant inflammatory myofibroblastic tumor.77 Interestingly, the F1174 residue is among the most commonly mutated sites in neuroblastoma.78
ALK secondary mutations in NSCLC are distributed throughout the kinase domain, including the solvent front (G1202R, S1206Y), gatekeeper residue (L1196M), ATP-binding pocket (G1269A), and N-terminal to the αC-helix (1151Tins, L1152R, and C1156Y).58–61,75 In vitro, ALK secondary mutations confer differential sensitivities to crizotinib and second-generation ALK TKIs.60 For example, the ALK S1206Y mutation confers lower degrees of crizotinib resistance compared with G1202R, L1196M, and 1151Tins mutations. It remains unclear, however, whether variations in ALK secondary mutations translate into different clinical responses to next-generation ALK inhibitors.
It is noteworthy that ALK-positive patients, like those with CML, develop multiple secondary mutations at the time of TKI resistance. This is in contrast to EGFR-mutant patients, in whom T790M is essentially the sole secondary mutation observed clinically. Although in vitro mutagenesis experiments identified several additional EGFR resistance mutations, T790M was the only mutation consistently identified in all screens.79 One potential explanation for this finding is that the EGFR-mutant kinase, which already possesses one alteration, may be unable to accommodate diverse drug-resistant, secondary mutations without compromising kinase function.61 Still another consideration is that gefitinib and erlotinib bind to the active conformation of EGFR, whereas crizotinib and imatinib bind to the inactive conformations of ALK and BCR-ABL, respectively.27 This may limit the spectrum of secondary EGFR mutations to those that influence drug binding in the ATP pocket.
Target Gene Amplification
Target gene amplification is another cause of acquired resistance that was first identified in CML.50,61,67,69,80 Gene amplification may shift the intracellular balance between kinase and TKI in favor of the kinase. Gene amplification may also augment the effects of secondary resistance mutations if both are present simultaneously. For example, in a report of 37 EGFR-mutant patients with resistance to TKIs, EGFR amplification was identified in three patients (8%).50 Interestingly, all three patients had simultaneous T790M mutations. Consistent with preclinical models, two of these patients seemed to have selective amplification of the T790M-containing allele.50,81
ALK fusion gene amplification has also been identified as a cause of crizotinib resistance.60,61,80 This was initially suggested by cell line models in which amplification of wild-type EML4-ALK was sufficient to confer crizotinib resistance.80 Subsequent studies have confirmed ALK fusion gene amplification in resistant clinical specimens.60,61 In one report, high-level amplification was identified in one (7%) of 15 specimens,60 whereas a separate study showed ALK copy number gain in two (18%) of 11 patients.61 One of these patients also had a secondary ALK G1269A mutation.
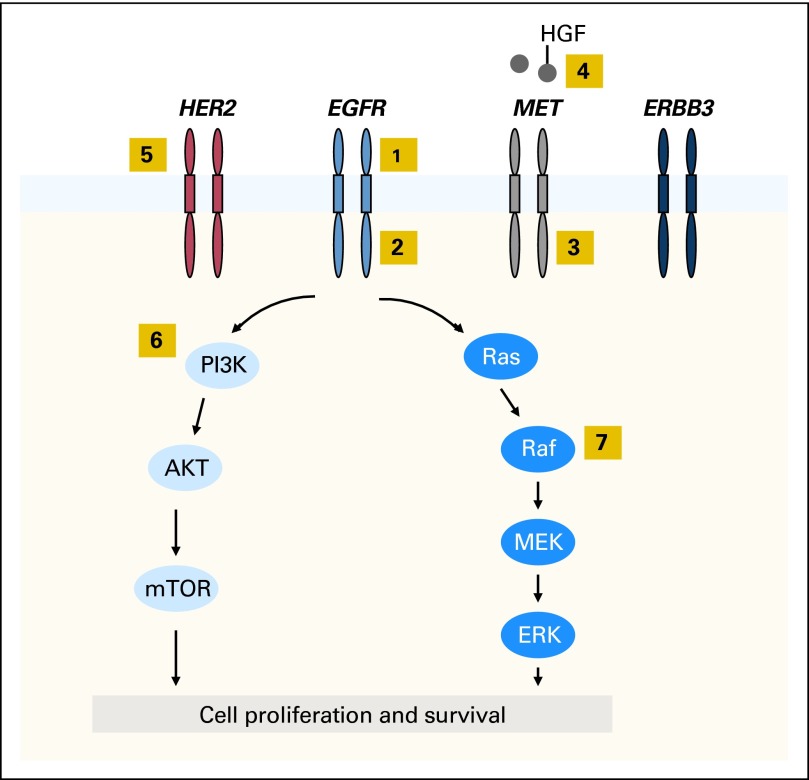
Bypass Signaling
TKI resistance can also develop through reactivation of downstream signaling pathways via bypass tracts (Fig 2). One well-described example in EGFR-mutant NSCLC is through MET amplification.35,82 Initially identified in 22% of EGFR TKI-resistant specimens, MET amplification confers resistance through ERBB3-mediated activation of downstream PI3K/AKT signaling, effectively bypassing the inhibited EGFR.35 Activation of MET through its ligand, hepatocyte growth factor, may also promote resistance.57,83 In more recent studies of EGFR TKI resistance, MET amplification has been identified in only 5% of specimens, perhaps reflecting differences in testing methodology and thresholds compared with those of earlier reports.50,51 Interestingly, MET-amplified subclones have been identified at low frequencies in untreated specimens.34 In a majority of these cases, the dominant mechanism of resistance at the time of disease progression was MET amplification, suggesting that these preexisting cells emerged as dominant clones as a result of selective pressure.
Fig 2.
Mechanisms of acquired resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors identified in EGFR-positive patient specimens. Activation of EGFR results in downstream signaling through the PI3K/AKT and RAS/RAF/MEK/ERK pathways, leading to cell proliferation and survival. The highlighted yellow boxes designate points along these pathways and others that have been associated with resistance to EGFR TKIs: (1) EGFR gene amplification, (2) secondary mutations in EGFR (eg, T790M), (3) MET amplification, (4) hepatocyte growth factor (HGF) –mediated MET activation, (5) HER2 amplification, (6) PIK3CA mutations, and (7) BRAF mutations.
Several additional bypass tracts have been implicated in EGFR TKI resistance.50,54–56 Recently, HER2 amplification was identified in three (12%) of 26 EGFR-mutant patients with acquired resistance to TKIs.54 When amplified, HER2 is believed to function in parallel to the inhibited EGFR to reactivate common downstream signaling pathways. Recently, genetic alterations in effectors downstream from EGFR have also been identified as potential mediators of resistance.50,55,84 For example, PIK3CA mutations have been identified in 5% of EGFR-mutant patients with acquired resistance.50 Preclinical studies suggest that these mutations confer resistance by activating downstream AKT.84 Recent studies have also focused on RAS/MAPK signaling as a source of EGFR TKI resistance because KRAS mutations have long been associated with primary resistance to EGFR inhibitors.85 Despite their role in primary resistance, no KRAS mutations have been detected in EGFR-mutant patients with acquired resistance.49,50,55 However, point mutations in BRAF, another member of the MAPK pathway, have been described.55
Bypass tracts also contribute to resistance in ALK-positive NSCLC. Preclinical studies identified EGFR coactivation as one potential mechanism of crizotinib resistance.59,60,86 In these models, EGFR activation did not occur through mutation. Instead, increases in EGFR ligands EGF86 and amphiregulin59 were observed. Recently, a collection of crizotinib-resistant, ALK-positive tumor specimens were examined for evidence of EGFR activation.60 Four (44%) of nine specimens demonstrated increased EGFR phosphorylation compared with precrizotinib samples, suggesting that EGFR upregulation may serve as a clinically relevant bypass track. A separate study also described one ALK-positive patient treated with crizotinib who was later found to have an EGFR mutation and a negative ALK FISH test in a repeat biopsy specimen.61 In this latter report, KRAS mutations were also identified in two ALK-positive patients after disease progression occurred during treatment with crizotinib, although one patient harbored this mutation pretreatment. Lastly, KIT gene amplification has been identified in two (15%) of 13 crizotinib-resistant specimens, suggesting that this signaling pathway may also be co-opted to mediate resistance.60
Phenotypic Alterations
Several groups have reported changes in tumor histology on development of resistance to EGFR TKIs. The most dramatic examples include transformation from NSCLC to small-cell lung cancer (SCLC) in a subset of patients.87,88 In one study of 37 EGFR-mutant patients with acquired resistance, repeat biopsies identified SCLC in five patients (14%), all of whom had adenocarcinoma at baseline.50 Interestingly, the original EGFR mutations were present in all SCLC specimens. Samples were negative for T790M and MET amplification, although one patient developed a new PIK3CA mutation. In a separate cohort of 106 EGFR-mutant patients, SCLC or high-grade neuroendocrine carcinoma were found in three TKI-resistant patients.51 The mechanistic basis for these observations remains unclear. Similar histologic changes have not been identified in ALK-positive patients.60
Another histologic change observed in EGFR TKI-resistant specimens is an epithelial to mesenchymal transition (EMT).50,89,90 EMT is characterized by loss of epithelial markers (eg, E-cadherin) and gain of mesenchymal features, including surface expression of vimentin.91 EMT is associated with enhanced motility, invasiveness, and in vitro EGFR TKI resistance.92–94 Clinically, EMT has been recognized in a subset of EGFR TKI-resistant specimens.50 The biology underlying this change and its impact on resistance remains unknown.
Other Mechanisms of Resistance
A subset of EGFR-mutant and ALK-positive patients have unknown mechanisms of TKI resistance. In preclinical models, EGFR TKI resistance has also been associated with insulin growth factor receptor signaling,95–97 nuclear factor κB activation,98 and loss of PTEN.99 Among ALK-positive patients, loss of the ALK fusion oncogene has also been raised as a potential mechanism of resistance.61 Additionally, resistance may be influenced by pharmacokinetic considerations. Gefitinib, erlotinib, and crizotinib are oral medications, which may be affected by absorption, patient compliance, drug–drug interactions, and metabolism. In CML, for example, plasma trough concentrations of imatinib have been correlated with response to therapy.100–102 Similarly, patient adherence rates to imatinib have been identified as independent predictors of response.103
Polyclonal Resistance
Occasionally, multiple different resistance mechanisms are found within the same biopsy specimen.50,51,60,61,82 Moreover, different resistance mechanisms may be found in separate tumor deposits within the same patient. For example, Yu et al104 recently identified an EGFR-mutant patient with T790M in one resistant sample and HER2 amplification in a separate specimen, underscoring the potential for clonal divergence across metastatic sites. Patients may have additional tumor heterogeneity in the form of TKI-sensitive cells admixed with resistant cells. Furthermore, the overall composition of such tumor populations may evolve with changes in therapy. For instance, EGFR-mutant and ALK-positive patients can experience a disease “flare” on TKI discontinuation, presumably because of accelerated growth of TKI-sensitive clones once selective pressure from the drug is removed.105,106 The dynamic nature of resistance underscores the value of repeat biopsies at each new phase of treatment to advance our understanding of resistance and guide clinical decision making. Nevertheless, care must be taken to balance biopsy-related risks and ensure adequate informed consent.107,108 This also emphasizes the need to develop noninvasive tools for monitoring resistance, such as mutational analysis of plasma DNA or circulating tumor cells.109,110
TREATMENT APPROACHES
Knowledge of the mechanisms underlying TKI resistance may inform new treatment strategies. We therefore conclude with an overview of treatment approaches for patients with TKI resistance. A more comprehensive discussion is beyond the scope of this review.
TKI Continuation Beyond Progression
In routine practice, oncologists typically discontinue a given therapy at the time of disease progression. It remains unclear, however, whether similar approaches should apply to TKIs in EGFR-mutant and ALK-positive patients, because resistance may be heterogeneous and TKI discontinuation may precipitate a disease flare.105,106 In cases of isolated progression (eg, CNS), local therapies followed by continuation of the relevant targeted therapy may be a viable approach in select patients.111–114 In EGFR-mutant patients for whom a switch to cytotoxic chemotherapy is ultimately deemed necessary, however, it remains unclear whether EGFR TKIs should be continued with standard chemotherapy. Several prospective clinical trials evaluating this question are currently ongoing (NCT01544179, NCT01310036).
Alternative Dosing
Another strategy is to use alternative doses or schedules of TKIs. In CML and GIST, for example, imatinib dose escalation has been effective in some patients experiencing disease progression at standard imatinib doses.24,115,116 In EGFR-mutant NSCLC, different dosing strategies may also be relevant because the FDA-approved dose of erlotinib was determined in unselected patients. Recent evolutionary mathematical modeling studies have proposed that alternative EGFR TKI doses and schedules may produce comparable results while delaying development of resistance.73,117
Next-Generation TKIs
Initial strategies to combat acquired resistance have centered on using next-generation TKIs. This approach has met with success in other oncogene-driven malignancies. In CML, four next-generation TKIs (dasatinib, nilotinib, bosutinib, and ponatinib) have shown activity in imatinib-resistant disease.118–123 One agent, ponatinib, is of particular note because it seems to overcome the gatekeeper T315I mutation.123 Unfortunately, similar strategies have been less successful in EGFR-mutant NSCLC. Second-generation EGFR TKIs, such as neratinib, dacomitinib, and afatinib, differ from gefitinib and erlotinib in that they form irreversible covalent bonds with EGFR.124 These agents also possess activity against other ERBB family members (eg, HER2). In preclinical models, irreversible EGFR TKIs demonstrated promising activity against T790M.125,126 Unfortunately, clinical trials of these agents in patients with acquired resistance have been largely disappointing, likely as a result of dose limitations from toxicity caused by inhibiting wild-type EGFR.127,128
Recently, third-generation EGFR inhibitors, such as WZ4002 and CO-1686, have been developed. In preclinical studies, these compounds are active against cell lines and murine models harboring T790M mutations.129,130 Moreover, WZ4002 and CO-1686 both seem to spare wild-type EGFR in vitro and in vivo. It is therefore hoped that these mutant-selective inhibitors will be able to overcome T790M-mediated resistance while producing less toxicity in the clinic.
Next-generation TKIs are also being investigated in ALK-positive patients. Indeed, five agents are currently in clinical testing131–135 (Table 3). In preliminary reporting, two of these compounds (LDK378 and AP26113) were associated with high ORRs in phase I trials of ALK-positive patients with crizotinib resistance.131,132 Both agents also demonstrated activity against brain metastases. This raises the possibility that next-generation ALK inhibitors may control disease in the CNS, which is among the most common sites of relapse among patients undergoing treatment with crizotinib. The mechanisms of crizotinib resistance for responders versus nonresponders to second-generation ALK TKIs have not been reported. Nevertheless, these encouraging early results suggest that use of more potent and/or structurally distinct ALK TKIs may be a promising strategy.
Table 3.
Ongoing Trials of Next-Generation ALK Inhibitors or ALK Inhibitor Combinations
| Compound(s) | Company | NCT Identifier | Description | Reference |
|---|---|---|---|---|
| AP26113 | Ariad | 01449461 | Phase I/II study of AP26113 in crizotinib-naïve and crizotinib-resistant, ALK-positive patients. | 132 |
| ASP3026 | Astellas | 01401504 | Phase I trial of ASP3026 in patients with solid tumors. | 133 |
| CH5424802 | Chugai | 01588028 | Phase I/II study of CH5424802 in crizotinib-naïve and crizotinib-resistant, ALK-positive patients. | 135 |
| LDK378 | Novartis | 01283516 | Phase I trial of LDK378 in crizotinib-naïve and crizotinib-resistant, ALK-positive patients. | 131 |
| 01685138 | Phase II trial of LDK378 in crizotinib-naïve, ALK-positive patients. | |||
| 01685060 | Phase II trial of LDK378 in ALK-positive patients previously treated with crizotinib and chemotherapy. | |||
| X-396 | Xcovery | 01625234 | Phase I study of X-396 in patients with solid tumors, including ALK-positive patients treated with crizotinib or other second-generation ALK TKIs. | NA |
| Crizotinib + STA-9090 | Pfizer/Synta | 01579994 | Phase I/II trial of combination crizotinib and STA-9090 (HSP90 inhibitor) in crizotinib-naïve, ALK-positive patients. | NA |
| Crizotinib + AT13387 | Pfizer/Astex | 01712217 | Phase I/II trial of AT13387 (HSP90 inhibitor) alone or in combination with crizotinib in ALK-positive patients. | NA |
Abbreviations: ALK, anaplastic lymphoma kinase; HSP90, Heat Shock Protein 90; NA, not applicable; NCT, National Clinical Trial.
Combinatorial Strategies
The diversity of resistance mechanisms in NSCLC provides a rationale for combinatorial approaches. Such strategies commonly aim to inhibit the primary oncogene in addition to compensatory signaling pathways. One such approach in EGFR-mutant NSCLC has been to use dual MET and EGFR TKIs in those patients with MET amplification.35,136 Similar combination strategies may have a role in ALK-positive NSCLC. As detailed earlier, KIT gene amplification and EGFR activation are possible mediators of crizotinib resistance. Given the availability of KIT and EGFR inhibitors already in clinical practice, KIT and EGFR may be effective targets for combination therapy. Indeed, crizotinib in combination with KIT inhibitors60 or EGFR inhibitors59,60,86 demonstrated activity in cell lines with upregulation of each respective kinase.
Combination strategies may also be considered when resistance arises through secondary mutations in the primary oncogene. In transgenic mouse models harboring EGFR T790M mutations, concurrent administration of the irreversible EGFR TKI afatinib and the EGFR monoclonal antibody cetuximab resulted in dramatic tumor shrinkage.137 In a phase I/II trial of this combination, responses were observed in 40% of patients with EGFR TKI resistance.138 Common adverse events were rash and diarrhea. The mechanisms underlying disease activity from this combination remain unclear.
Additional combinations are currently being investigated. Given the association between BIM levels and apoptotic response to TKIs, proposed strategies include combinations of EGFR TKIs and modulators of apoptosis.42–45 Still another approach to combat resistance in NSCLC is to target the molecular chaperone heat shock protein 90 (HSP90) because certain oncogenic kinases rely on this protein for proper folding. HSP90 inhibitors have demonstrated efficacy in EGFR-mutant cell lines and murine models harboring secondary mutations, including T790M.139 Clinical studies using HSP90 inhibitors in patients with EGFR TKI resistance are ongoing, but responses have been reported.140 Preclinical and early clinical findings suggest that HSP90 inhibitors may also have activity in ALK-positive patients.60,80,140–143 HSP90 inhibitors may therefore be attractive options for use alone or in combination with TKIs in the management of resistance.
In summary, although kinase-directed therapies have reshaped treatment approaches in oncogene-driven NSCLC, these therapies have been universally limited by the development of resistance. It is therefore vital to develop new paradigms for understanding the mechanisms driving TKI resistance. EGFR-mutant and ALK-rearranged lung cancers offer instructive conceptual frameworks. Both highlight common principles of resistance, such as the development of secondary mutations in the target kinase, target gene amplification, and activation of bypass tracts. However, experiences in both malignancies also demonstrate the complexity, heterogeneity, and dynamic nature of resistance, suggesting that resistance will need to be approached on a truly “personalized” basis. Although future directions include development of noninvasive genotyping tools, the dynamic nature of resistance highlights the current importance of serial biopsies at the time of disease progression. Such reassessments of the changing molecular profiles of tumors may influence the development of novel therapeutic strategies and inform rational trial design.
Footnotes
See accompanying article on page 3926
Supported by National Institutes of Health Grant No. 5R01CA164273-02, by a V Foundation Translational Research grant, and by the Evan Spirito Memorial Foundation for support of lung cancer research. A.T.S. is the Charles W. and Jennifer C. Johnson MIT Koch Institute Clinical Investigator.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) and/or an author's immediate family member(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Alice T. Shaw, Pfizer (C), Novartis (C), Ariad (C), Chugai (C), Daiichi-sankyo (C) Stock Ownership: None Honoraria: None Research Funding: None Expert Testimony: None Patents: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Weinstein IB. Cancer: Addiction to oncogenes—The Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 2.Druker BJ, Guilhot F, O'Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 3.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 4.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 7.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 8.Apperley JF. Part I: Mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8:1018–1029. doi: 10.1016/S1470-2045(07)70342-X. [DOI] [PubMed] [Google Scholar]
- 9.Gramza AW, Corless CL, Heinrich MC. Resistance to tyrosine kinase inhibitors in gastrointestinal stromal tumors. Clin Cancer Res. 2009;15:7510–7518. doi: 10.1158/1078-0432.CCR-09-0190. [DOI] [PubMed] [Google Scholar]
- 10.Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 13.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 14.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 15.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 16.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 17.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi K, Choi YL, Soda M, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res. 2008;14:6618–6624. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- 19.Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: Updated results from a phase 1 study. Lancet Oncol. 2012;13:1011–1019. doi: 10.1016/S1470-2045(12)70344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D, Ahn M, Shi Y, et al. Results of a global phase II study with crizotinib in advanced ALK-positive non-small cell lung cancer (NSCLC) J Clin Oncol. 2012;30(suppl):488s. abstr 7533. [Google Scholar]
- 24.Baccarani M, Cortes J, Pane F, et al. Chronic myeloid leukemia: An update of concepts and management recommendations of European LeukemiaNet. J Clin Oncol. 2009;27:6041–6051. doi: 10.1200/JCO.2009.25.0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Glabbeke M, Verweij J, Casali PG, et al. Initial and late resistance to imatinib in advanced gastrointestinal stromal tumors are predicted by different prognostic factors: A European Organisation for Research and Treatment of Cancer-Italian Sarcoma Group-Australasian Gastrointestinal Trials Group study. J Clin Oncol. 2005;23:5795–5804. doi: 10.1200/JCO.2005.11.601. [DOI] [PubMed] [Google Scholar]
- 26.Jackman D, Pao W, Riely GJ, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol. 2010;28:357–360. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: Preclinical data and clinical implications. Lancet Oncol. 2012;13:e23–e31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 29.Wu JY, Wu SG, Yang CH, et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res. 2008;14:4877–4882. doi: 10.1158/1078-0432.CCR-07-5123. [DOI] [PubMed] [Google Scholar]
- 30.Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inukai M, Toyooka S, Ito S, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66:7854–7858. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 32.Tokumo M, Toyooka S, Ichihara S, et al. Double mutation and gene copy number of EGFR in gefitinib refractory non-small-cell lung cancer. Lung Cancer. 2006;53:117–121. doi: 10.1016/j.lungcan.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 34.Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 36.Heuckmann JM, Balke-Want H, Malchers F, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res. 2012;18:4682–4690. doi: 10.1158/1078-0432.CCR-11-3260. [DOI] [PubMed] [Google Scholar]
- 37.Ou SH, Bartlett CH, Mino-Kenudson M, et al. Crizotinib for the treatment of ALK-rearranged non-small cell lung cancer: A success story to usher in the second decade of molecular targeted therapy in oncology. Oncologist. 2012;17:1351–1375. doi: 10.1634/theoncologist.2012-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw AT, Solomon B, Kenudson MM. Crizotinib and testing for ALK. J Natl Compr Canc Netw. 2011;9:1335–1341. doi: 10.6004/jnccn.2011.0115. [DOI] [PubMed] [Google Scholar]
- 39.Camidge DR, Kono SA, Flacco A, et al. Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatment. Clin Cancer Res. 2010;16:5581–5590. doi: 10.1158/1078-0432.CCR-10-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuribara R, Honda H, Matsui H, et al. Roles of Bim in apoptosis of normal and Bcr-Abl-expressing hematopoietic progenitors. Mol Cell Biol. 2004;24:6172–6183. doi: 10.1128/MCB.24.14.6172-6183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuroda J, Puthalakath H, Cragg MS, et al. Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukemic cells, and resistance due to their loss is overcome by a BH3 mimetic. Proc Natl Acad Sci U S A. 2006;103:14907–14912. doi: 10.1073/pnas.0606176103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costa DB, Halmos B, Kumar A, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 4:1669–1679. doi: 10.1371/journal.pmed.0040315. discussion 1680, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cragg MS, Kuroda J, Puthalakath H, et al. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med. 4:1681–1689. doi: 10.1371/journal.pmed.0040316. discussion 1690, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong Y, Somwar R, Politi K, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faber AC, Corcoran RB, Ebi H, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takezawa K, Okamoto I, Nishio K, et al. Role of ERK-BIM and STAT3-survivin signaling pathways in ALK inhibitor-induced apoptosis in EML4-ALK-positive lung cancer. Clin Cancer Res. 2011;17:2140–2148. doi: 10.1158/1078-0432.CCR-10-2798. [DOI] [PubMed] [Google Scholar]
- 47.Ng KP, Hillmer AM, Chuah CT, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 49.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res. 2011;17:1169–1180. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 53.Bean J, Riely GJ, Balak M, et al. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clin Cancer Res. 2008;14:7519–7525. doi: 10.1158/1078-0432.CCR-08-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 amplification: A potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2:922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohashi K, Sequist LV, Arcila ME, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109:E2127–2133. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheung HW, Du J, Boehm JS, et al. Amplification of CRKL induces transformation and epidermal growth factor receptor inhibitor resistance in human non-small cell lung cancers. Cancer Discov. 2011;1:608–625. doi: 10.1158/2159-8290.CD-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yano S, Wang W, Li Q, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–9487. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- 58.Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 59.Sasaki T, Koivunen J, Ogino A, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res. 2011;71:6051–6060. doi: 10.1158/0008-5472.CAN-11-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med. 2012;4:120ra17. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18:1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–79. doi: 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 63.Engelman JA, Jänne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 64.Sierra JR, Cepero V, Giordano S. Molecular mechanisms of acquired resistance to tyrosine kinase targeted therapy. Mol Cancer. 2010;9:75. doi: 10.1186/1476-4598-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ellis LM, Hicklin DJ. Resistance to targeted therapies: Refining anticancer therapy in the era of molecular oncology. Clin Cancer Res. 2009;15:7471–7478. doi: 10.1158/1078-0432.CCR-09-1070. [DOI] [PubMed] [Google Scholar]
- 66.Bixby D, Talpaz M. Mechanisms of resistance to tyrosine kinase inhibitors in chronic myeloid leukemia and recent therapeutic strategies to overcome resistance. Hematology Am Soc Hematol Educ Program. 2009:461–476. doi: 10.1182/asheducation-2009.1.461. [DOI] [PubMed] [Google Scholar]
- 67.Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 68.Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, et al. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100:1014–1018. doi: 10.1182/blood.v100.3.1014. [DOI] [PubMed] [Google Scholar]
- 69.Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li D, Shimamura T, Ji H, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12:81–93. doi: 10.1016/j.ccr.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 71.Godin-Heymann N, Bryant I, Rivera MN, et al. Oncogenic activity of epidermal growth factor receptor kinase mutant alleles is enhanced by the T790M drug resistance mutation. Cancer Res. 2007;67:7319–7326. doi: 10.1158/0008-5472.CAN-06-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mulloy R, Ferrand A, Kim Y, et al. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res. 2007;67:2325–2330. doi: 10.1158/0008-5472.CAN-06-4293. [DOI] [PubMed] [Google Scholar]
- 73.Chmielecki J, Foo J, Oxnard GR, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oxnard GR, Arcila ME, Sima CS, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: Distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17:1616–1622. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lovly CM, Pao W. Escaping ALK inhibition: Mechanisms of and strategies to overcome resistance. Sci Transl Med. 2012;4:120ps2. doi: 10.1126/scitranslmed.3003728. [DOI] [PubMed] [Google Scholar]
- 76.Zhang S, Wang F, Keats J, et al. Crizotinib-resistant mutants of EML4-ALK identified through an accelerated mutagenesis screen. Chem Biol Drug Des. 2011;78:999–1005. doi: 10.1111/j.1747-0285.2011.01239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sasaki T, Okuda K, Zheng W, et al. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer Res. 2010;70:10038–10043. doi: 10.1158/0008-5472.CAN-10-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.George RE, Sanda T, Hanna M, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Avizienyte E, Ward RA, Garner AP. Comparison of the EGFR resistance mutation profiles generated by EGFR-targeted tyrosine kinase inhibitors and the impact of drug combinations. Biochem J. 2008;415:197–206. doi: 10.1042/BJ20080728. [DOI] [PubMed] [Google Scholar]
- 80.Katayama R, Khan TM, Benes C, et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc Natl Acad Sci U S A. 2011;108:7535–7540. doi: 10.1073/pnas.1019559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ercan D, Zejnullahu K, Yonesaka K, et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene. 2010;29:2346–2356. doi: 10.1038/onc.2009.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamada T, Matsumoto K, Wang W, et al. Hepatocyte growth factor reduces susceptibility to an irreversible epidermal growth factor receptor inhibitor in EGFR-T790M mutant lung cancer. Clin Cancer Res. 2010;16:174–183. doi: 10.1158/1078-0432.CCR-09-1204. [DOI] [PubMed] [Google Scholar]
- 84.Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tanizaki J, Okamoto I, Okabe T, et al. Activation of HER family signaling as a mechanism of acquired resistance to ALK inhibitors in EML4-ALK-positive non-small cell lung cancer. Clin Cancer Res. 2012;18:6219–6226. doi: 10.1158/1078-0432.CCR-12-0392. [DOI] [PubMed] [Google Scholar]
- 87.Zakowski MF, Ladanyi M, Kris MG, et al. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med. 2006;355:213–215. doi: 10.1056/NEJMc053610. [DOI] [PubMed] [Google Scholar]
- 88.Morinaga R, Okamoto I, Furuta K, et al. Sequential occurrence of non-small cell and small cell lung cancer with the same EGFR mutation. Lung Cancer. 2007;58:411–413. doi: 10.1016/j.lungcan.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 89.Uramoto H, Iwata T, Onitsuka T, et al. Epithelial-mesenchymal transition in EGFR-TKI acquired resistant lung adenocarcinoma. Anticancer Res. 2010;30:2513–2517. [PubMed] [Google Scholar]
- 90.Chung JH, Rho JK, Xu X, et al. Clinical and molecular evidences of epithelial to mesenchymal transition in acquired resistance to EGFR-TKIs. Lung Cancer. 2011;73:176–182. doi: 10.1016/j.lungcan.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 91.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 92.Thomson S, Buck E, Petti F, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–9462. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 93.Rho JK, Choi YJ, Lee JK, et al. Epithelial to mesenchymal transition derived from repeated exposure to gefitinib determines the sensitivity to EGFR inhibitors in A549, a non-small cell lung cancer cell line. Lung Cancer. 2009;63:219–226. doi: 10.1016/j.lungcan.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 94.Suda K, Tomizawa K, Fujii M, et al. Epithelial to mesenchymal transition in an epidermal growth factor receptor-mutant lung cancer cell line with acquired resistance to erlotinib. J Thorac Oncol. 2011;6:1152–1161. doi: 10.1097/JTO.0b013e318216ee52. [DOI] [PubMed] [Google Scholar]
- 95.Guix M, Faber AC, Wang SE, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gong Y, Yao E, Shen R, et al. High expression levels of total IGF-1R and sensitivity of NSCLC cells in vitro to an anti-IGF-1R antibody (R1507) PLoS One. 2009;4:e7273. doi: 10.1371/journal.pone.0007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bivona TG, Hieronymus H, Parker J, et al. FAS and NF-κB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yamasaki F, Johansen MJ, Zhang D, et al. Acquired resistance to erlotinib in A-431 epidermoid cancer cells requires down-regulation of MMAC1/PTEN and up-regulation of phosphorylated Akt. Cancer Res. 2007;67:5779–5788. doi: 10.1158/0008-5472.CAN-06-3020. [DOI] [PubMed] [Google Scholar]
- 100.Larson RA, Druker BJ, Guilhot F, et al. Imatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia: A subanalysis of the IRIS study. Blood. 2008;111:4022–4028. doi: 10.1182/blood-2007-10-116475. [DOI] [PubMed] [Google Scholar]
- 101.Picard S, Titier K, Etienne G, et al. Trough imatinib plasma levels are associated with both cytogenetic and molecular responses to standard-dose imatinib in chronic myeloid leukemia. Blood. 2007;109:3496–3499. doi: 10.1182/blood-2006-07-036012. [DOI] [PubMed] [Google Scholar]
- 102.Singh N, Kumar L, Meena R, et al. Drug monitoring of imatinib levels in patients undergoing therapy for chronic myeloid leukaemia: Comparing plasma levels of responders and non-responders. Eur J Clin Pharmacol. 2009;65:545–549. doi: 10.1007/s00228-009-0621-z. [DOI] [PubMed] [Google Scholar]
- 103.Marin D, Bazeos A, Mahon FX, et al. Adherence is the critical factor for achieving molecular responses in patients with chronic myeloid leukemia who achieve complete cytogenetic responses on imatinib. J Clin Oncol. 2010;28:2381–2388. doi: 10.1200/JCO.2009.26.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chaft JE, Oxnard GR, Sima CS, et al. Disease flare after tyrosine kinase inhibitor discontinuation in patients with EGFR-mutant lung cancer and acquired resistance to erlotinib or gefitinib: Implications for clinical trial design. Clin Cancer Res. 2011;17:6298–6303. doi: 10.1158/1078-0432.CCR-11-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pop O, Pirvu A, Toffart AC, et al. Disease flare after treatment discontinuation in a patient with EML4-ALK lung cancer and acquired resistance to crizotinib. J Thorac Oncol. 2012;7:e1–e2. doi: 10.1097/JTO.0b013e318257fc1d. [DOI] [PubMed] [Google Scholar]
- 107.Overman MJ, Modak J, Kopetz S, et al. Use of research biopsies in clinical trials: Are risks and benefits adequately discussed? J Clin Oncol. 2013;31:17–22. doi: 10.1200/JCO.2012.43.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Peppercorn J. Toward improved understanding of the ethical and clinical issues surrounding mandatory research biopsies. J Clin Oncol. 2013;31:1–2. doi: 10.1200/JCO.2012.44.8589. [DOI] [PubMed] [Google Scholar]
- 109.Kuang Y, Rogers A, Yeap BY, et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res. 2009;15:2630–2636. doi: 10.1158/1078-0432.CCR-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Weickhardt AJ, Scheier B, Burke JM, et al. Local ablative therapy of oligoprogressive disease prolongs disease control by tyrosine kinase inhibitors in oncogene-addicted non-small-cell lung cancer. J Thorac Oncol. 2012;7:1807–1814. doi: 10.1097/JTO.0b013e3182745948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shukuya T, Takahashi T, Naito T, et al. Continuous EGFR-TKI administration following radiotherapy for non-small cell lung cancer patients with isolated CNS failure. Lung Cancer. 2011;74:457–461. doi: 10.1016/j.lungcan.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 113.Yu H, Sima C, Drilon A, et al. Local therapy as a treatment strategy in EGFR-mutant advanced lung cancers that have developed acquired resistance to EGFR tyrosine kinase inhibitors. J Clin Oncol. 2012;30(suppl):486s. doi: 10.1097/JTO.0b013e31827e1f83. abstr 7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oxnard G, Lo P, Jackman D, et al. Delay of chemotherapy through use of post-progression erlotinib in patients with EGFR-mutant lung cancer. J Clin Oncol. 2012;30(suppl):491s. abstr 7547. [Google Scholar]
- 115.Kantarjian HM, Talpaz M, O'Brien S, et al. Dose escalation of imatinib mesylate can overcome resistance to standard-dose therapy in patients with chronic myelogenous leukemia. Blood. 2003;101:473–475. doi: 10.1182/blood-2002-05-1451. [DOI] [PubMed] [Google Scholar]
- 116.Blanke CD, Rankin C, Demetri GD, et al. Phase III randomized, intergroup trial assessing imatinib mesylate at two dose levels in patients with unresectable or metastatic gastrointestinal stromal tumors expressing the kit receptor tyrosine kinase: S0033. J Clin Oncol. 2008;26:626–632. doi: 10.1200/JCO.2007.13.4452. [DOI] [PubMed] [Google Scholar]
- 117.Foo J, Chmielecki J, Pao W, et al. Effects of pharmacokinetic processes and varied dosing schedules on the dynamics of acquired resistance to erlotinib in EGFR-mutant lung cancer. J Thorac Oncol. 2012;7:1583–1593. doi: 10.1097/JTO.0b013e31826146ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Talpaz M, Shah NP, Kantarjian H, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 119.Hochhaus A, Kantarjian HM, Baccarani M, et al. Dasatinib induces notable hematologic and cytogenetic responses in chronic-phase chronic myeloid leukemia after failure of imatinib therapy. Blood. 2007;109:2303–2309. doi: 10.1182/blood-2006-09-047266. [DOI] [PubMed] [Google Scholar]
- 120.Kantarjian H, Giles F, Wunderle L, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 121.Kantarjian HM, Giles F, Gattermann N, et al. Nilotinib (formerly AMN107), a highly selective BCR-ABL tyrosine kinase inhibitor, is effective in patients with Philadelphia chromosome-positive chronic myelogenous leukemia in chronic phase following imatinib resistance and intolerance. Blood. 2007;110:3540–3546. doi: 10.1182/blood-2007-03-080689. [DOI] [PubMed] [Google Scholar]
- 122.Cortes JE, Kantarjian HM, Brümmendorf TH, et al. Safety and efficacy of bosutinib (SKI-606) in chronic phase Philadelphia chromosome-positive chronic myeloid leukemia patients with resistance or intolerance to imatinib. Blood. 2011;118:4567–4576. doi: 10.1182/blood-2011-05-355594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cortes JE, Kantarjian H, Shah NP, et al. Ponatinib in refractory Philadelphia chromosome-positive leukemias. N Engl J Med. 2012;367:2075–2088. doi: 10.1056/NEJMoa1205127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ou SH. Second-generation irreversible epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs): A better mousetrap? A review of the clinical evidence. Crit Rev Oncol Hematol. 2012;83:407–421. doi: 10.1016/j.critrevonc.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 125.Kwak EL, Sordella R, Bell DW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Engelman JA, Zejnullahu K, Gale CM, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007;67:11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- 127.Sequist LV, Besse B, Lynch TJ, et al. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: Results of a phase II trial in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:3076–3083. doi: 10.1200/JCO.2009.27.9414. [DOI] [PubMed] [Google Scholar]
- 128.Miller VA, Hirsh V, Cadranel J, et al. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): A phase 2b/3 randomised trial. Lancet Oncol. 2012;13:528–538. doi: 10.1016/S1470-2045(12)70087-6. [DOI] [PubMed] [Google Scholar]
- 129.Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Walter A, Tjin R, Haringsma H, et al. CO-1686, an orally available, mutant-slective inhibitor of the epidermal growth factor receptor (EGFR), causes tumor shrinkage in non-small cell lung cancer (NSCLC) with T790M mutations. Presented at the International Conference of the American Association for Cancer Res-National Cancer Institute-European Organization for Research and Treatment of Cancer, San Francisco, CA, November. 2011:12–16. [Google Scholar]
- 131.Shaw A, Camidge R, Felip E, et al. Results of a first-in-human phase I study of the ALK inhibitor LDK378 in advanced solid tumors. Ann Oncol. 2012;23(suppl):153. abstr 440O. [Google Scholar]
- 132.Gettinger S, Weiss G, Salgia R, et al. A first-in-human dose-finding study of the ALK/EGFR inhibitor AP26113 in patients with advanced malignancies. Ann Oncol. 2012;23(suppl):152. abstr 439O. [Google Scholar]
- 133.Kuromitsu S, Mori M, Shimada I, et al. Antitumor activities of ASP3026 against EML4-ALK-dependent tumor models. Presented at the International Conference of the American Association for Cancer Res-National Cancer Institute-European Organization for Research and Treatment of Cancer, San Francisco, CA, November. 2011:12–16. [Google Scholar]
- 134.Sakamoto H, Tsukaguchi T, Hiroshima S, et al. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell. 2011;19:679–690. doi: 10.1016/j.ccr.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 135.Nishio M, Kiura K, Nakagawa K, et al. A phase I/II study of ALK inhibitor CH5424802 in patients with ALK-positive NSCLC; safety and efficacy interim results of the phase II portion. Ann Oncol. 2012;23(suppl):153. abstr 441O. [Google Scholar]
- 136.Xu L, Kikuchi E, Xu C, et al. Combined EGFR/MET or EGFR/HSP90 inhibition is effective in the treatment of lung cancers codriven by mutant EGFR containing T790M and MET. Cancer Res. 2012;72:3302–3311. doi: 10.1158/0008-5472.CAN-11-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Regales L, Gong Y, Shen R, et al. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J Clin Invest. 2009;119:3000–3010. doi: 10.1172/JCI38746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Janjigian Y, Smit E, Horn L, et al. Activity of afatinib/cetuximab in patients (pts) with EGFR mutant non-small cell lung cancer (NSCLC) and acquired resistance (AR) to EGFR inhibitors. Ann Oncol. 2012;23(suppl):401. abstr 12270. [Google Scholar]
- 139.Shimamura T, Li D, Ji H, et al. Hsp90 inhibition suppresses mutant EGFR-T790M signaling and overcomes kinase inhibitor resistance. Cancer Res. 2008;68:5827–5838. doi: 10.1158/0008-5472.CAN-07-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Garon E, Moran T, Barlesi F, et al. Phase II study of the HSP90 inhibitor AUY922 in patients with previously treated, advanced non-small cell lung cancer (NSCLC) J Clin Oncol. 2012;30(suppl):490s. abstr 7543. [Google Scholar]
- 141.Chen Z, Sasaki T, Tan X, et al. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res. 2010;70:9827–9836. doi: 10.1158/0008-5472.CAN-10-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Normant E, Paez G, West KA, et al. The Hsp90 inhibitor IPI-504 rapidly lowers EML4-ALK levels and induces tumor regression in ALK-driven NSCLC models. Oncogene. 2011;30:2581–2586. doi: 10.1038/onc.2010.625. [DOI] [PubMed] [Google Scholar]
- 143.Sequist LV, Gettinger S, Senzer NN, et al. Activity of IPI-504, a novel heat-shock protein 90 inhibitor, in patients with molecularly defined non-small-cell lung cancer. J Clin Oncol. 2010;28:4953–4960. doi: 10.1200/JCO.2010.30.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]