Abstract
Ubiquitin carboxyl terminal hydrolase 1 (UCHL1) catalyzes the hydrolysis of COOH-terminal ubiquityl esters and amides. It has been reported as either an oncogene or a tumor suppressor in cancers. However, UCHL1's role in ovarian cancer is still unclear. Therefore, we conducted an analysis to understand the role of UCHL1 in ovarian cancer. Firstly, we detected UCHL1 promoter methylation status in 7 ovarian cancer cell lines. 4 of them with UCHL1 silencing showed heavy promoter methylation while the other 3 with relative high UCHL1 expression showed little promoter methylation. Then we reduced UCHL1 expression in ovarian cancer cell line A2780 and IGROV1 and found that inhibition of UCHL1 promoted cell proliferation by increasing cells in S phases of cell cycle. Knockdown of UCHL1 also reduced cell apoptosis and contributed to cisplatin resistance. Furthermore, the expression level of UCHL1 in several ovarian cancer cell lines correlated negatively with their cisplatin resistance levels. Microarray data revealed that UCHL1 related genes are enriched in apoptosis and cell death gene ontology (GO) terms. Several apoptosis related genes were increased after UCHL1 knockdown, including apoptosis regulator BCL2, BCL11A, AEN and XIAP. Furthermore, we identified up-regulation of Bcl-2 and pAKT as well as down-regulation of Bax in UCHL1 knockdown cells, while no significant alteration of p53 and AKT1 was found. This study provides a new and promising strategy to overcome cisplatin resistance in ovarian cancer via UCHL1 mediated pathways.
Keywords: Ubiquitin carboxyl terminal hydrolase 1, tumor suppressor
1. Introduction
Ovarian cancer is a serious health problem for women all over the world. Due to non-obvious early symptoms, about 70% of all cases are diagnosed at advanced stages 1. In the USA, estimated new cases of ovarian cancer in 2012 account for only 3% of all kinds of female cancer patients and it's estimated death rate doubles, reaching 6% 2, which is higher than other gynecological malignancies.
UCHL1, short for ubiquitin carboxyl-terminal hydrolase L1, is a member of ubiquitin carboxy terminal hydrolase (UCH) family that catalyzes the hydrolysis of COOH-terminal ubiquityl esters and amides. UCHL1 was initially found to be expressed specifically in testis/ovary and brain3, 4. UCHL1 mutations are related to neurodegenerative diseases such as Parkinson's diseases (PD) and Alzheimer disease (AD) 5. UCHL1 has been reported as either an oncogene or a tumor suppressor gene. UCHL1 has been reported to be up-regulated in a variety of cancers. For example, over expression UCHL1 in lymphoma strongly accelerated lymphomagenesis by boosting signaling through the AKT pathway via down regulation of the antagonistic phosphatase PHLPP1 6. In non-small lung cancer cell line H157, UCHL1 promotes invasion by upstream activation of Akt 7. In colorectal cancer, UCHL1 acts as an oncogene via activation of the beta-catenin/TCF pathway through its deubiquitinating activity8. In contrary, promoter hypermethylation that leads to silencing or low expression of UCHL1 were also found in several types of cancers, including nasopharyngeal carcinoma 9, melanoma 10, hepatocellular carcinoma 11, esophageal squamous cell carcinoma 12, renal cancer 13, gastric cancer 14 and prostate cancer 15.
Cisplatin is a common reagent used in chemotherapy for ovarian cancers with an initial response rate of about 70%, yet recurrence is common as patients often develop resistance to cisplatin therapy 16. Proteins modulating cisplatin responses are of great importance to overcome cisplatin resistance. UCHL1 was a protein identified as differentially expressed proteins between the cisplatin-sensitive (A2780S) and cisplatin-resistant (A2780-CP) ovarian cancer cell lines17. However, the mechanism of how UCHL1 modulating cisplatin resistance has not been established. Here we describe our study in helping understanding the role of UCHL1 in chemotherapy response of ovarian cancers. We first showed that silencing of UCHL1 in ovarian cancer cell lines were caused by its promoter methylation and treatment of demethylation reagent could restore its expression. Furthermore, UCHL1 knockdown in ovarian cancer cell lines A2780 and IGROV1 promoted cell growth while causing reduction of apoptosis. Microarray analysis demonstrated that UCHL1 knockdown induced alterations in apoptosis and cell death. UCHL1 knockdown ovarian cancer cells (A2780-shUCHL1 and IGROV1-shUCHL1) became more resistant to cisplatin than their corresponding controls. Moreover, we found that UCHL1 expression level was negatively correlated with cisplatin resistance in ovarian cancer cells. We then detected up-regulation of Bcl-2 and down-regulation of Bax in UCHL1 knockdown cells, however, no significant alteration of p53 was detected. Also we detected up-regulation of phospho AKT. These results suggest that UCHL1 is mediator of apoptosis and targeting UCHL1 might be of great potential to overcome cisplatin resistance in ovarian cancer.
2. Materials and Methods
2.1 Cell lines and cell culture
Ovarian cancer cell lines A2780, A2780CP, SKOV3 and IGROV1 were kind gifts from Dr. Stephen Collins from UC San Diego (CA, USA). ES-2, OVCAR3 and CAOV3 were purchased from Cellbank, Chinese Academy of Sciences, Shanghai, China. ES-2 and SKOV3 were cultured in McCoy's 5A medium (Invitrogen, Carlsbad, CA) with 10% FBS, while other cell lines were cultured in RPMI 1640 (Invitrogen) medium supplemented with 10% FBS at 37 ºC and 5% CO2.
2.2 Lentiviral vector and infection
UCHL1 was knocked down by pLKO.1-puro lentiviral Vector cloned with UCHL1-target shRNA (CCGGGTGTGAGCTTCAGATGGTGAACTCGAGTTCACCATCTGAAGCTCACACTTTTT), purchased from Sigma-Aldrich. A Non-Mammalian shRNA (SHC002V) was used as a negative control. The lentivirus or the control was added to A2780 and IGROV1 cells with 8 µg/ml (final concentration) hexadimethrine bromide (H9268, Sigma-Aldrich) for infection. 500ng/ml (final concentration) puromycin was added to select positive clones. Stable transfectants were isolated after 3 weeks selection.
2.3 MTT assay and Cytotoxicity assay
Growth was measured by MTT assay. Briefly, 3,000 cells were collected and seeded to each well of 96-well plate. After 1, 3, 5 days, MTT was added to cells (final concentration was 0.5mg/ml). After 4 hours incubation, medium was replaced by 100ul DMSO and OD value at 490nm was measured.
To measure cisplatin sensitivity, 8,000 cells were seeded to each well in 5 replicates of 96-well plate. Cells were treated with a series dose of cisplatin from 0 µg/ml to 16 µg/ml and 48 hours later MTT assay was performed to measure cell viability. To induce apoptosis to measure active caspase-3 protein level, 20µg/ml cisplatin (final concentration) was added and 16 hours later cells were harvested to perform western blot analysis.
2.4 RNA isolation and Real-time PCR
Total RNAs were extracted from cells using Trizol reagent (Invitrogen) according to the manufacturer's instructions. 2 µg RNA was reverse transcribed in a final volume of 25µl using M-MLV reverse transcriptase (Promega, USA). Real-time PCR were performed using SYBR-Green PCR kit (Takara, Japan) in Bio-RAD CFX96 Real-Time System. All measurements were performed in triplicate following the cycle scheme: 95 ºC for 2 min, 40 cycles of 95 ºC for 15s, 58 ºC for 30s and 72ºC for 15s. Primers for real-time PCR were designed as follows: UCHL1, forward, 5'- CCGAGATGCTGAACAAAG-3', and reverse, 5'-CAGAGACTCCTCTTCCAG-3'; BCL2, forward, 5'-ATAACGGAGGCTGGGATG-3', reverse, 5'-GCATGTTGACTTCACTTGTG-3'; GAPDH, forward, 5'-GGAGATTGTTGCCATCAACG-3', and reverse, 5'-TTGGTGGTGCAGGATGCATT-3'.
2.5 5-Aza-2'-deoxycytidine treatment
A2780CP cell line was treated with 10 µM 5-Aza-dC (Sigma-Aldrich) for 5 days. Medium was replaced with fresh 5-Aza-dC every 24 hours.
2.6 DNA extraction, methylation specific PCR and bisulfite-modified DNA sequencing
Genomic DNA was extracted from cultured cells using DNeasy Blood & tissue Kit (Qiagen, Germany). Then genomic DNA was quantified and 250 ng of DNA was bisulfite-treated with EZ DNA Methylation-GoldTM Kit (Zymo Research, CA, USA) according to the manufacturer's protocol. Methylation specific PCR (MSPCR) for UCHL1 was performed using Zymo Taq™ DNA polymerase (Zymo Research) with primers as follows: M, forward, 5'-TTTATTTGGTCGCGATCGT TC-3', reverse, 5'-AAACTACATCTTCGCGAAACG-3'; U, forward, 5'-GTATTTA TTTGGTTGTGATTGTTT-3', reverse, 5'-CTTAAACTACATCTTCACAAAAC A-3'. The MSPCR was carried out following the polymerase's protocol with annealing temperature 57 ºC.
For bisulfite-modified DNA sequencing, Zymo Taq™ DNA polymerase was used to amplify a fragment of the UCHL1 promoter CpG islands containing 34 CpG sites with primers as follows: forward, 5'-GTTTGGTTTTGTTTTT GTTTTTTTT-3', reverse, 5'-ACTTAATCCCTACCAACAACC-3'. The PCR cycle scheme was the same as MSPCR. PCR products were then cloned into pMD® 18-T Vector (Takara) and 6-8 colonies were randomly chosen for sequencing. Primers for both MSPCR and bisulfite PCR were designed by web tools MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) 18.
2.7 Flow cytometry analysis of cell cycle and apoptosis
For cell cycle assay, cells were cultured in 12-well plates to about 70% in reduced serum (1%) for 24 hours to get synchronized. Then cells were trypsinized and harvested, and washed with cold PBS. Next, cells were fixed with 80% cold ethanol over night at 4 ℃. Fixed cells were centrifuged and resuspended in propidium iodide solution, and subjected to flow cytometry analysis after 30 minutes' incubation at 37℃.
For apoptosis analysis, cells (10,000/well) were cultured in 6-well plates to about 70% confluence then were cultured for 24 hours in reduced serum (1%). Then cells were collected and Annexin V-FITC/PI staining was performed according to standard protocols. After staining, cells were analyzed using BD FACS Array™ (Becton-Dickinson, San Jose, CA) according to manufacturer's guidelines.
2.8 Western blot
Protein was extracted using RIPA [50mM Tris(pH 7.4), 150mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 2mM sodium pyrophosphate, 25mM β-glycerophosphate, 1mM EDTA, 1mM Na3VO4, 0.5µg/ml leupeptin] and concentration was measured with BCA Protein Assay Kit (23225, Thermo Scientific). Equal amounts of protein (20µg) were added to 12% SDS-PAGE gel for electrophoresis. After that, proteins were transferred to polyvinylidene fluoride (PVDF) membranes and blocked in 5% non-fat milk in TBST for 1 hour. Antibodies used were UCHL1 (PAB3903, Abnova), Bax (Epitomics), BCL2 (Epitomics), GAPDH (Epitomics). GAPDH was used as internal control.
2.9 Microarray and Gene Ontology analysis
To identify genes regulated by UCHL1, we performed a microarray assay using Affymetrix U133 plus 2 genechip (Affymetrix, Santa Clara, CA, USA). After RMA normalization, data were exported to the GeneSpring program (Agilent, Inc.) to filter out lowly expressed and unreliably detected genes with probe intensity <100. After that, genes with fold change <2 was furthered eliminated. Finally, Gene Ontology analysis was performed on line using the GoMiner program 19.
2.10 Statistical analysis
All data were presented with mean ± SD. Cell survival curves were fitted to the data points with nonlinear regression analysis using GraphPad Prism 5.0. Two-tailed Student's t-test was used to compare mean values. p<0.05 was considered statistically significant.
3. Results
3.1 UCHL1 expression was correlated with its promoter methylation and could be restored with 5-Aza-dC treatment
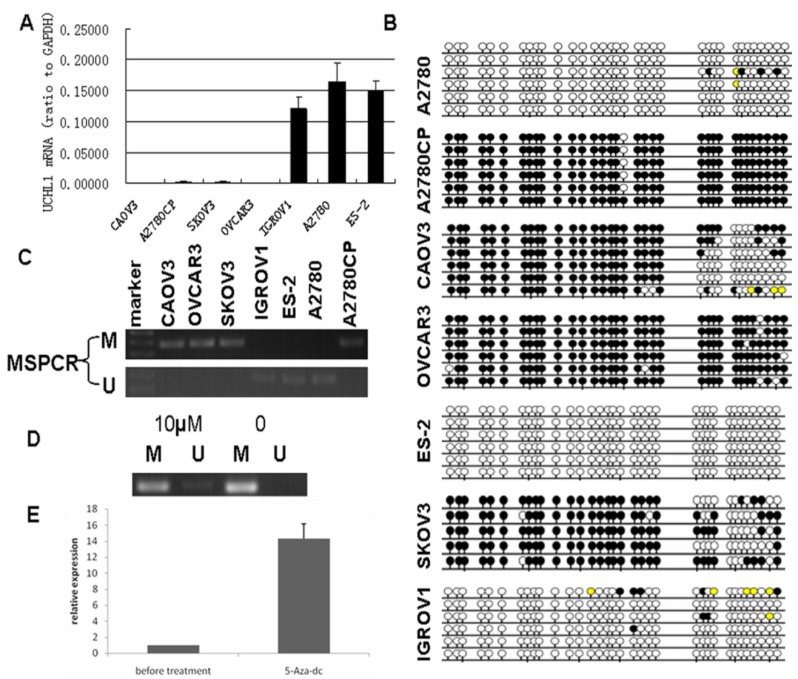
UCHL1 was reported frequently methylated in various cancers, including ovarian cancer20. To validate its promoter methylation status, we performed MS-PCR in 7 ovarian cancer cell lines (ES-2, A2780, IGROV1, OVCAR3, SKOV3, A2780CP and CAOV3). We found that the methylation status of UCHL1 in the promoter region correlated negatively with UCHL1 expression: those cell lines with very low UCHL1 expression showed positive methylation while those with relatively high UCHL1 expression showed negative methylation (Fig.1A). We next performed bisulfite genomic sequencing (BGS) of all 7 ovarian cancer cell lines (Fig. 1B). BGS data matched perfectly with MS-PCR results (Fig. 1C): The MS-PCR positive cell lines showed heavy promoter methylation while MS-PCR negative cell lines showed very weak or no promoter methylation (Fig.1B-C).
Fig 1.
Silencing of UCHL1 was related to its promoter hypermethylation and UCHL1 expression could be restored by 5-Aza-dc treatment. (A) UCHL1 expression in seven ovarian cancer cell lines. (B) Bisulfite-modified DNA sequencing of UCHL1 promoter in seven ovarian cancer cell lines. Open circles, unmethylated; closed circles, methylated; yellow, unknown methylation status. (C) Methylation status of UCHL1 promoter in seven ovarian cancer cell lines. M, methylated; U, unmethylated. (D) Methylation status of UCHL1 in A2780CP cells treated with/without 10 µM 5-Aza-dc. M, methylated; U, unmethylated. (E) UCHL1 expression could be restored by 5-Aza-dc treatment in A2780CP cells.
We then chose A2780CP, an isogenic cell line of A2780 that differ in cisplatin resistance, and treated A2780CP cells with demethylation reagent 5-Aza-dC for 5 days to confirm whether silencing of UCHL1 is caused by its promoter hypermethylation. After 5-Aza-dC treatment, A2780CP showed a weak band of unmethylation (Fig.1D). Furthermore, UCHL1 expression was significantly induced after 5-Aza-dC treatment by RT-PCR analysis (Fig.1E). These results suggest that hypermethylation of UCHL1 in promoter region is responsible for its transcriptional silencing.
3.2 Reduced expression of UCHL1 in A2780 and IGROV1 reduced cell apoptosis but increases cell proliferation
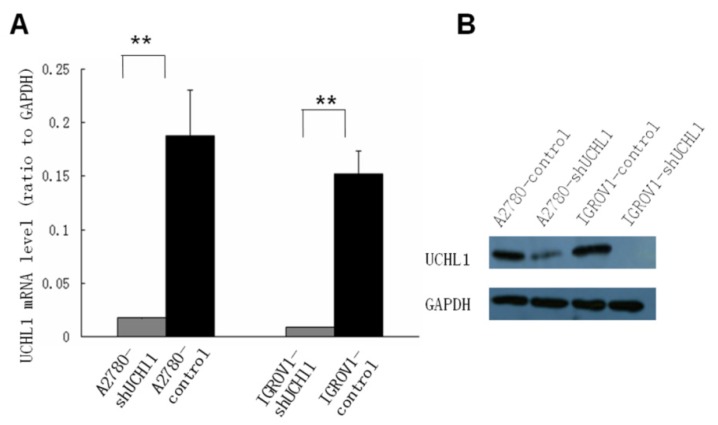
To study the function of UCHL1 in ovarian cancer, two ovarian cancer cell lines, A2780 and IGROV1, which express relatively high level UCHL1, were chosen to perform UCHL1 knockdown. Lentivirus infection led to a high and stable knockdown-efficiency in both A2780 and IGROV1 cells. UCHL1 expression was significantly reduced at mRNA level as well as protein level (Fig. 2).
Fig 2.
Suppression of UCHL1 expression by shRNA. Lentivirus harboring shRNA against UCHL1 or Non-Mammalian shRNA was used to infect UCHL1-positive A2780 and IGROV1 cell lines. Expression of UCHL1 was determined by quantitative real-time polymerase chain reaction (qRT-PCR) (A) and Western Blot (B). Both A2780-shUCHL1 and IGROV1-shUCHL1 showed a significant reduction of UCHL1 expression. **, p <0.01.
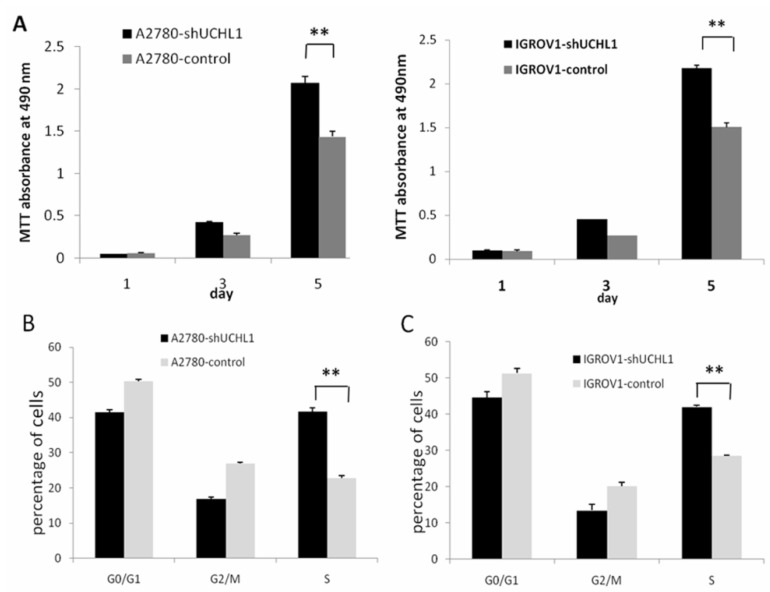
We performed a cell growth curve assay to determine the effect of UCHL1-knockdown in A2780 and IGROV1 cells. As shown in Fig. 3A, MTT absorbance at 490 nm of A2780-shUCHL1 and A2780-control differed significantly on the 5th day, indicating that reduced expression of UCHL1 in A2780 cells increased cell proliferation. The same situation was detected between IGROV1-shUCHL1 and IGROV1-control. Furthermore, cell cycle analysis revealed up-regulation of S phase both in A2780-shUCHL1 and IGROV1-shUCHL1 (Fig. 3B-C), indicating that UCHL1 knockdown promoted cell proliferation by progressing cell cycle.
Fig 3.
Effect of UCHL1 knockdown on cell proliferation and cell cycle. (A) Cell proliferation ability was determined by MTT assay. Data was shown by mean of spectrometric absorbance ±SD of four independent experiments. (B) Cell cycle of A2780-shUCHL1 and A2780-control was analyzed by flow cytometry. (C) Cell cycle of IGROV1-shUCHL1 and IGROV1-control was analyzed by flow cytometry.
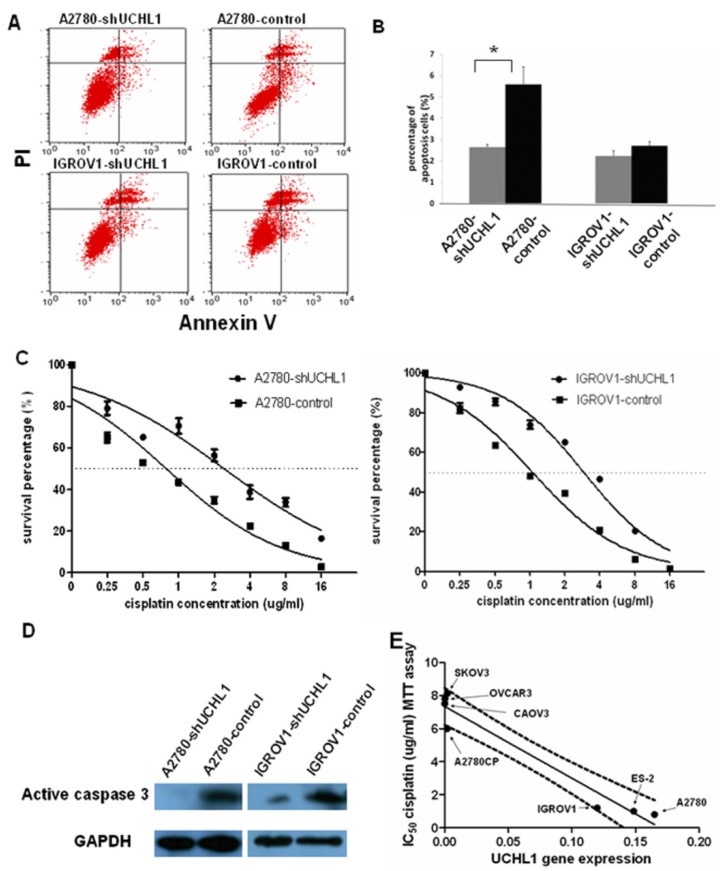
Then we assessed the effects of UCHL1 knockdown on apoptosis. We found that reduced UCHL1 expression resulted in reduction of apoptosis. In A2780-shUCHL1 cells, this effect was especially significant (p<0.05) (Fig. 4A). IGROV1-shUCHL1 cells also showed a similar trend while the effect was not statistically significant (Fig. 4B).
Fig 4.
Knockdown of UCHL1 reduced cell apoptosis and contributed to cisplatin resistance. (A) Cell apoptosis ratio was analyzed by flow cytometry. Shown were representative apoptosis figures of each cell line and data was summarized in (B). (C) UCHL1 knockdown induced cisplatin resistance in ovarian cancer cell lines. A2780-shUCHL1, A2780-control, IGROV1-shUCHL1 and IGROV1-control were treated with cisplatin in different doses from 0µg /ml to 16µg/ml for 48 hours. Then MTT assay was performed to measure the cell survival. Dotted line, 50% survival. (D) Total protein was extracted from the four cell lines which were treated with 20µg/ml cisplatin for 16 hours and analyzed with active caspase 3 antibody, with GAPDH as internal control. (E) Expression of UCHL1 in diverse ovarian cancer lines negatively correlates with cisplatin resistance. UCHL1 mRNA expression and cisplatin resistance in a panel of 7 ovarian cancer cell lines were negatively correlated, with correlation coefficient -0.96 (p<0.001). Dotted line, 95% confidence interval.
3.3 Reduced expression of UCHL1 in A2780 and IGROV1 induces cisplatin resistance
Most anti-cancer reagents work by triggering apoptosis in tumor cells and the ability to induce apoptosis determines the therapeutic efficacy. Since suppression UCHL1 reduced apoptosis, we performed MTT assay to determine whether UCHL1 knockdown in ovarian cancer cells influences resistance to cisplatin, a common reagent used to treat ovarian cancer. As shown in Fig.4C, A2780-shUCHL1 and IGROV1-shUCHL1 showed relatively high cisplatin resistance compared to their controls, with IC50 increasing from 0.8µg/ml to 2.4µg/ml for A2780-control and A2780-shUCHL1, and from 1.2µg/ml to 3.2µg/ml for IGROV1-control and IGROV1-shUCHL1. Furthermore, we conducted western blot to determine cleaved caspase 3 expression after cisplatin treatment of these cells. As expected, there was a decrease in cleaved caspase 3 expression in UCHL1 knockdown cells (Fig.4D), indicating that reduced expression of UCHL1 in A2780 and IGROV1 made the cells become resistant to cisplatin-induced apoptosis.
We questioned whether UCHL1 expression level correlates with cisplatin resistance in ovarian cancer. We measured UCHL1 mRNA expression level and determined cisplatin resistance with MTT assay in a panel of ovarian cancer cell lines. SKOV3, OVCAR3 are derived from metastatic tumors of patients treated with chemotherapy and are resistant to cisplatin. CAOV3 also exhibited cisplatin resistance although it's sensitive to other chemotherapy agent 21. A2780, ES-2, and IGROV1, derived from untreated patients, are sensitive to cisplatin 22, 23. A2780CP, a parental cell line of A2780, is resistant to cisplatin 24. We found that UCHL1 expression levels were negatively correlated with IC50 values from cisplatin MTT assay with a correlation coefficient of -0.96 (**, p<0.001), which was statistically significant (Fig. 4E).
3.4 Identification of biological process regulated by UCHL1 suppression
To explore the mechanisms of how reduced expression of UCHL1 promotes cell proliferation, reduces apoptosis and confers cisplatin resistance, we performed a microarray analysis to identify different genes regulated by UCHL1-knockdown. The array data was submitted to the GEO database (GSE43884) (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43884). Comparing gene expression profile between A2780-shUCHL1 and A2780-control, we identified 1549 differentially expressed genes (724 down-regulated and 825 up-regulated) with fold change >2 (Supplementary Table 1). We noted up-regulation of several apoptosis related genes including apoptosis regulator BCL2 (B-cell CLL/lymphoma 2) (up 4.1 folds), BCL11A (B-cell CLL/lymphoma 11A) (up 29 folds), AEN (apoptosis enhancing nuclease) (up 2.0 folds) and XIAP (X-linked inhibitor of apoptosis) (up 2.6 folds).
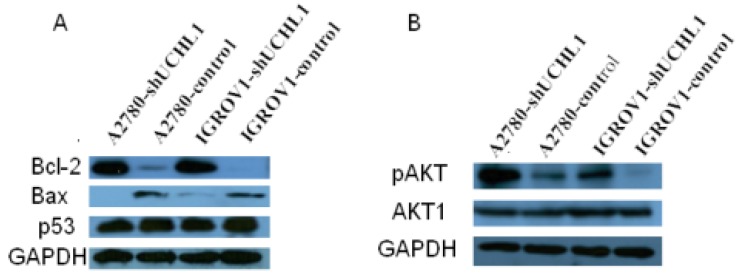
We next focused on BCL2, an important regulator of apoptosis, for validation. First, we validated that BCL2 also was up-regulated at protein levels in knock-down cell lines A2780-shUCHL1 and IGROV1-shUCHl1 (Fig. 5A). Another member of BCL2 family Bax, which heterodimerizes with Bcl-2 to accelerate cell apoptosis 25, was down-regulated (Fig. 5A). As p53 expression was reported to be altered in UCHL1 over-expression or knockdown cells 9, 11, 26, we also determined p53 level using western blot. However, we did not observe any significant alteration of its expression (Fig. 5A). Moreover, we detected higher pAKT expression in A2780-shUCHL1 and IGROV1-shUCHl1 while no significant alteration of AKT1 was detected, suggesting that the AKT pathway, rather than p53 signaling pathway might be involved in UCHL1 regulated apoptosis in these ovarian cancer cells.
Fig 5.
Detection of apoptosis related proteins by western blot. (A) Up-regulation of Bcl-2/Bax but no significant alteration of p53 were detected in UCHL1 knockdown cells. (B) Up-regulation of AKT pathway activity was found in UCHL1 knockdown cells.
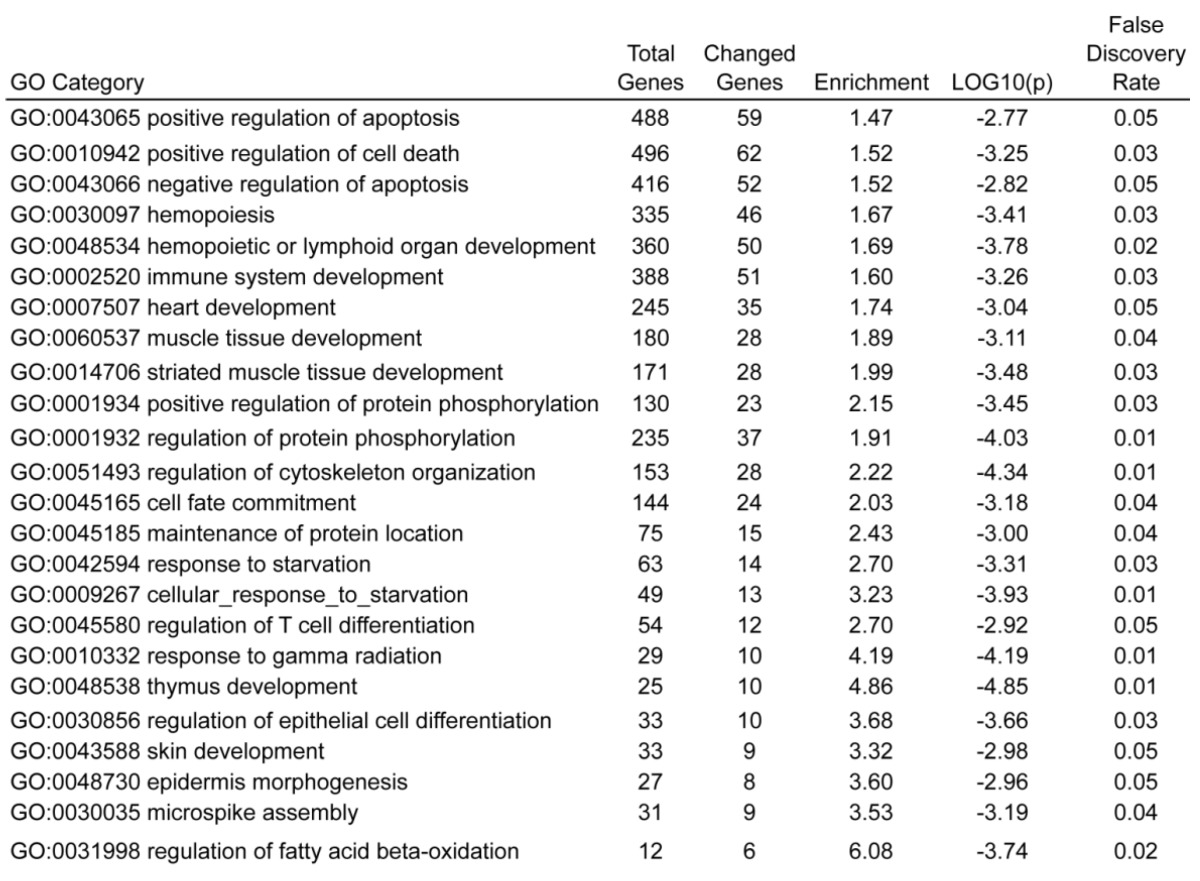
Gene ontology analysis by GoMiner of these differentially expressed genes, with all gene list as total file, showed that 24 GO terms were found to be enriched, including positive regulation of apoptosis (GO:0043065), regulation of cell death (GO:0010942), negative regulation of apoptosis (GO:0043066), which were top three enriched GO terms (Table 1). This result was consistent with our observations, strongly indicating that UCHL1 is likely to be involved in the regulation of apoptosis.
Table 1.
Enriched GO terms in biological process for UCHL1 regulated genes.
4. Discussion
In this study, we described, for the first time, how UCHL1 modulates cisplatin resistance in ovarian cancers. We showed that knockdown of UCHL1 in ovarian cancer cells promoted cell proliferation (Fig. 3A) by inducing S phase cell cycle (Fig. 3B-C) and reduced apoptosis (Fig. 4A-B). Caspase 3, a frequently activated apoptosis protease and a typical hallmark of apoptosis 27, is reduced upon UCHL1 knockdown (Fig. 4D). Our data is consistent with previous reports of UCHL1 in other types of cancers. UCHL1 over-expression has been reported to induce cell growth arrest in prostate cancer 15 and breast cancer 26, 28. In breast cancer, ectopic expression of UCHL1 is able to induce apoptosis by stabilizing p53. In nasopharyngeal carcinoma, UCHL1 was found to form a complex with p53/MDM2/ARF to promote p53 signaling 9. In hepatocellular carcinoma, it was reported that UCHL1 serves as a tumor suppressor gene by directly interacting with p53 and stabilizing p53 11. Yet in our study, no significant alteration of p53 was found (Fig. 5A), suggesting UCHL1 mediated apoptosis in other ways rather than by regulating p53.
We initially observed that UCHL1 expression is anti-correlated with cisplatin resistant levels (as measured by IC50) (Fig. 4E). This is consistent with a UCHL1 expression at the GEO database showing that UCHL1 was reduced to no expression in the tissues from carboplatin resistant ovarian cancer patients (http://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS1381:36990_at) compared to the carboplatin sensitive tissues (Supplementary Fig. 1)29. Carboplatin, a parental platinum compound of cisplatin, shares similar biochemical characters as well as similar anti-cancer mechanisms with cisplatin 30. The fact that cross resistance between carboplatin and cisplatin 31, 32 ensure us making use of the GEO dataset.
We further observed that methylation of the UCHL1 promoter correlated negatively to the expression of UCHL1 (Fig. 1A-C). Demethylation of A2780CP with Aza restored UCHL1 expression, confirming promoter methylation was responsible for its silencing (Fig. 1D-E). This is consistent with the previous observation by Okochi-Takada et al. who showed that UCHL1 was silenced in about half of the ovarian cancer cell lines that they tested and the expression was also correlated with promoter methylation status 33. Moreover, we have extended this finding to further experimentation and showed that knockdown of UCHL1 increased cisplatin resistance in both A2780 and IGROV1 ovarian cancer cells (Fig. 4C). Our data demonstrated, for the first time, that UCHL1 expression directly regulated cisplatin resistance in ovarian cancers.
We also performed microarray expression profiling to understand the global picture of UCHL1's effects on ovarian cancer cells. GoMiner analysis showed that apoptosis and cell death related biological processes are enriched in UCHL1 regulated genes. In particular, we found that several apoptosis related genes including apoptosis regulator BCL2 (B-cell CLL/lymphoma 2), BCL11A (B-cell CLL/lymphoma 11A), AEN (apoptosis enhancing nuclease) and XIAP (X-linked inhibitor of apoptosis). AEN was recently identified as a direct target gene of p53 34. Bcl-2/Bax ratio is a determinant of cell fate and was considered to be a “rheostat” that regulates apoptosis 35, 36. We found up-regulated Bcl-2/Bax ratio in UCHL1-knockdown cells (Fig. 5A), however, no significant alteration of p53 was found (Fig. 5A).
AEN induces apoptosis by itself, and more importantly, it is also required for efficient DNA fragmentation in p53-dependent apoptosis 34. The role of AEN in UCHL1 knockdown induced apoptosis remains to be studied. XIAP regulates AKT activity and caspase-3-dependent cleavage during cisplatin-induced apoptosis in human ovarian epithelial cancer cells 37. Interestingly, in our study we found up-regulated AKT activity in UCHL1-knockdown ovarian cancer cells (Fig. 5B). Downregulation of XIAP inhibits proliferation, induces apoptosis, and reverses the cisplatin resistance of ovarian carcinoma 38. Here we demonstrated that UCHL1 regulated the expression of AEN and XIAP, suggesting that UCHL1 might be a key master regulator of cisplatin resistance in ovarian cancers. Further experimentation is warranted.
In summary, we present data showing that UCHL1 might be a tumor suppressor in ovarian cancers. Furthermore, UCHL1 expression level is negatively related to cisplatin resistance in ovarian cancer and knockdown of UCHL1 increased cisplatin resistance. GoMiner analysis showed that apoptosis and cell death related biological processes are enriched in UCHL1 regulated genes. Our study provides a foundation to develop promising strategy to overcome cisplatin resistant ovarian cancer via UCHL1 mediated pathways.
Supplementary Material
Acknowledgments
This research was supported by grants from MOST, China (2012AA022705 and 2007DFC30360) and a grant from the Shaoxing Major Scientific and Technological Projects (2011A11013).
References
- 1.Naora H, Montell DJ. Ovarian cancer metastasis: integrating insights from disparate model organisms. Nat Rev Cancer. 2005;5:355–66. doi: 10.1038/nrc1611. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: A Cancer Journal for Clinicians. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Sekiguchi S, Yoshikawa Y, Tanaka S. et al. Immunohistochemical analysis of protein gene product 9.5, a ubiquitin carboxyl-terminal hydrolase, during placental and embryonic development in the mouse. Exp Anim Tokyo. 2003;52:365–9. doi: 10.1538/expanim.52.365. [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson KD, Lee K, Deshpande S, Duerksenhughes P, Boss JM, Pohl J. The Neuron-Specific Protein Pgp-9.5 Is a Ubiquitin Carboxyl-Terminal Hydrolase. Science. 1989;246:670–3. doi: 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- 5.Setsuie R, Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem Int. 2007;51:105–11. doi: 10.1016/j.neuint.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Hussain S, Foreman O, Perkins SL. et al. The de-ubiquitinase UCH-L1 is an oncogene that drives the development of lymphoma in vivo by deregulating PHLPP1 and Akt signaling. Leukemia. 2010;24:1641–55. doi: 10.1038/leu.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HJ, Kim YM, Lim S, Nam YK, Jeong J, Lee KJ. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2008;28:117–27. doi: 10.1038/onc.2008.364. [DOI] [PubMed] [Google Scholar]
- 8.Zhong J, Zhao M, Ma Y. et al. UCHL1 acts as a colorectal cancer oncogene via activation of the beta-catenin/TCF pathway through its deubiquitinating activity. Int J Mol Med. 2012;30:430–6. doi: 10.3892/ijmm.2012.1012. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Tao Q, Jin H. et al. The Tumor Suppressor UCHL1 Forms a Complex with p53/MDM2/ARF to Promote p53 Signaling and Is Frequently Silenced in Nasopharyngeal Carcinoma. Clinical Cancer Research. 2010;16:2949–58. doi: 10.1158/1078-0432.CCR-09-3178. [DOI] [PubMed] [Google Scholar]
- 10.Bonazzi VF, Nancarrow DJ, Stark MS. et al. Cross-platform array screening identifies COL1A2, THBS1, TNFRSF10D and UCHL1 as genes frequently silenced by methylation in melanoma. PLoS One. 2011;6:e26121. doi: 10.1371/journal.pone.0026121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, Tao Q, Cheung KF. et al. Epigenetic identification of ubiquitin carboxyl-terminal hydrolase L1 as a functional tumor suppressor and biomarker for hepatocellular carcinoma and other digestive tumors. Hepatology. 2008;48:508–18. doi: 10.1002/hep.22343. [DOI] [PubMed] [Google Scholar]
- 12.Mandelker DL, Yamashita K, Tokumaru Y. et al. PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer Res. 2005;65:4963–8. doi: 10.1158/0008-5472.CAN-04-3923. [DOI] [PubMed] [Google Scholar]
- 13.Seliger B, Handke D, Schabel E, Bukur J, Lichtenfels R, Dammann R. Epigenetic control of the ubiquitin carboxyl terminal hydrolase 1 in renal cell carcinoma. Journal of Translational Medicine. 2009;7:90. doi: 10.1186/1479-5876-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita K, Park HL, Kim MS. et al. PGP9.5 methylation in diffuse-type gastric cancer. Cancer Res. 2006;66:3921–7. doi: 10.1158/0008-5472.CAN-05-1511. [DOI] [PubMed] [Google Scholar]
- 15.Ummanni R, Jost E, Braig M. et al. Ubiquitin carboxyl-terminal hydrolase 1 (UCHL1) is a potential tumour suppressor in prostate cancer and is frequently silenced by promoter methylation. Mol Cancer. 2011;10:129. doi: 10.1186/1476-4598-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Einhorn LH. Testicular cancer: An oncological success story. Clinical Cancer Research. 1997;3:2630–2. [PubMed] [Google Scholar]
- 17.Gong F, Peng X, Zeng Z, Yu M, Zhao Y, Tong A. Proteomic analysis of cisplatin resistance in human ovarian cancer using 2-DE method. Mol Cell Biochem. 2011;348:141–7. doi: 10.1007/s11010-010-0648-6. [DOI] [PubMed] [Google Scholar]
- 18.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 19.Zeeberg BR, Feng W, Wang G. et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okochi-Takada E, Nakazawa K, Wakabayashi M. et al. Silencing of the UCHL1 gene in human colorectal and ovarian cancers. Int J Cancer. 2006;119:1338–44. doi: 10.1002/ijc.22025. [DOI] [PubMed] [Google Scholar]
- 21.Petru E, Sevin BU, Perras J. et al. Comparative Chemosensitivity Profiles in 4 Human Ovarian-Carcinoma Cell-Lines Measuring Atp Bioluminescence. Gynecologic Oncology. 1990;38:155–60. doi: 10.1016/0090-8258(90)90032-g. [DOI] [PubMed] [Google Scholar]
- 22.Katano K, Kondo A, Safaei R. et al. Acquisition of resistance to cisplatin is accompanied by changes in the cellular pharmacology of copper. Cancer Research. 2002;62:6559–65. [PubMed] [Google Scholar]
- 23.Wang Y, Niu XL, Qu Y. et al. Autocrine production of interleukin-6 confers cisplatin and paclitaxel resistance in ovarian cancer cells. Cancer Letters. 2010;295:110–23. doi: 10.1016/j.canlet.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 24.Yu W, Jin CM, Lou XY, Global Analysis of DNA Methylation by Methyl-Capture Sequencing Reveals Epigenetic Control of Cisplatin Resistance in Ovarian Cancer Cell. Plos One. 2011. 6. [DOI] [PMC free article] [PubMed]
- 25.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–19. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 26.Xiang T, Li L, Yin X. et al. The Ubiquitin Peptidase UCHL1 Induces G0/G1 Cell Cycle Arrest and Apoptosis Through Stabilizing p53 and Is Frequently Silenced in Breast Cancer. PLoS One. 2012;7:e29783. doi: 10.1371/journal.pone.0029783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porter AG, Janicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6:99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- 28.Liu XP. et al. Over-expression of ubiquitin carboxy terminal hydrolase-L1 induces apoptosis in breast cancer cells. International Journal of Oncology. 2008;33:1037–1045. doi: 10.3892/ijo_00000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters D, Freund J, Ochs RL. Genome-wide transcriptional analysis of carboplatin response in chemosensitive and chemoresistant ovarian cancer cells. Mol Cancer Ther. 2005;4:1605–16. doi: 10.1158/1535-7163.MCT-04-0311. [DOI] [PubMed] [Google Scholar]
- 30.Knox RJ, Friedlos F, Lydall DA, Roberts JJ. Mechanism of cytotoxicity of anticancer platinum drugs: evidence that cis-diamminedichloroplatinum(II) and cis-diammine-(1,1-cyclobutanedicarboxylato)platinum(II) differ only in the kinetics of their interaction with DNA. Cancer Res. 1986;46:1972–9. [PubMed] [Google Scholar]
- 31.Gore ME, Fryatt I, Wiltshaw E, Dawson T, Robinson BA, Calvert AH. Cisplatin/carboplatin cross-resistance in ovarian cancer. Br J Cancer. 1989;60:767–9. doi: 10.1038/bjc.1989.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boven E, van der Vijgh WJ, Nauta MM, Schluper HM, Pinedo HM. Comparative activity and distribution studies of five platinum analogues in nude mice bearing human ovarian carcinoma xenografts. Cancer Res. 1985;45:86–90. [PubMed] [Google Scholar]
- 33.Okochi-Takada E, Nakazawa K, Wakabayashi M. et al. Silencing of theUCHL1 gene in human colorectal and ovarian cancers. International Journal of Cancer. 2006;119:1338–44. doi: 10.1002/ijc.22025. [DOI] [PubMed] [Google Scholar]
- 34.Kawase T, Ichikawa H, Ohta T. et al. p53 target gene AEN is a nuclear exonuclease required for p53-dependent apoptosis. Oncogene. 2008;27:3797–810. doi: 10.1038/onc.2008.32. [DOI] [PubMed] [Google Scholar]
- 35.Korsmeyer SJ, Shutter JR, Veis DJ, Merry DE, Oltvai ZN. Bcl-2/Bax: a rheostat that regulates an anti-oxidant pathway and cell death. Semin Cancer Biol. 1993;4:327–32. [PubMed] [Google Scholar]
- 36.Chresta CM, Masters JR, Hickman JA. Hypersensitivity of human testicular tumors to etoposide-induced apoptosis is associated with functional p53 and a high Bax:Bcl-2 ratio. Cancer Res. 1996;56:1834–41. [PubMed] [Google Scholar]
- 37.Asselin E, Mills GB, Tsang BK. XIAP regulates Akt activity and caspase-3-dependent cleavage during cisplatin-induced apoptosis in human ovarian epithelial cancer cells. Cancer Res. 2001;61:1862–8. [PubMed] [Google Scholar]
- 38.Ma JJ, Chen BL, Xin XY. XIAP gene downregulation by small interfering RNA inhibits proliferation, induces apoptosis, and reverses the cisplatin resistance of ovarian carcinoma. Eur J Obstet Gynecol Reprod Biol. 2009;146:222–6. doi: 10.1016/j.ejogrb.2009.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.