Abstract
Although peptides are well recognised biological molecules in vivo, their selection from libraries is challenging because of relative low affinity whilst in linear conformation. We hypothesized that multiplexed peptides and DNA on the surface of beads would provide a platform for enhanced avidity and the selection of relevant peptides from a library (ORBIT bead display). Using human immunodeficiency virus (HIV-1) gp120 as a target, we identify peptides that inhibit HIV-1 replication in vitro through blocking of protein:protein interaction with the co-receptor CCR5. The bead display approach has many potential applications for probing biological systems and for drug lead development.
Peptides represent important biological molecules in vivo and indeed several peptides have been taken forward into therapeutic use1. However uptake of therapeutic peptide technology has been slow compared to antibody therapeutics despite advantages in cost, tissue penetration and possible non-parenteral route of administration eg inhaled. This is related to a number of problems including the difficulty of screening for peptides of interest from libraries because of their relative low affinity in linear conformation. Secondly, conventional peptide libraries generated using F-moc chemistry are prohibitively expensive for the library sizes required for drug discovery. Thirdly problems have included peptide stability in vivo, but this can be addressed through the use of unnatural amino acids or structured/stapled peptides eg ciclosporin, or other conjugations eg pegylation. We sought to develop a system that would directly address the first two of these issues by multiplexing a library of translated peptides and their encoding DNA on the surface of beads (ORBIT bead display). The DNA serves the dual purposes of acting as a library source for the translated peptides and for being the bar code for identification of selected peptides.
Existing approaches that are amenable to protein library screening and that link genotype to phenotype include phage-display, yeast-display, ribosomal and mRNA displays2,3,4,5,6. The latter in vitro library approaches are challenging for peptides as it is difficult to multiplex expressed peptides to the numbers required for detection of low affinity interactions. In contrast, the in vivo methods of phage and yeast display allow multiplexing of proteins or peptides on their surface, but they share the same disadvantage that in addition to the proteins under study there are other proteins that are on both the phage and/or the yeast which can interfere with binding. This can lead to non-specific interactions which can be difficult to distinguish from interactions of interest which is particularly problematic for peptide screening. Furthermore, these methods commonly use coat fusion proteins which can alter the conformation of both interacting partners and influence binding. The produced proteins may be toxic to the yeast or the phage or influence their replication which can select out library bias. The use of unnatural amino acids and post-translational modifications may provide significant affinity and stability advantages and these are difficult to achieve using phage or yeast during the selection step. Lastly, in vivo systems have associated limitations in library size that can be overcome using in vitro expression systems.
Griffiths et al. established emulsion micro-compartments as a means to isolate reactions7. It was reported that in a 1 ml reaction volume, more than 1010 water-in-oil emulsion micro-compartments can be created, with each having a mean diameter in the range of 2–3 μm and mean volume of 5 femtolitres. At this volume, a single molecule achieves a concentration of approximately 0.5 nM, thus enabling a single DNA molecule to be transcribed and translated8. With appropriate dilution of DNA molecules, it is possible to create individual water-in-oil emulsions in which only one DNA molecule is present in a microcompartment, and the protein expressed is trapped in a single confined physical space, i.e. creating 1010 unique directed evolution reactions. Compared to the current library display technology, emulsions provide the convenience of a cell-free environment, preventing the interference of toxic substrates or unwanted cellular interactions9. Nakano and colleagues extended the technology to combine emulsion PCR with emulsion in vitro transcription/translation, to generate beads combining protein and the DNA encoding the said protein. Beads binding to the selected target were obtained using flow cytometric guided cell sorting and multiple rounds of selection and bead re-derivation10,11,12. However flow cytometric based approaches associate with loss of sensitivity and specificity secondary to the requirement to express threshold levels of fluorescence before selection which is not amenable to linear peptide selection with relative low affinity. As our hypothesis required selection of peptides from an in vitro library with relatively low affinity we could not use flow cytometry as monomeric ligand:target pairs will be missed. We therefore had to develop a novel multiplexed peptide expression and selection system (ORBIT, figure 1a). We further hypothesized that the use of a protein scaffold (beta-2-microglobulin) with known ability to deliver peptides to a tertiary molecule (human leucocyte antigen, HLA) would allow us to present and screen peptides that bind to our gp120 target of interest. Beta-2-microglobulin has a number of other advantages as a carrier molecule including survival at the relatively low endosomal pH which may be important for screening peptides that might modify protein:protein interactions that occur in vivo at low pH. This bead system has a number of further advantages which we have found to be of value, including the ability to easily handle the beads for transfer to sequential binding steps with different conditions and requirements (eg positive/negative selection). For example, beads can be transferred sequentially to binding cells with different targets or conditions to select for beads with multiple characteristics without the need for bead re-derivation between each round of selection.
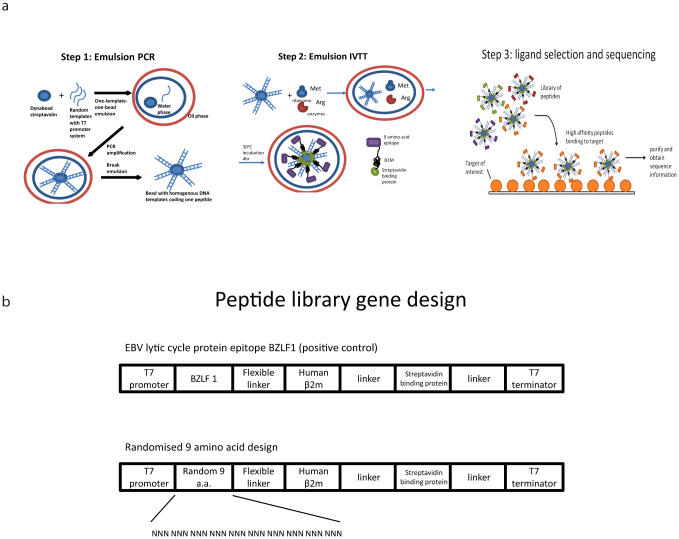
Figure 1.
(a) Overview of the ORBIT system. Step 1: Each bead is coated with homogenous DNA coding a unique peptide. A water-in-oil emulsion is formed for each bead, with each emulsion containing a single template with PCR components, generating bead-DNA complexes after PCR. Step 2: each bead-DNA complex forms a new emulsion with in vitro transcription/translation (IVTT) components for protein synthesis. Since the protein translated contains Streptavidin binding peptide (SBP), it will attach to the bead as soon as it is translated, resulting the formation of a protein-DNA-bead complex. Step 3: the protein-DNA-bead complexes form a library of variable peptides, which are challenged with any target of interest (e.g. gp120), the complexes will bind to the target, and the encoding templates can be sequenced by conventional or next generation sequencing. Figure 1(b) Peptide library gene design. This 782 base-pair DNA template acts as the starting template for the synthesis of the randomised peptide library.
Using HIV-1 gp120 as a model system, we screened a small peptide library for low affinity interactions with gp120 and identified peptides that despite low affinity could inhibit HIV-1 replication through blocking protein:protein interaction between gp120 and the co-receptor CCR5.
Results
Library structure and generation
It was hypothesized that beta-2-microglobulin (β2m) would act as a good protein scaffold to present peptide on the surface of beads as it is known to be able to deliver peptides for binding to associated proteins (HLA). Figure 1a shows a schematic of the bead structure and selection approach. Multiple DNA constructs with variant sizes of T7 promoter regions, peptide, linkers, beta-2-microglobulin and SBP (streptavidin – binding peptide) tag were tested (data not shown) but the positive control (“BZLF1” RAKFKQLL) and the corresponding random construct in figure 1b proved to be optimal in terms of consistent expression. We used random peptide libraries containing nine and fifteen amino acids.
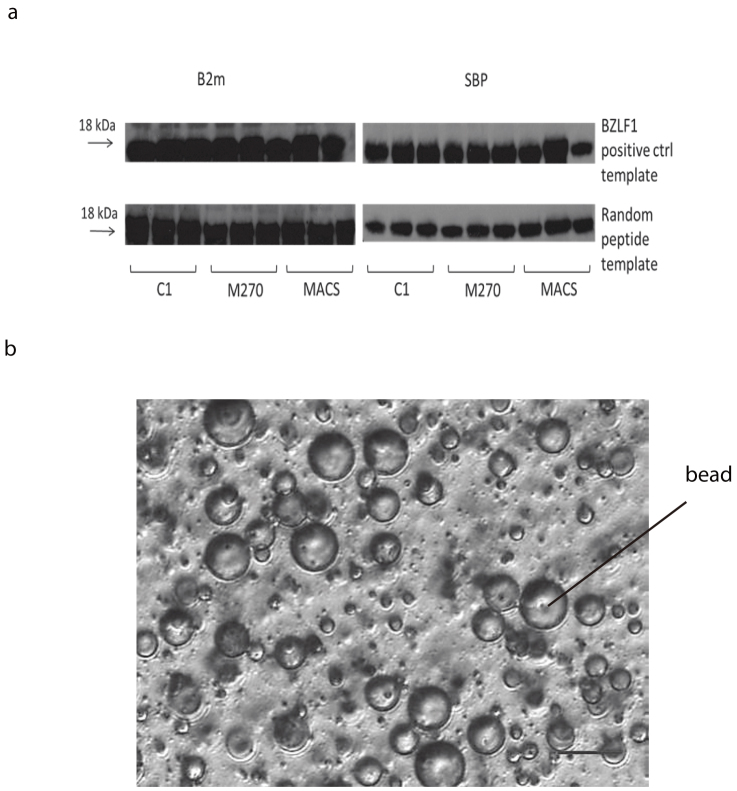
Emulsion PCR was undertaken using streptavidin magnetic beads labelled with biotinylated primers (forward primer: 5’- biotin -GATCTCGATCCCGCGAAATT; reverse primer: unmodified, 5’- TCCGGATATAGTTCCTCCTT -3’). The emulsion oil for each PCR reaction was prepared in a universal tube using surfactant and mineral oil. To equilibrate the emulsion oil, a magnetic stirrer (VWR UK) was used. Each aqueous PCR reaction (150 μl) was prepared as follows: 6 μl of primer-coupled beads, 3 μl of complement unmodified primers (400 μM), 3 μl of unmodified primer (2.5 μM), 3 μl of 10 mM PCR grade dNTPs, 4.5 μl of 50 mM MgCl2, 10 ul of 20 pM DNA template, 15 μl 10× PCR buffer, and 9 μl of Taq DNA polymerase (5 U/μl). The water-in-oil emulsion was prepared by slow addition of the aqueous PCR mixture into the spinning emulsion oil. The emulsions were then aliquoted into 100 μl each, and PCR initiated. The emulsion was broken by adding 1 ml breaking buffer (10 mM Tris-HCl pH7.5, 1% Triton-X100, 1% SDS, 100 mM NaCl, 1 mM EDTA). Bead bound DNA could be detected by using magnetic bead separation from the supernatant and then PCR directly from the surface of the beads. Figure 2 shows DNA product from 8 independent emulsion PCR products with Dynabead C1. We undertook deep sequencing of the libraries and were able to show that >98% of the sequences were unique, suggesting the library was diverse. Furthermore, as expected the libraries showed random incorporation of stop codons which will not show full length translation from those beads.
Figure 2. Post-emulsion PCR gel examinations performed with Dynabead C1.

The DNA gel represents 8 independent emulsion PCR reactions. The emulsions were broken after PCR, and the DNA region containing random amino acids (approximately 450 bps, template = 9 random amino acids) were amplified by standard PCR.
DNA-bound beads were then transferred to a new emulsion for in vitro transcription translation. Emulsion oils were prepared for a single reaction using mineral oil, Span-80, Tween-80, Triton X-100. Each in vitro transcription/translation reaction (IVTT) was prepared (RTS 100 E. coli, Roche) and the beads were added and mixed thoroughly before adding to the spinning emulsions as described above. The reaction was incubated at 30°C for 4 hours and the emulsions broken as described above. The integrity of the translated proteins was examined by Western blot (figure 3a). Different bead sizes were tested and each showed successful expression of β2m and streptavidin binding peptide (SBP) epitopes, but with variable levels of expression (figure 3b). After confirmation of the protein expression and ligation onto the beads, libraries of randomised 9/15-amino-acid peptides were generated.
Figure 3.
(a) emIVTT on different types of beads. Cropped western blot results showing protein-DNA-beads attaching with either EBV BZLF1 positive control template (known peptide sequence) on upper row, or templates containing randomised peptide sequences (lower row). Full gel is available as supplementary information. The left column shows the results of human β-2-microglobulin (β2m) staining and the right shows SBP staining. Each type of beads was labelled with three different concentrations of biotin labelled primers attaching on the beads, e.g. for C1 column, 1000 pmol, 500 pmol, 250 pmol primers were attached on 1 mg of beads (6–7 × 107). Figure 3(b) The 40× inverted microscopic image of a water-in-oil emulsion mixture with Dynabead C1. A successful water-in-oil emulsion has diameter of 10-15 μm and contains a single magnetic bead in the aqueous phase. The scale bar represents 20 μm.
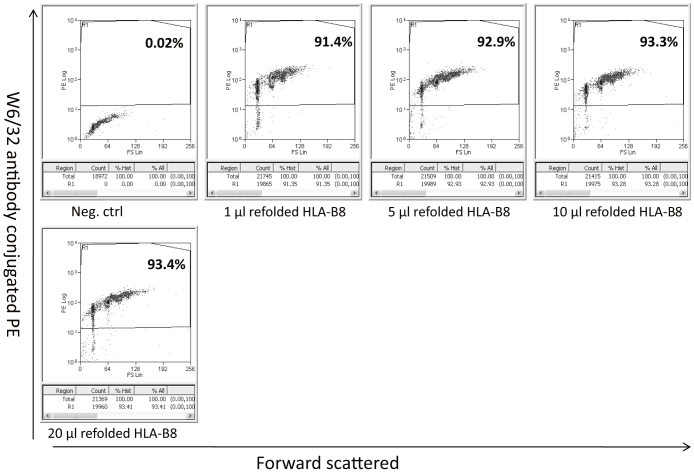
Beta-2-microglobulin is correctly folded on the bead surface and presents peptides for tertiary interactions
In order to confirm that the β2m was correctly refolded and able to present peptides for delivery to other molecules, we refolded the beads containing the fixed BZLF1 peptide (known HLA-B*0801 binding peptide) with exogenous HLA-B*0801 to generate the complete HLA, peptide, β2m complex on the surface of beads. As shown in figure 4, the beads were able to bind to W6/32 which is a conformationally dependent antibody. This suggests that the β2m is not only refolded correctly on the beads but that the peptide is available for interaction with tertiary molecules. To next confirm expression of the random library of peptides on the beads, we screened for binding partners of bovine serum albumin (BSA). 210 sequences were identified without pre-clearing and five peptides were synthesized. All 5 peptides bound BSA on ELISA (supplementary figure 1) confirming that the random peptide library system was functional. We felt then able to proceed to identify peptides with potential biological function, using HIV-1 gp120 as the target.
Figure 4. Beads coated with b2m and the BZLF1 peptide were refolded with recombinant HLA-B*0801 and stained for reactivity with W6/32 (a conformation dependent antibody).
Validating the binding of predicted peptides to gp120
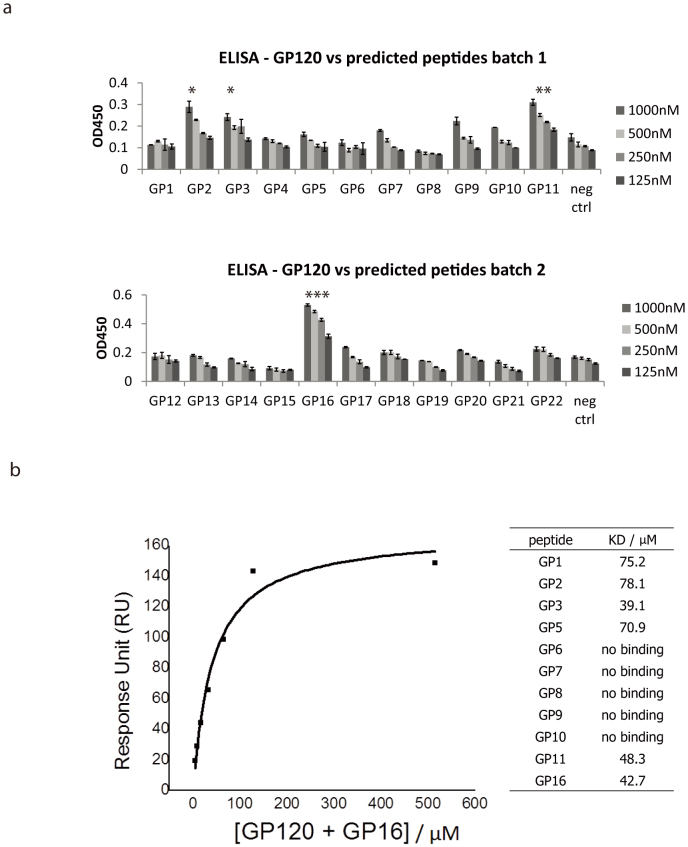
Having successfully generated the random peptide library on the surface of the beads together with the encoding DNA, we used both the 9 amino acid and the 15 amino acid library to select peptides binding to HIV-1 viral coat protein gp120. After selection, the DNA templates of the beads remaining on the gp120-coated plate were amplified by PCR and the sequences of these peptides were obtained by sequencing. The most repeated peptide sequences are presented in Table 1 and show no similarities with the bovine serum albumin binding peptides (supplementary figure 1). Highly ranked gp120-binding peptides were synthesised by F-moc chemistry with purity confirmed by HPLC. Figure 5a showed the result of ELISA tests of the twenty two peptides. An irrelevant peptide was used as a reference for the peptides which has no binding to gp120. As shown in Figure 5a, GP2, GP3, GP11 and GP16 had significantly higher binding to gp120 than the control hemagglutinin-binding peptide HA2 in a dose-dependent manner. However, while being a potentially useful screening tool, ELISA is susceptible to detection of non-specific binding. We therefore proceeded to determine specific binding affinities using surface plasmon resonance.
Table 1. Peptide sequences for 22 GP120-binding peptides selected by ORBIT derived from the 9 amino acid and 15 amino acid libraries. All peptides have glycine-serine repeats (GGGGGGSGGSG) at C-terminals for flexibility, followed by a lysine K residue for biotinylation.
| peptide | sequence |
|---|---|
| GP1 | MSTSCCLPS |
| GP2 | RRRNVPLIL |
| GP3 | SYSFCPRRQ |
| GP4 | AVNIVGYSN |
| GP5 | LVLLSSASS |
| GP6 | LRPLLSSMR |
| GP7 | LVLLLSSMS |
| GP8 | EMANRRSGS |
| GP9 | WFFTSLVTC |
| GP10 | SILSSCWQS |
| GP11 | LLALSAYMV |
| GP12 | RPLPILAPC |
| GP13 | GTRVFMRSL |
| GP14 | LSVLSRGML |
| GP15 | WHVLIWLLF |
| GP16 | LWCRRLNLL |
| GP17 | LWHTRRGPG |
| GP18 | SSCAGNLPREDQECC |
| GP19 | SSCQRLFVIAYPFKC |
| GP20 | SSCCFLPASWLKSLC |
| GP21 | SSCSVRSSCRLYYSC |
| GP22 | SSCLLFTSMLPDKTC |
Figure 5.
(a) ELISA for the binding between predicted peptides and GP120 protein. Some predicted peptides (GP2, GP3, GP11, GP16) showed higher absorbance, i.e. peptides bind to GP120 in a dose-dependent manner, compared to the negative control (HA2, hemagglutinin-binding peptide). While most peptides bind to GP120, GP16 showed the highest binding affinity to GP120. (Paired t-test against negative controls, *p = 0.02, **p = 0.003, ***p = 0.005; n = 3). Figure 5(b) Equilibrium dissociation constant KD calculation using surface plasmon resonance (SPR). An illustrated example of the KD calculation for GP16 peptide. The left of the figure is the non-linear fit of the Langmuir binding isotherm for GP16 binding. The table summarises the KD of the peptides tested.
In order to obtain quantitative measurements of binding reactions between predicted peptides and gp120, surface plasmon resonance (BIAcore) was used to measure the equilibrium dissociation constant KD of the predicted peptides and gp120. Four peptides which showed higher absorbance in the ELISA experiments were selected for BIAcore experiments, together with five predicted to have intermediate or no binding from the ELISA. Since the KD to gp120 is relative to different analytes, purified protein HLA-A*0201 heavy chain was used as a reference control to measure the relative binding affinity of peptides to gp120. Purified gp120 and HLA-A*0201 were immobilised on chips, and the peptides were passed through both flow cells at a constant rate of 10 μl/min. An illustrated example of the KD calculation for GP16 peptide and the KD for different peptides is shown in Figure 5b. In general the peptides with positive binding on ELISA had a KD value ranging from approximately 20 to 80 μM, indicating that these peptides had the ability to bind gp120 protein, but with relative weak affinity. The BIAcore experiment acted as a second confirmation that the randomized peptide library had successfully captured peptides which bind to gp120 in a single round of selection, and that the sequence information coding the binding peptides was correctly preserved and sequenced. Although binding affinity was in the micromolar range, we were aware that this was also the case of Enfuvirtide T20 which is a licensed therapeutic for HIV-1, and so proceeded to determine whether the peptides had HIV-1 replication inhibition activity either as monomers or as multimeric complexes.
HIV-1 viral inhibition with streptavidin-conjugated peptides
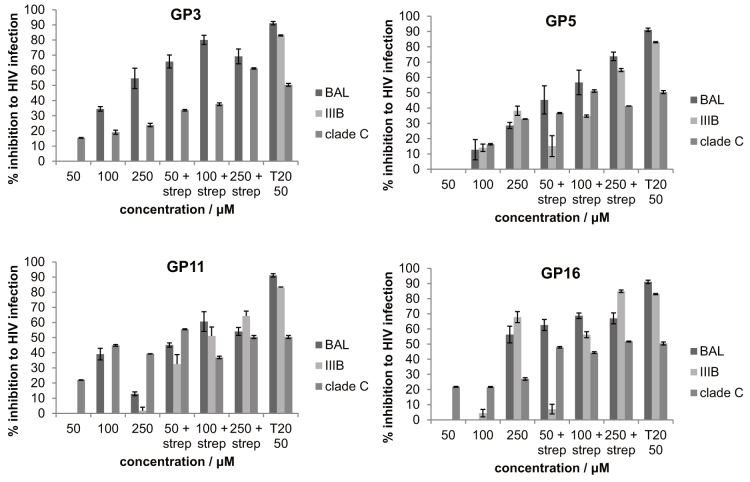
Figure 6 shows the HIV-1 replication assay using biotinylated forms of the four peptides of highest affinity, with or without multiplexing using streptavidin conjugation (GP3, GP5, GP11, and GP16). In general, peptides administered directly without streptavidin conjugation had a very low inhibitory effect on HIV-1 replication, e.g. GP16 inhibited HIV-1 BaL infection by less than 1% at 50 and 100 μM, and 50% at 250 μM; GP3 had no inhibitory effect at 50 μM, and 34.5% and 54.7% inhibition at 100 μM and 250 μM respectively. Without streptavidin conjugation, peptides only exhibited viral inhibition at high concentration, and the effect was weak compared to the positive control Enfuvirtide (T20) (91.1% inhibition at 50 μM). When the streptavidin conjugated peptides were used, the inhibition was efficient and required concentrations comparable to Enfuvirtide T20, e.g. GP16 achieved 62.6% viral inhibition at 50 μM when conjugated with streptavidin. Similar patterns could be observed in different peptides and different HIV-1 clades. GP16, GP3 and GP11 exhibited cross clade HIV-1 inhibitory activity, and the effect was augmented with streptavidin conjugation. Of note, GP21 did not show any binding on ELISA and did not show any HIV-1 inhibition, consistent with the ELISA being useful screening tool for subsequent biological activity.
Figure 6. HIV-1 (HIV BAL, IIIB, clade C) inhibition assay.
Peptides GP3, 5, 11, 16 were administered in 3 concentrations with or without streptavidin conjugation. T20 (Enfurvitide) was used as positive control. At 50 μM peptide with strep, GP5, 11, 16 showed significant (P<0.01) inhibition of BAL, IIIB, clade C; and GP3 showed significant (P<0.01) inhibition of BAL and clade C HIV-1.
As gp120 is a highly polymorphic protein, HIV-1 neutralising antibodies are an invaluable tool to assess binding sites within the GP120 molecule. Three commercially available HIV-1 broadly neutralising antibodies V3 (anti-V3 447-52D, CCR5/CXCR4 binding sites), VRC01 (discontinuous CD4 binding site) and F105 (CD4 binding site) were prepared to test the binding sites of GP3 and GP16 peptides as these showed the greatest inhibition of HIV-1 replication. Peptide GP3 showed significant reduction (p = 0.025) in absorbance when gp120 was pre-blocked with the V3 antibody, whereas GP16 had no reduction in gp120 blocked with any of the three antibodies, suggesting that GP16 bound to gp120 on other domains. Our data showed that peptide GP3 exhibited inhibition of replication of the HIV-1 clade C isolate and HIV-1 BaL, which are CCR5 tropic viruses; however no inhibition was observed when GP3 was co-incubated with HIV-1 IIIB, which is a CXCR4 tropic virus. This evidence suggested that peptide GP3 may be binding to gp120 CCR5 binding region. Indeed we observed a significant reduction in recombinant CCR5 binding to gp120 that had been pre-incubated with GP3 (65.8% reduction in absorbance at 125 ng peptide, p = 0.0038).
Discussion
The development and use of peptides as tools to target protein:protein interactions to probe biological processes or as the basis for future drugs or drug leads has been hampered by the lack of good cost-effective technologies to screen peptide libraries for binding activity. Conventional peptide libraries generated using F-moc chemistry are prohibitively expensive for library sizes of sufficient numbers required for drug discovery. Given that linear peptides will have relative low binding affinity, it is likely that multiplexing the peptides will be an important prerequisite for success. Of the existing systems of linking genotype:phenotype, only phage and yeast display can really achieve the levels of multiplicity required. However they too are not ideally suited for longer term systems of peptide library generation as they have associated problems of non-specific interactions with other coat proteins and are not easy to use with unnatural amino acids and for chemical modification before screening due phage/yeast toxicity. We therefore established a novel in vitro bead display approach (ORBIT) for displaying peptides using β2m as a scaffold structure. The use of β2m as a scaffold allows effective translation and is known to deliver linked peptides to tertiary molecules which we confirm here.
Further advantages of using an in vitro system include the possibility of using extremely large library sizes that are not limited by cloning efficiency into organisms. In vitro expression does not risk bias away from variants that confer growth disadvantage to particular phage or advantage to non-recombined phage. The beads are very simple, containing just peptide/protein and the encoding DNA; this means that there are fewer opportunities for non-specific interactions and that selection rounds can be reduced. Furthermore selection against a target can be undertaken in specific milieu that may not be possible using in vivo presentation, e.g. low pH that may be relevant for viral membrane fusion inhibitors or gastric environments for oral absorption. Lastly, beads are potentially amenable to binding to targets on cells which adds further applications which are currently being explored.
Instead of using fluorescence based bead selection, we have used a binding system based on molecular interactions to retain the selected beads. This has many advantages, as small numbers of beads can be selected from large libraries without dependence on threshold levels of fluorescence for detection which will be low for individual peptide:ligand interactions. Furthermore the beads can be readily isolated after each round of selection and transferred to sequential flow cells with different targets or conditions (eg target concentration, temperature, pH, buffer constituents) in order to undertake serial positive and/or negative selection to define multiple characteristics. Beads do not necessarily have to be re-derived between each sequential step and so even multiple selections can be undertaken quickly. However re-derivation of beads between selection steps would be expected to increase affinity of peptide ligands identified.
The peptides may be of value themselves, but in addition, they may provide structural data that are of value to the design of small drugs or other peptide mimetics. Although we wished to use peptides defined after a single round of selection to assess whether the micromolar affinity peptides could have HIV-1 replication inhibition activity, it is possible to use multiple rounds of selection to pick out peptides with higher affinity. Peptides (and peptide drug precursors) offer potential advantages over antibody approaches, as the inclusion of certain sequence and structural components can allow the absorption from the gut or lungs and the targeting of intracellular proteins as shown by ciclosporin. Lastly, peptides are readily amenable to multimerisation and modulation of pharmacokinetics such as through the use of pegylation with multiple peptide binding sites.
HIV-1 shows enormous sequence diversity which is believed to contribute to evasion of the humoral immune response. Until relatively recently, the robust isolation of cross-clade neutralizing antibodies has been rare, with only a handful of well documented antibodies in the literature. However with advances in antibody isolation technology, it is becoming apparent that infected individuals not only develop neutralizing antibodies, but these can be isolated and characterized13,14,15. With high levels of somatic mutation, it seems likely that parrying rounds of mutation and selection occur between Env and the evolving humoral immune response. There are some common features of the emerging Env target sites, including the CD4-binding site, quaternary V2/V3 loops, gp41 Membrane Proximal External Region and Env carbohydrates16. Indeed Env acquisition of an extensive evolving glycan shield is believed to restrict access to conserved protein backbone amino acids17. Interestingly many of the isolated antibodies show poly-reactivity including self-reactivity, which may be important for neutralization capacity18. Understanding of the characteristics of neutralizing antibodies is important for the development of vaccination strategies, but is also of importance for the development of HIV-1-binding or HIV-1-fusion inhibitors. For example, enfuvirtide (T20) is a licensed peptide HIV-1 fusion inhibitor that through binding to gp41 inhibits HIV-1 fusion with the target cell membrane. Given the glycan shield protection, we hypothesized that peptides may offer advantages over the relatively large Env-specific antibodies, by targeting small accessible niches on the protein backbone.
The binding of the GP3 peptide to GP120 can be partially blocked by anti-V3 antibody. This gave an indication that anti-V3 antibody might share the same binding site to GP3. In addition, we showed reduction in binding between GP3-gp120 and recombinant CCR5, suggesting the possibility that a part of the CCR5 binding domain (possibly the V3 loop residues) on gp120 could bind to GP3 peptide, preventing the access of recombinant CCR5 protein. Recent studies also suggested the possibility that small peptides derived from CCR5 are capable of binding to HIV-1 gp120 and have neutralising effects19,20. Furthermore, it has been known that small cyclic peptides such as β-defensin 2 and 3 have anti-HIV-1 activity21. The gp120-binding peptides themselves, their variants or small mimetic drugs may therefore have diagnostic, monitoring or therapeutic uses.
In summary, ORBIT bead display of peptides using β2m as a scaffold base is a powerful approach to the identification of peptide ligands of disease relevant targets. We used a multiplex in vitro translation system that is able to pick up low affinity peptides in a cost-effective approach. Although these peptides may be of value themselves, they can form the basis of further modifications or structural studies to inform the development of small drugs and other peptide mimetics for use to probe protein:protein interactions to understand biological process and for future therapeutic development.
Methods
Gp120 synthesis
Gp120 was produced by transient transfection of 293 T cells using the JRFL gp120 – pLex construct. HIV-1 JRFL gp120 protein was captured from the supernatant by Co2+ affinity chromatography to the C-terminal his6 tag. The protein was further purified by size exclusion chromatography on a SD200 16/60 column and the protein purity was verified to be >98% pure by polyacrylamide SDS-PAGE. Conformation and binding function was confirmed by binding with recombinant human CD4 and a panel of HIV-1 Env-specific monoclonal antibodies in ELISA assays.
Beads selection
Gp120 at a concentration of 100 μg/ml was coated to an immuno 96 micro-well plate (Nunc, UK) at 4°C overnight. After washing in PBST (0.05% Tween 20) 6 times, 50 μl PBS containing approximately 10 million beads collected after IVTT was added and incubated at 4°C for 1 hr. There was no pre-clearing of the beads. After the incubation, the supernatant was collected and the wells were washed with PBST for 6 times. The beads remained on the plate were then collected in TE buffer for subsequent PCR to pull down the DNA fragment containing the random nucleotide region.
Peptide analysis and synthesis
The sequence of 9 amino acids or 15 amino acids (cys) was translated from DNA sequencing data. The sequence analysis is performed with software R to characterise the frequency of different amino acids in each position and the frequency of 3 or longer amino-acid fragments. The sequences containing a high frequency of certain amino acids or fragments were chosen for synthesis. A GS-linker (GGGGSGG) and a biotin were added at the C terminal of the peptides. The purity was established by high-pressure liquid chromatography, and the individual peptides were dissolved at 20 mg/ml in dimethyl sulfoxide and diluted at 1 mg/ml with PBS.
Peptide selection via amino acid sequence analysis
The 9 mer peptides were broken into 3 mer fragments and the frequency of these 3 mers was analysed with scripts written in R software. The peptides contain higher-frequency 3 mers appeared in independent replicates were chosen to synthesis. For example, GP16 (LWCRRLNLL) was the peptide containing the NLL repeated for 6 times in 4 replicates and LWC repeated 2 times in 2 replicates.
ELISA (antibody blocking)
ELISA plate (Greiner, 655061) was coated with 1 μg/ml purified gp120 in PBS and incubated at 4°C overnight. The plate was washed with ELISA wash buffer (PBS, 0.05% Tween20, VWR) followed by 100 μl blocking buffer (PBS, 0.05% Tween20, 2% BSA powder, Sigma) for 1 hr at room temperature. For antibody blocking experiment, V3/VRC01/F105 antibodies (NIBSC, CFAR3219/3291/3115) were used in 1 μg/ml for 1 hr at room temperature. After blocking, the plates were washed and peptides were added at different concentrations and incubate for 1 hr at room temperature. After washes, the detection of the binding was done by the addition of streptavidin conjugated peroxidase (Sigma, at 1:2500 dilution in PBS for 1 hr at room temperature and developed by TMB substrate (Thermo, 8008743723). After colour change was observed, stop buffer (0.16 M sulphuric acid, Sigma) was added to stop the reaction, and the absorbance was read at OD450 with ELISA plate reader.
ELISA (peptide blocking)
JRFL gp120 protein was coated on ELISA plate at 1 μg/ml 4°C overnight and blocking with BSA as described above. Peptides were prepared in 4 dilutions and added to the plate and incubated for 1 hr at room temperature. The plates were washed and recombinant CCR5 GST tag (H00001234-P01, ABNOVA) was added to the plate at 0.1 μg/ml and incubated for 1 hr at room temperature. The plates were washed and the detection antibody anti-GST HRP conjugated was added at 1:2000, 1 hr, room temperature. The developing process was identical as described above. Paired t-test was used to compare between different concentrations of peptides (GP1-22) and the negative control peptide (hemagglutinin-binding peptide, 9 amino acids) in the ELISA absorbance reading, 95% confidence level.
Surface plasmon resonance (BIAcore)
Ligand immobilisation
Immobilisation of purified gp120 protein (1 mg/ml) and purified HLA-A2 heavy chain (reference control, 1 mg/ml) onto the surface of individual flow-cells (Fc1 and Fc2 respectively) of CM5 sensor chip (GE Healthcare) was performed under conditions of 20 μl/min flow rate and 25°C, with HBS-EP buffer (0.01 M HEPES pH 7.4, 0.15 M NaCl, 0.005% v/v Surfactant P20; GE Healthcare) as the running buffer. After initiating the system, 100 μl 0.05 M solution of EDC [1-ethyl-3-(3-dimethylaminopropyl) carbodiimide]/NHS (N-Hydroxysuccinimide) at 1:1 ratio was injected to activate the dextran matrix of the sensor chip surface. After immobilisation, 150 μl of 1 M ethanolamine hydrochloride was injected to deactivate the unreacted carboxylmethyl groups on the dextran matrix. Finally, 30 μl of Glycine pH2.5 was injected to remove any non-covalently bound proteins.
Measurement of the binding affinity of the interaction between predicted peptides and gp120
The affinity analyses of the interaction between 9-amino-acid predicted peptides and purified gp120 were performed using the BIAcore T200 (GE Healthcare), under conditions of 10 μl/min flow rate and 25°C and using HBS-EP buffer (0.01 M HEPES pH 7.4, 0.15 M NaCl, 0.005% v/v Surfactant P20; GE Healthcare) as the running buffer. Prior to experiment, the 9-amino-acid peptides were diluted to 1 mg/ml with HBS-EP buffer. A 2-fold serial dilution of totally 8 concentrations was used to pass through the immobilised peptides. The data were analysed using Microcal Origin version 7.5 (Microcal Software Limited).
HIV-1 inhibition assay
Intracellular p24 antigen expression in CD4+ cells were used to quantify HIV-1 infection. Viral inhibition was assessed by the HIV-1 infection rate after the peptide incubation. Cryopreserved PBMCs were thawed and stimulated with PHA (5 μg/mL) in RPMI 1640 medium (Sigma, UK) supplemented with 10% fetal calf serum (R10) for 3 days. Biotinylated peptides were incubated with streptavidin for 10 min in room temperature. The peptide tetramers (10 μM) and HIV-1BaL (National Institute for Biological Standards and Control, United Kingdom), HIV-1 IIIB and HIV-1 clade C (ES-X1936-C) were incubated at 37°C for 30 min. Licensed HIV-1 fusion inhibitor T20 (5 μM) and HIV-1 without peptides were used as positive and negative controls. After the incubation, PBMCs were infected with HIV-1 isolates (with or without peptides incubation) at a multiplicity of infection (MOI) of 0.002. HIV-1-infected PBMC (2 × 105) were cultured in triplicate in R10 with interleukin 2 (20 IU/mL) in 96-well round-bottomed plates. Cells were harvested at day 3, stained with Aqua Live/Dead Fixable stain (Invitrogen), fixed with 1% paraformaldehyde/20 μg/mL lysolecithin at RT, permeabilized with cold 50% methanol followed by 0.1% Nonidet P-40, finally stained with p24 antibody (KC-57-FITC; Beckman Coulter) and antibodies to CD3, CD4, and CD8 (conjugated to APC-Cy7, PE and APC, respectively; BD Biosciences) and analyzed by flow cytometry. HIV-1 infection rate was expressed as percentage of HIV-1-infected CD4+ T cells (identified by gating on CD3+ CD8- cells, to allow for HIV-induced CD4 downregulation) and determined as follows: [(p24+, CD4+ T cells) + (p24+, CD4-T cells)]/[(p24+, CD4+ T cells) + (p24+, CD4-T cells) + (P24-, CD4+ T cells)] × 100. The protocols were described previously by Dorrell et al.22.
Author Contributions
L.H., X.P. and G.O. designed the experiments and undertook experimental work. H.Y., L.W., G.S.-J., L.D. also designed experiments and carried out experimental work. All authors have reviewed the manuscript.
Supplementary Material
Supplementary information
Acknowledgments
We are grateful for funding support from the Medical Research Council and NIHR Biomedical Research Centre programme and ISIS Innovation. L.D. is a Jenner Investigator.
Footnotes
A patent has been filed on the technology but there are no financial arrangements beyond that. There are no other financial conflicts of interest.
References
- Vlieghe P., Lisowski V., Martinez J. & Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug discovery today 15, 40–56 (2010). [DOI] [PubMed] [Google Scholar]
- McCafferty J., Griffiths A. D., Winter G. & Chiswell D. J. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348, 552–554 (1990). [DOI] [PubMed] [Google Scholar]
- Georgiou G. et al. Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nature biotechnology 15, 29–34 (1997). [DOI] [PubMed] [Google Scholar]
- Boder E. T. & Wittrup K. D. Yeast surface display for screening combinatorial polypeptide libraries. Nature biotechnology 15, 553–557 (1997). [DOI] [PubMed] [Google Scholar]
- Roberts R. W. & Szostak J. W. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proceedings of the National Academy of Sciences of the United States of America 94, 12297–12302 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovsek D. & Pluckthun A. In-vitro protein evolution by ribosome display and mRNA display. J Immunol Methods 290, 51–67 (2004). [DOI] [PubMed] [Google Scholar]
- Williams R. et al. Amplification of complex gene libraries by emulsion PCR. Nat Meth 3, 545–550 (2006). [DOI] [PubMed] [Google Scholar]
- Tawfik D. S. & Griffiths A. D. Man-made cell-like compartments for molecular evolution. Nature biotechnology 16, 652–656 (1998). [DOI] [PubMed] [Google Scholar]
- Lu W. C. & Ellington A. D. In vitro selection of proteins via emulsion compartments. Methods (2012). [DOI] [PubMed] [Google Scholar]
- Kojima T. et al. PCR amplification from single DNA molecules on magnetic beads in emulsion: application for high-throughput screening of transcription factor targets. Nucleic Acids Res 33, e150 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan R., Yamanaka Y., Kojima T. & Nakano H. Microbeads display of proteins using emulsion PCR and cell-free protein synthesis. Biotechnol Prog 24, 1107–1114 (2008). [DOI] [PubMed] [Google Scholar]
- Paul S., Stang A., Lennartz K., Tenbusch M. & Uberla K. Selection of a T7 promoter mutant with enhanced in vitro activity by a novel multi-copy bead display approach for in vitro evolution. Nucleic Acids Res (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L. M. et al. Broad and Potent Neutralizing Antibodies from an African Donor Reveal a New HIV-1 Vaccine Target. Science 326, 285–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L. M. et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElrath M. J. & Haynes B. F. Induction of Immunity to Human Immunodeficiency Virus Type-1 by Vaccination. Immunity 33, 542–554 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X. et al. Antibody neutralization and escape by HIV-1. Nature 422, 307–312 (2003). [DOI] [PubMed] [Google Scholar]
- Mouquet H. et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 467, 591–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogo-Isonagie C. et al. Peptides from second extracellular loop of C-C chemokine receptor type 5 (CCR5) inhibit diverse strains of HIV-1. The Journal of biological chemistry 287, 15076–15086 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnur E. et al. The conformation and orientation of a 27-residue CCR5 peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide. Journal of molecular biology 410, 778–797 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones-Mateu M. E. et al. Human epithelial beta-defensins 2 and 3 inhibit HIV-1 replication. AIDS 17, F39–48 (2003). [DOI] [PubMed] [Google Scholar]
- Yang H. et al. Antiviral inhibitory capacity of CD8+ T cells predicts the rate of CD4+ T-cell decline in HIV-1 infection. The Journal of infectious diseases 206, 552–561 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information