Abstract
Purpose
KU7 is a popular urothelial carcinoma cell line that was isolated from the bladder of a patient at the Keio University (KU) in 1980. It has subsequently been widely used in laboratories around the world. Here we describe how routine cell line authentication has revealed that KU7 was cross-contaminated almost 30 years ago with HeLa, a cervical carcinoma cell line.
Materials and Methods
Presumed KU7 clones dating from 1984 to 1999 were provided by MD Anderson Cancer Center (MDACC), the Vancouver Prostate Centre (VPC), Kyoto University, Tokyo Medical University and KU, and HeLa was purchased from the ATCC. Genomic DNA was isolated and short tandem repeat (STR) analysis was performed by the Characterized Cell Line Core Facility at MDACC, the Fragment Analysis Facility at Johns Hopkins University and the RIKEN Bioresource Center (Japan). Comparative genomic hybridization (CGH) was performed on the Agilent platform at the VPC.
Results
The STR profile of all KU7 clones was an exact match with HeLa. The CGH of all samples revealed an abundance of shared chromosomal aberrations. Slight differences in some genomic areas are explained by genomic drift occurring in different KU7 clones separated by many years.
Conclusions
Our analysis identified that a cross-contamination of KU7 with HeLa occurred prior to 1984 at the source institution. All KU7 clones in the urologic literature should be considered HeLa and the experimental results should be viewed in this light. Our results emphasize the need to authenticate cell lines in oncologic research.
Keywords: Bladder cancer cell lines, cross-contamination, KU7, HeLa
Introduction
Bladder cancer is the sixth most common cancer in the North America.1,2 Approximately 75% of cases are non-muscle invasive bladder tumors that have a high propensity for recurrence and a variable risk of progression to more invasive disease.3,4 The remaining 25% of cases are muscle invasive at the time of diagnosis. These tumors are at risk for local progression as well as nodal and systemic metastasis.5 Laboratory research to identify new treatment paradigms for both disease scenarios lags behind many other cancers, including a number of less common cancers.
Human cancer cell lines are an essential tool in furthering our understanding of cancer biology and developing novel therapies. The first cell line which was successfully propagated and permanently cultivated in vitro was HeLa, derived from an epidermoid carcinoma of the human cervix at Johns Hopkins University (Baltimore, MD) in 1951.6,7,8 In the subsequent decades, numerous cancer cell lines from various tumors including bladder cancer were described and established.9,10 Compared to other organ sites such as prostate cancer, we have a relative luxury of multiple cell lines for bladder cancer research. One of the most popular bladder cancer cell lines has been KU7, which was isolated from a patient with low grade papillary bladder cancer at the Keio University (KU) in 1980.11 KU7 has been widely used due to its robust growth in vitro, its amenability to molecular manipulation12,13, and its reliable growth characteristics in xenograft models.14,15
Cross-contamination and misidentification of human cancer cell lines was first reported for HeLa by Gartler in 196716 and subsequently was recognized as a major issue in academic research. In 1983 a contamination of putative independent bladder cancer cell lines by T24, a cell line established in 1970 in Prague9, was revealed by analysis of HLA (human leukocyte antigen) and isoenzyme patterns.17 Despite common knowledge about existing cross-contamination18 a notice of the National institutes of Health (NIH) was not sent out until 2007.19 As a consequence routine cell line authentication has become common practice recently20,21, and has been mandated by major cancer research journals. The most established technique for cell line authentication is Short Tandem Repeat (STR) analysis, which involves the measurement of the number of specific short repetitive sequences at specific loci throughout the genome.20,21 Through initiation of such practices at The University of Texas, MD Anderson Cancer Center (MDACC, Houston, Texas), cross-contamination of KU7 cells with HeLa was revealed. Since KU7 has been used frequently throughout the world in the past 30 years, we aimed to trace the roots of KU7 clones from different centers in order to investigate the extent of KU7 cross-contamination.
Material and Methods
Cell lines and cell culture
Presumed KU7 clones were provided from the Keio University and research institutions which received frozen stocks from Keio University in the 1980s, 1990s and 2000s (Pathology Core of the GU SPORE in Bladder Cancer at MDACC, Kyoto University, Tokyo Medical University). The clone at Vancouver Prostate Centre (VPC) originally was shipped from MDACC in 2005. HeLa was commercially purchased from the American Type Culture Collection (ATCC). Frozen stocks were thawed and maintained as monolayer cultures on 10cm dishes in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum at 37°C in humidified 5% CO2 atmosphere.
DNA isolation
At cell confluence genomic DNA of all clones was isolated by the DNeasy® Tissue Kit (QIAGEN, Valencia, CA) in a clean environment. DNA purity was validated by measuring the ratio of 260nm/280nm absorbance with a Nanodrop 2000® spectrophotometer (Thermo Scientific; Wilmington, DE). Samples with a ratio under 1.87 were excluded from further analysis.
Short Tandem Repeat (STR) profile
In order to reveal possible genetic relationships among the available clones genomic DNA was analyzed by short tandem repeats (STR) DNA fingerprinting in the Fragment Analysis Facility at Johns Hopkins University. STRs are short sequences of DNA, normally 2–5 base pairs in length, that are repeated numerous times at specific sites in the genome. The number of repeats at different sites is specific to an individual in a population and can be used for genetic identification (e.g. paternity testing). For this purpose, DNA was isolated from the cell lines and residual RNA was removed by RNase prior to submission of samples. The Identiflier® kit (Applied Biosystems, Carlsbad, CA) was used to analyse STRs of either 10 (CSF1PO, D13S317, D16S539, D21S11, D5S818, D7S820, FGA, THO1, TPOX, vWA for KU7 Keio 1984) or 15 microsatellite regions (CSF1PO, D13S317, D16S539, D18S51, D19S433, D21S11, D2S1338, D3S1358, D5S818, D7S820, D8S1179, FGA, THO1, TPOX, vWA for all other clones). The difference in the number of regions analyzed was based strictly on a change in routine methodology at the reference laboratory. A polymerase chain reaction (PCR) was performed using three-color detection fluorescent dye-linked primers. After amplification the PCR products were separated by size and color using the capillary electrophoresis instrument ABI PRISM® 3730xl (Applied Biosystems). Analysis of the amplifications was subsequently performed with the GeneMapper® v 4.0 software (Applied Biosystems). For comparison, the amplifications of all samples were compared to 9 specific microsatellite regions (CSF1PO, D13S317, D16S539, D21S11, D5S818, D7S820, THO1, TPOX, vWA) in the ATCC database and percent match of STR was calculated. Amelogenin was analyzed for gender identification.
Independent STR fingerprint analysis of KU7 clones was performed by the Characterized Cell Line Core Facility at the MDACC (for KU7 MDACC 1999) and at the RIKEN Bioresource Center in Ibaraki, Japan (for KU7 Tokyo).
Array comparative genomic hybridization
In order to reveal larger scale chromosomal aberrations the whole genome was analysed by array comparative genomic hybridization (aCGH) on the Agilent Human Genome CGH Microarray® platform (Agilent, Santa Clara, CA) at the VPC. Array CGH reveals copy number gains and losses in the genome that are specific to an individual in a population. Genomic DNA from each KU7 clone was quantified by the Nanodrop 2000® spectrophotometer (ThermoScientific)and a quantity of 0.5µg was fluorescently labeled according to the NimbleGen enzymatic labeling protocol (NimbleGen Arrays User Guide CGH Analysis v6.0, Roche NimbleGen, Madison, WI). 5µg of each Cy5-labeled sample was co-hybridized with 5µg of gender matched Cy3-labeled human female reference DNA (Promega, Madison, WI) on Agilent SurePrint G3 Human® CGH 4×180K microarrays. Arrays were scanned with the Agilent DNA Microarray Scanner at a 3µm scan resolution, and quantified with Feature Extraction® 10.10.1.1 software (Agilent). CGH processed signal was then uploaded into Nexus CGH® software (Biodiscovery, Hawthorne, CA) where the quality was assessed and data was visualized and analyzed.
Results
All analyzed KU7 clones showed a match of their STR profile to HeLa in the ATCC database and on direct comparison to cultured HeLa (HeLa ATCC) [Tab. 1, Fig. 1]. These observations were confirmed with the independent STR fingerprint analysis at the Characterized Cell Line Core Facility (MDACC) and the RIKEN Bioresource Center (Ibaraki, Japan) [Tab.1].
Table 1.
STR Profile of all analysed cell clones
| Cell line | HeLa | KU7 | |||||
|---|---|---|---|---|---|---|---|
| Origin | ATCC | Keio | Kyoto | Tokyo | MDACC (1999) |
MDACC (2008) |
VPC |
| Loci | Quantity of repeating units | ||||||
| AMEL | X | X | X | X | X | X | X |
| CSF1PO | 9,10 | 9,10 | 9,10 | 9,10 | 9,10 | 9,10 | 9,10 |
| D13S317 | 12,13.3 | 12,13.3 | 12,13.3 | 12 | 12 | 12,13.3 | 12,13.3 |
| D16S539 | 9,10 | 9,10 | 9,10 | 9,10 | 9,10 | 9,10 | 9,10 |
| D18S51 | 16 | No analysis | 16 | 16 | 16 | 16 | 16 |
| D19S433 | 13,14 | No analysis | 13,14 | No analysis | 13,14 | 13,14 | 13,14 |
| D21S11 | 27,28 | 27,28 | 27,28 | 27,28 | 27,28 | 27,28 | 27,28 |
| D2S1338 | 17 | No analysis | 17 | No analysis | 17 | 17 | 17 |
| D3S1358 | 15,18 | No analysis | 15,18 | 15,18 | 15,18 | 15,18 | 15,18 |
| D5S818 | 11,12 | 11,12 | 11,12 | 11,12 | 11,12 | 11,12 | 11,12 |
| D7S820 | 8,12 | 8,12 | 8,12 | 8,12 | 8,12 | 8,12 | 8,12 |
| D8S1179 | 12,13 | No analysis | 12,13 | 12 | 12,13 | 12,13 | 12,13 |
| FGA | 18,21 | 18,21 | 18,21 | 18,21 | 18,21 | 18,21 | 18,21 |
| TH01 | 7 | 7 | 7 | 7 | 7 | 7 | 7 |
| TPOX | 8,12 | 8,12 | 8,12 | 8,12 | 8,12 | 8,12 | 8,12 |
| vWA | 16,18 | 16,18 | 16,18 | 16,18 | 16,18 | 16,18,19 | 16,18 |
Figure 1.
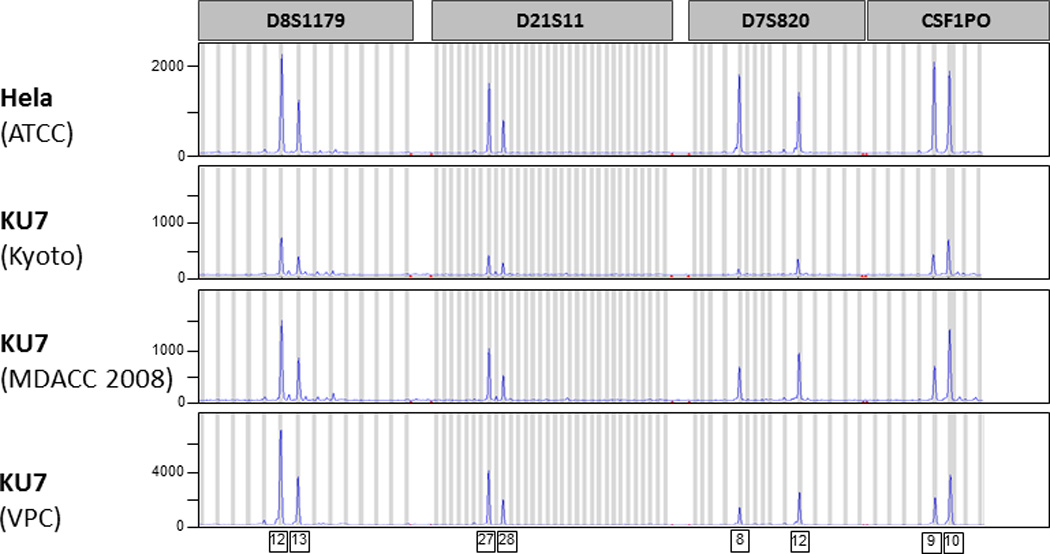
Short tandem repeat (STR) fingerprint analysis of HeLa and putative KU7 clones. Four representative microsatellite regions are illustrated (D8S1179, D21S11, D7S820, CSF1PO). Each peak on the curve illustrates the amplification of a specific number of repeating units.
Accordingly, aCGH of all samples revealed an abundance of shared chromosomal aberrations [Fig. 2]. To illustrate these similarities in detail, the 11q14.1 – 11q22.3 region was analyzed more closely [Fig. 3]. All clones shared similar copy number aberrations (gain or loss of chromosomal regions). The breakpoints within the resolution of the technology were located in identical locations. Slight differences in aberrations in some genomic areas (especially chromosome 1, 2 and 6) are explained by genomic drift occurring in different KU7 clones that were separated many years ago and cultured in different research institutions.
Figure 2.
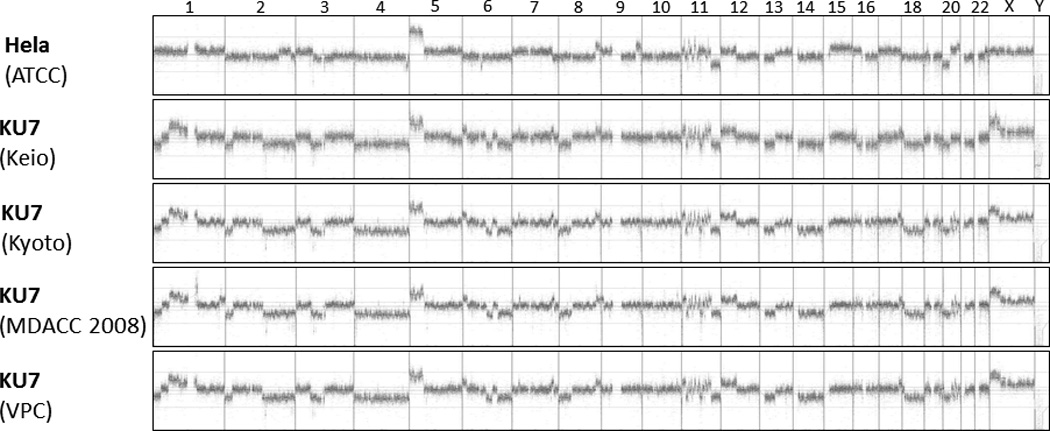
Array comparative genomic hybridisation (aCGH) of the whole genome. In this depiction, each vertical line separates one chromosome from the next, with the chromosomes lined up in numeric order from left to right. The X and Y chromosomes are at the right end of the curves. Any elevation above the baseline represents a copy number gain of that segment of the chromosome, and any depression below the baseline represents a loss.
Figure 3.
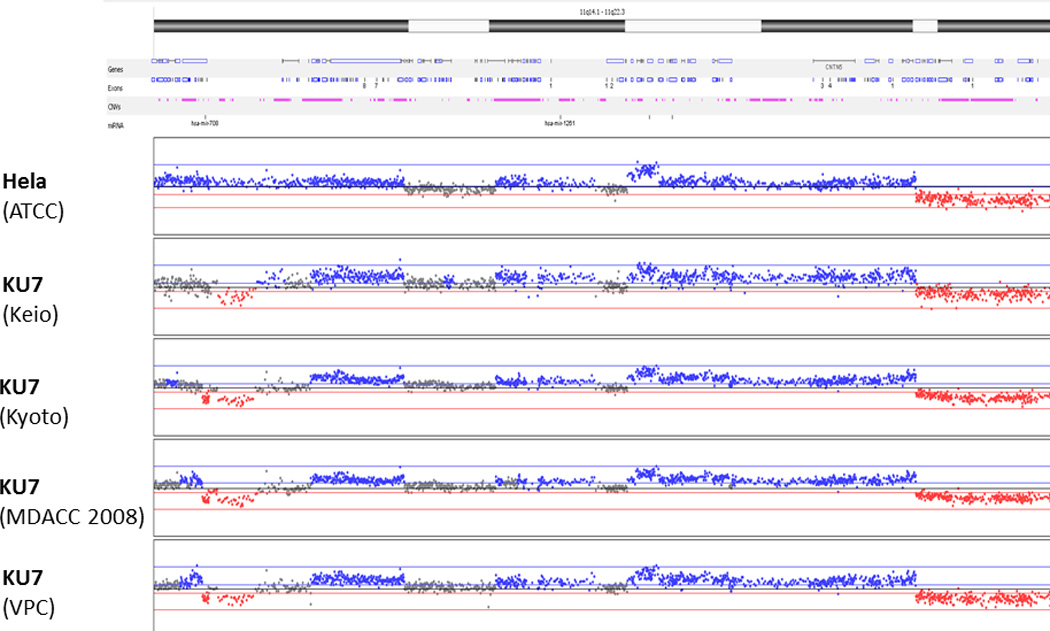
Magnified representation of chromosome 11 (region q14.1 – q22.3) by array CGH. Slight differences in some genomic areas are explained by genomic drift usually occurring in highly proliferative cell lines under cultivation in different institutes.
Regarding the gender, STR analysis of Amelogenin (AMEL) revealed a female gender of all analysed cancer cells which was confirmed by the homozygous deletion of the Y-chromosome in the aCGH. HeLa was isolated from a female patient. The original source for KU7 (Keio University) states a male gender of the patient from which it was isolated.
Taken together, our analysis reveals that KU7was likely contaminated in the source institution before 1984 by Hela, and was subsequently distributed to all other institutions (Fig. 4).
Figure 4.
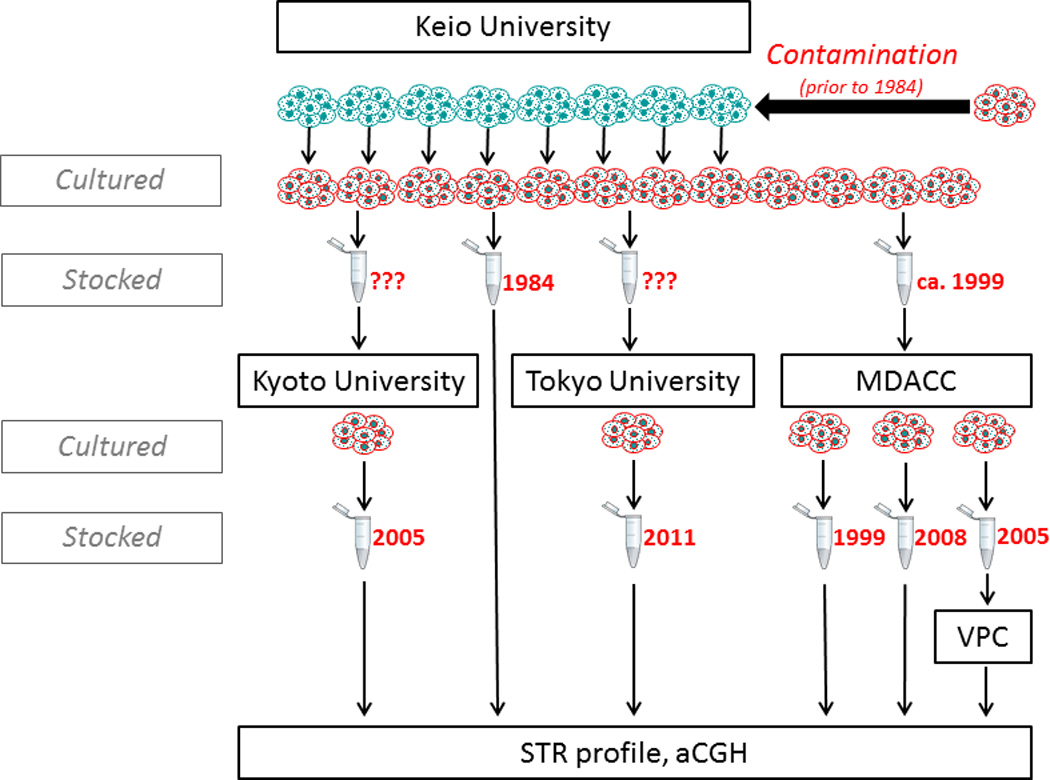
Genealogical tree of sampled KU7 clones. KU7 was established in 1981 at Keio University (green cells), and the oldest available stock from that institution from 1984 was found to be replaced by HeLa (red cells). Subsequent clones sent to MDACC, Kyoto University, Tokyo Medical College and the Vancouver Prostate Centre were all shown to be HeLa.
Discussion
Human cancer cell lines are an essential tool in oncologic research and their authentication should be mandatory in order to ensure that experimental results are reliable. While the establishment and propagation of many cancer cell lines around the world has been a key piece in the enormous expansion of cancer research in the past 60 years, unfortunately the phenomenon of cross-contamination and misidentification of cell lines seems to be almost as old as their existence.16 Numerous cell lines have likely been used for decades without recognition of their real identity. Technological innovations have simplified cell line authentication by STR fingerprint analysis, which is now the gold standard for this process.20,21
Our analysis of putative KU7 clones by STR identified that all worldwide available clones are likely identical to HeLa. Since it is common for such highly proliferative cancer cells to undergo genetic drift in culture22, we undertook aCGH to investigate subtle genetic aberrations beyond what is seen in STR analysis. The aCGH results confirmed the STR results and clearly demonstrate a common lineage of these cell lines, but slight differences in copy number variants were observed, thereby demonstrating the genetic drift.
Our experience with KU7 highlights specifically for researchers of genitourinary cancers how imperative it is for all laboratories to validate the identity of all cell lines. The ATCC Standards Development Organization (SDO) developed ANSI Standard ASN-0002 (“Authentication of Human Cell Lines: Standardization of STR Profiling”), which recommends that cell lines should be tested when a cell line is first grown in a laboratory, when preparing the initial frozen cell stock, after two or three passages while cells are expanded during the course of experimentation and at the time the research using the cell samples is published.20
Interestingly, KU7 has behaved over many years as a highly invasive and rapidly growing cell line14,15,23, which is quite disparate to its original description as being derived from a low grade papillary tumor.10 This discrepancy, however, was not recognized until we had already determined the true identity of KU7. This highlights the importance of bearing the disease context of cell lines in mind when using them for pre-clinical modeling.
The widespread contamination of KU7 clones determined here makes us believe that all KU7 in the urologic literature since at least 1984 is likely HeLa. As both cell lines were co-cultured at Keio University in the early 80s, the cross-contamination likely occurred at that time at the institution of origin.24 It is important to view past studies in this light, but it does not necessarily invalidate all of the work with this cell line. Laboratory results obtained in human cell culture and their derived xenografts are generally limited in their clinical applicability due to inherent shortcomings in this type of pre-clinical modeling. Any robust analysis is dependent on multiple cell lines, so that results with KU7 can be seen as one element of the overall experimental evidence. Ultimately KU7 is reflective of a highly aggressive, poorly differentiated epithelial carcinoma that likely retains little organ-specificity.
Our group and others have been particularly interested in KU7 for its utility in an orthotopic xenograft model of intravesical bladder tumor growth.15,23,25–28 Only when bladder cancer cells are instilled into the lumen of the bladder, as opposed to direct injection into the bladder wall, will they grow in such a manner that they are accessible to intravesical instillation of therapeutic agents. Tumors inoculated in this fashion are exposed at the luminal surface of the bladder and have limited penetration through the bladder wall for 3–4 weeks after inoculation.15 Modeling the intravesical instillation of drugs is critical in bladder cancer, as this route of administration is used routinely in clinical practice and allows for high local dose application with minimal systemic toxicity. The intravesical model, however, has proven to be limited by the number of different cell lines that will grow in this context. KU7 has been the only cell line that has been uniformly reliable in multiple centers.15,23,25–28 UM-UC3 and UC14 have been used with varying success.25,29,30 Anecdotally we can report that UM-UC3 at one point in our hands was KU7 (and therefore HeLa; true UM-UC3 clones are readily available), and MGHU3 has been shown to be the myeloma cell line KMS-18 (data not shown), so that a certain degree of skepticism is necessary for historical papers unless cell line authentication is described. The loss of KU7 as a model for testing intravesical drug delivery has left a significant gap in our modeling ability. As discussed above, however, the intravescial growth of HeLa can still be used as a model for testing intravesical therapy as pertains to drug delivery, tumor uptake, and biologic endpoints, provided it is recognized that the observed effects are not necessarily specific to bladder cancer.
Conclusions
Routine authentication of bladder cancer cell lines has revealed that KU7 was contaminated and replaced by the cervical carcinoma cell line HeLa almost 30 years ago at the institution of origin. Review of prior research using this cell line needs to be viewed in this light, although many of the scientific conclusions likely remain valid with HeLa. Our elucidations once again underline the necessity of validating tumor cell line identification as a routine practice in cancer research.
List of abbreviations
- aCGH
Array comparative genomic hybridisation
- ATCC
American Type Culture Collection
- KU
Keio University
- MDACC
MD Anderson Cancer Center
- STR
Short tandem repeat
- VPC
Vancouver Prostate Centre
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Canadian Cancer Statistics 2012. Toronto, ON: Canadian Cancer Society; 2012. Canadian Cancer Society’s Steering Committee on Cancer Statistics. [Google Scholar]
- 3.Babjuk M, Oosterlinck W, Sylvester R. EAU guidelines on non–muscle-invasive urothelial carcinoma of the bladder, the 2011 update. Eur Urol. 2011;59:997. doi: 10.1016/j.eururo.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 4.Sylvester RJ, van der Meijden AP, Oosterlinck W. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006;49:466. doi: 10.1016/j.eururo.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 5.Shariat SF, Karakiewicz PI, Palapattu GS. Outcomes of radical cystectomy for transitional cell carcinoma of the bladder: a contemporary series from the Bladder Cancer Research Consortium. J Urol. 2006;176:2414. doi: 10.1016/j.juro.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Scherer WF, Syverton JT, George GO. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med. 1953;97:695. doi: 10.1084/jem.97.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones HW., Jr Record of the first physician to see Henrietta Lacks at the Johns Hopkins Hospital: history of the beginning of the HeLa cell line. Am J Obstet Gynecol. 1997;176:227. doi: 10.1016/s0002-9378(97)70379-x. [DOI] [PubMed] [Google Scholar]
- 8.Lucey BP, Nelson-Rees WA, Hutchins GM. Henrietta Lacks, HeLa cells, and cell culture contamination. Arch Pathol Lab Med. 2009;133:1463. doi: 10.5858/133.9.1463. [DOI] [PubMed] [Google Scholar]
- 9.Bubeník J, Barešová M, Viklický V. Established cell line of urinary bladder carcinoma (T24) containing tumour-specific antigen. Int J Cancer. 1973;11:765. doi: 10.1002/ijc.2910110327. [DOI] [PubMed] [Google Scholar]
- 10.Tachibana M. Studies on cellular adhesiveness in five different culture cell lines derived from carcinoma of the urinary bladder. Keio J. Med. 1982;31:127. doi: 10.2302/kjm.31.127. [DOI] [PubMed] [Google Scholar]
- 11.Ieda K. Studies on cellular adhesiveness in five different culture cell lines derived from carcinoma of the urinary bladder. Keio J. Med. 1982;31:127. doi: 10.2302/kjm.31.127. [DOI] [PubMed] [Google Scholar]
- 12.Tazaki H, Tachibana M. Studies on KU-1 and KU-7 cells as an in vitro model of human transitional cell carcinoma of urinary bladder. Hum Cell. 1988;1:78. [PubMed] [Google Scholar]
- 13.Miyajima A, Nakashima J, Yoshioka K. Role of reactive oxygen species in cis-dichlorodiammineplatinum-induced cytotoxicity on bladder cancer cells. Br J Cancer. 1997;762:206. doi: 10.1038/bjc.1997.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibayama T, Tachibana M, Deguchi N. SCID mice: a suitable model for experimental studies of urologic malignancies. J Urol. 1991;146:1136. doi: 10.1016/s0022-5347(17)38025-4. [DOI] [PubMed] [Google Scholar]
- 15.Hadaschik BA, Black PC, Sea JC. A validated mouse model for orthotopic bladder cancer using transurethral tumour inoculation and bioluminescence imaging. BJU Int. 2007;100:1377. doi: 10.1111/j.1464-410X.2007.07165.x. [DOI] [PubMed] [Google Scholar]
- 16.Gartler SM. Genetic markers as tracers in cell culture. Natl Cancer Inst Monogr. 1967;26:167. [PubMed] [Google Scholar]
- 17.O'Toole CM, Povey S, Hepburn P. Identity of some human bladder cancer cell lines. Nature. 1983;301:429. doi: 10.1038/301429a0. [DOI] [PubMed] [Google Scholar]
- 18.Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nat Rev Cancer. 2002;2:315. doi: 10.1038/nrc775. [DOI] [PubMed] [Google Scholar]
- 19.Bravo NR, Gottesman M. Notice Regarding Authentication of Cultured Cell Lines. National Institutes of Health (NIH); 2007. Notice Number:NOT-OD-08-017. [Google Scholar]
- 20.American Type Culture Collection Standards Development Organization Workgroup ASN-0002. Cell line misidentification: the beginning of the end. Nat Rev Cancer. 2010;10:441. doi: 10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- 21.Capes-Davis A, Theodosopoulos G, Atkin I. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127:1. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- 22.Masramon L, Vendrell E, Tarafa G. Genetic instability and divergence of clonal populations in colon cancer cells in vitro. J Cell Sci. 2006;119:1477. doi: 10.1242/jcs.02871. [DOI] [PubMed] [Google Scholar]
- 23.Hadaschik BA, Zhang K, So AI. Oncolytic vesicular stomatitis viruses are potent agents for intravesical treatment of high-risk bladder cancer. Cancer Res. 2008;68:4506. doi: 10.1158/0008-5472.CAN-08-0238. [DOI] [PubMed] [Google Scholar]
- 24.Hagiwara M. Regrowth assay method for quantitative evaluation of drug-induced cell damage in vitro: clarification of cell reduction kinetics of anticancer drugs and differential drug sensitivities of genitourinary cancer cell lines. Keio J Med. 1981;30:141. doi: 10.2302/kjm.30.141. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka M, Gee JR, de la Cerda J. Noninvasive Detection of Bladder Cancer in an Orthotopic Murine Model With Green Fluorescence Protein Cytology. J Urol. 2003;170:975. doi: 10.1097/01.ju.0000073209.65128.c1. [DOI] [PubMed] [Google Scholar]
- 26.Benedict WF, Tao Z, Kim CS. Intravesical Ad-IFNalpha causes marked regression of human bladder cancer growing orthotopically in nude mice and overcomes resistance to IFN-alpha protein. Mol Ther. 2004;10:525. doi: 10.1016/j.ymthe.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 27.Adam L, Black PC, Kassouf W. Adenoviral mediated interferon-alpha 2b gene therapy suppresses the pro-angiogenic effect of vascular endothelial growth factor in superficial bladder cancer. J Urol. 2007;177:1900. doi: 10.1016/j.juro.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Mugabe C, Matsui Y, So AI. In vivo evaluation of mucoadhesive nanoparticulate docetaxel for intravesical treatment of non-muscle-invasive bladder cancer. Clin Cancer Res. 2011;17:2788. doi: 10.1158/1078-0432.CCR-10-2981. [DOI] [PubMed] [Google Scholar]
- 29.van der Horst G, van Asten JJ, Figdor A. Real-time cancer cell tracking by bioluminescence in a preclinical model of human bladder cancer growth and metastasis. Eur Urol. 2011;60:337. doi: 10.1016/j.eururo.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Nogawa M, Yuasa T, Kimura S. Intravesical administration of small interfering RNA targeting PLK-1 successfully prevents the growth of bladder cancer. J Clin Invest. 2005;115:978. doi: 10.1172/JCI23043. [DOI] [PMC free article] [PubMed] [Google Scholar]


