Abstract
High quality nanocrystals have demonstrated substantial potential for biomedical applications. However, being generally hydrophobic, their use has been greatly limited by complicated and inefficient surface engineering that often fails to yield biocompatible nanocrystals with minimal aggregation in biological fluids and active targeting toward specific biomolecules. Using chimeric DNA molecules, we developed a one-step facile surface engineering method for hydrophobic Nanocrystals. The procedure is simple and versatile, generating individual nanocrystals with multiple ligands. In addition, the resulting nanocrystals can actively and specifically target various molecular addresses, varying from nucleic acids to cancer cells. Together, the strategy developed here holds great promise in generating critical technologies needed for biomedical applications of nanocrystals.
Nanometer-scale crystallites, which possess unique size- or shape-dependent physical and chemical properties, have demonstrated substantial potential for biomedical applications, including molecular imaging,1,2 disease diagnostics,3,4 cancer therapy,5-7 etc. For example, quantum dots (QDs) have been used to sensitize photodynamic therapy (PDT) agents, leading to a novel class of PDT sensitizers with tunable optical properties for treating both shallow- and deep-seated tumors.8 However, high quality nanocrystals are typically synthesized in organic solvents at elevated temperatures, resulting in bioincompatible nanocrystals coated with hydrophobic surfactant stabilizers (such as oleylamine, oleic acid and any other hydrocarbon chain-containing ligands).9,10 To address this issue, two major strategies have been devised for hydrophobic nanocrystal surface engineering to generate soluble and stable nanocrystals in aqueous solutions: (1) ligand exchange with thiol-, phosphine-, or dopamine-containing molecules; and (2) ligand encapsulation by a layer of amphiphilic polymers or silica shell.11-13 Unfortunately, despite recent advances, these surface engineering approaches often fail to produce individually dispersed nanocrystals in various biological fluids. In addition, complicated procedures and intensive energy input (e.g., heat and sonication) are generally required.14 Most importantly, additional steps are needed to incorporate biological moieties for specific molecular recognition, making the entire process time-and labor-consuming.
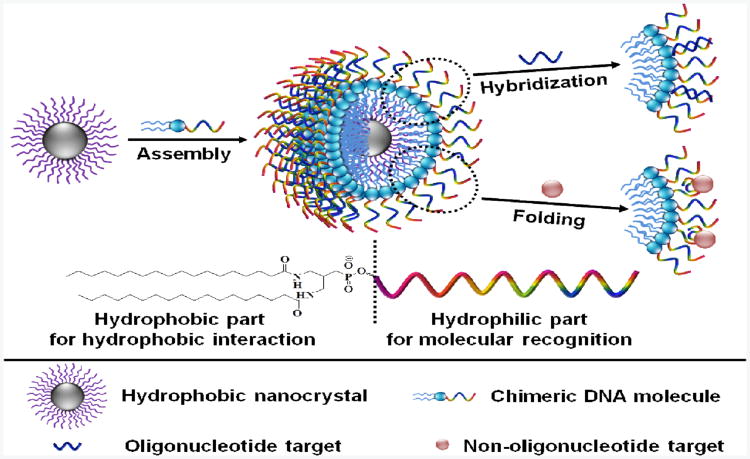
Here, we demonstrate a novel one-step method for hydrophobic nanocrystal surface engineering to produce different types of water soluble nanocrystals with tunable molecular recognition, using chimeric DNA molecules containing both hydrophobic (diacyllipid) and hydrophilic (oligonucleotide) parts. Through hydrophobic interactions, whereby relatively apolar molecules aggregate in aqueous solutions,15 these chimeric DNA molecules spontaneously intercalate in the surfactant layer of hydrophobic nanocrystals using their hydrophobic parts, encapsulating an individual nanocrystal inside the diacyllipid core and leaving an oligonucleotide corona outside (Scheme 1). The resulting water-soluble nanocrystals have a relatively narrow hydrodynamic size distribution and long-term stability in various biological media. In addition, since DNA can specifically recognize their targets by either Watson-Crick base pairing or by folding into distinct tertiary structures, these functionalized nanocrystals possess excellent selectivity to a variety of biomolecular targets, varying from nucleic acids to cancer cells.
Scheme 1.
Strategy for one-step surface engineering of hydrophobic nanocrystals with designer recognition. Hydrophobic interactions between the surfactant ligands (purple) on the nanocrystal surface and the diacyllipids (blue) in the chimeric DNA molecules lead to spontaneous assembly, encapsulating individual nanocrystal inside the diacyllipid core and leaving the oligonucleotide (multi-color) corona outside to confer excellent water solubility and tunable molecular recognition. The molecular structure of the chimeric DNA molecule is shown at the bottom.
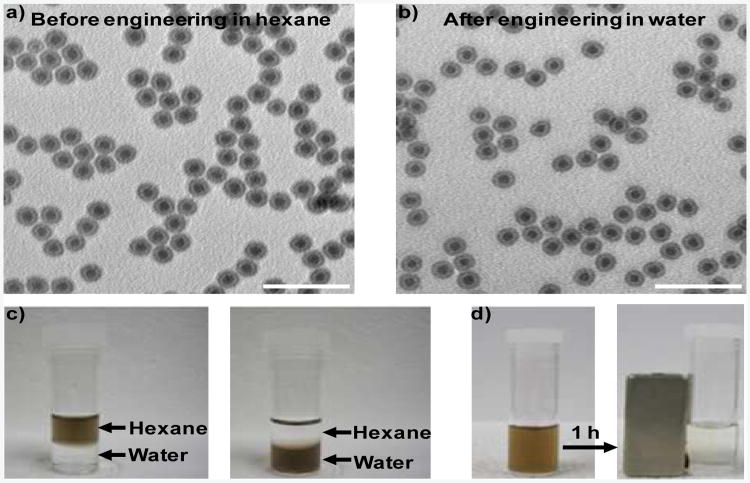
The chimeric DNA molecules are synthesized by efficiently incorporating a diacyllipid at the 5′ end of oligonucleotides through solid phase DNA synthesis on a fully automated DNA/RNA synthesizer, according to our previously reported procedure.16,17 To test the feasibility of this facile surface engineering method, oleylaminecoated Fe-Fe3O4 core-shell nanoparticles (CSNPs) (13 nm) are first used. The as-prepared Fe-Fe3O4 CSNPs were spherical and fairly monodisperse, as shown in a transmission electron microscopy (TEM) image (Figure 1a). For functionalization, Fe-Fe3O4 CSNPs in tetrahydrofuran, a water-miscible organic solvent, were mixed with chimeric DNA molecules in water. The reaction was conducted under ambient atmosphere while shaking. After washing to remove excess chimeric DNA molecules, the resulting Fe-Fe3O4 CSNPs were readily dispersed in water (Figure 1c) with negligible aggregation (Figure 1b). Dynamic light scattering (DLS) and zetapotential measurements indicated that the as-prepared Fe-Fe3O4 CSNPs in hexane had a diameter of 16.2 nm and a zeta-potential of 6.06 mV (Figure S1 in Supporting Information), respectively. While the modified Fe-Fe3O4 CSNPs in water had a diameter of 27.2 nm and a zetapotential of -30.17 mV (Figure S1 and Table S2 in Supporting Information), respectively. These results suggested that hydrophobic Fe-Fe3O4 CSNPs were stabilized by chimeric DNA molecules in water and formed uniformly distributed nanoparticles. The maximal concentration that functionalized Fe-Fe3O4 CSNPs can reach in aqueous solution is more than 5 mg/mL, which is sufficient for most of their biomedical applications. In addition, the engineered Fe-Fe3O4 CSNPs preserved the magnetic properties of the original nanocrystals (Figure 1d). Both UV-visible and FT-IR spectroscopies verified the presence of chimeric DNA molecules on the surfaces of functionalized Fe-Fe3O4 CSNPs (Figure S2 and S3 in Supporting Information): the characteristic UV absorption peak around 260 nm and vibrational band between 750 and 1750 cm−1 belonging to DNA were easily identified.
Figure 1.
Characterization of chimeric DNA molecule-engineered Fe-Fe3O4 CSNPs. TEM images of Fe-Fe3O4 CSNPs a) before and b) after chimeric DNA molecule modification in hexane and water, respectively. c) Solvent dispersity of Fe-Fe3O4 CSNPs before and after chimeric DNA molecule functionalization. The as prepared Fe-Fe3O4 CSNPs are hydrophobic and disperse in hexane, while the functionalized Fe-Fe3O4 CSNPs are hydrophilic and disperse in water. d) Magnetic separation of engineered Fe-Fe3O4 CSNPs. Lipid-T20 was used as the chimeric DNA molecule here. Detailed sequence information for all the chimeric DNA molecules can be find in Table S1 (SI). Scale bars: 50 nm.
The surface density of chimeric DNA molecules on the modified Fe-Fe3O4 CSNPs was estimated by fluorescence measurements (See Supporting Information for detailed procedures). With the newly developed surface engineering method, the number of chimeric DNA molecule per functionalized nanoparticle increased with ligand concentration and reached a plateau at the saturation concentration (Figure S4 in Supporting Information). Using fluorescently labeled lipid-T20, the saturation concentration for 0.5 mg/mL Fe-Fe3O4 CSNPs was 25 μM. On average, there were 48 chimeric DNA molecules per modified nanoparticle at that concentration. In order to obtain functionalized nanoparticles with desired solubility in aqueous environments, enough chimeric DNA molecules were needed – Fe-Fe3O4 CSNPs modified with adequate chimeric DNA molecules (≥ 10 μM) are soluble upon adding water, whereas these ones engineered with insufficient ligands (≤ 5 μM) need vigorous votex or even sonication (Figure S5 in Supporting Information). In addition, chimeric DNA molecules with varying lengths (from 5 to 60 nt) and distinct sequence information were all found to generate functionalized nanoparticles with excellent water dispersity (Figure S6 in Supporting Information). Moreover, the hydrodynamic diameter and zeta-potential of modified nanoparticles increased with the length of chimeric DNA molecules (Table S2 in Supporting Information).
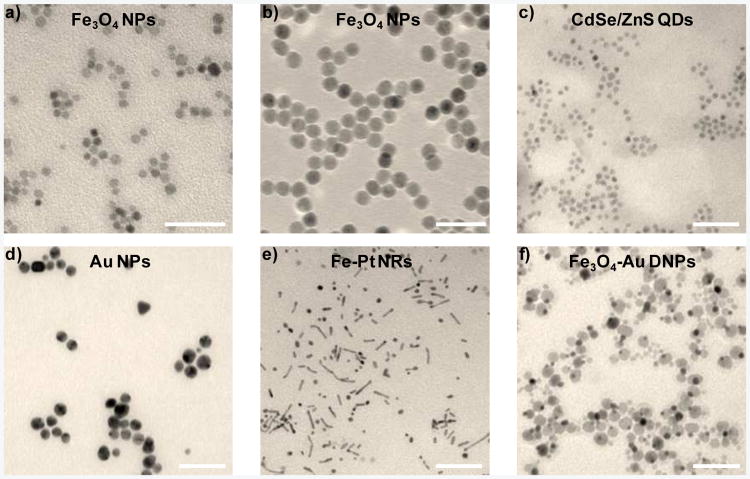
Since this novel surface engineering approach does not rely on the properties of the nanocrystal core or the reactivity of the nanocrystal surface, it could be generalized for many hydrophobic nanocrystals with variable size, composition, and morphology. To verify this point, chimeric DNA molecules were used to engineer Fe3O4 nanoparticles (NPs) with two different sizes (7 and 15 nm diameter). Water-soluble Fe3O4 NPs with minimal aggregation were obtained under both circumstances (Figure 2a-b). In addition, this method worked equally well for nanocrystals with other compositions (CdS/ZnS QDs and Au NPs) and morphologies (Fe-Pt nanorods (NRs) and Au-Fe3O4 dimer nanoparticles (DNPs)) (Figure 2c-f). Detailed synthesis procedure for these hydrophobic nanocrystals can be found in Supporting Information. Therefore, the surface engineering method demonstrated here is independent of nanocrystal size, composition, and morphology. In addition, it is highly efficient with only a few empty micelles remaining (Figure S7 in Supporting Information).
Figure 2.
Surface engineering of various hydrophobic nanocrystals with different size, composition, and morphology with chimeric DNA modification. TEM images of a) Fe3O4 NPs (7 nm diameter), b) Fe3O4 NPs (15 nm diameter), c) CdS/ZnS QDs, d) Au nanoparticles, e) Fe-Pt NRs, and f) Au-Fe3O4 DNPs, after chimeric DNA molecule engineering in water. Lipid-T20 was used as the chimeric DNA molecule here. Scale bars: 50nm.
In order to determine their utility in biomedical applications, the stability of engineered nanocrystals was evaluated in various solution environments. No obvious aggregation was observed in water, phosphate buffered saline, or cell culture medium, even after more than 6 months.
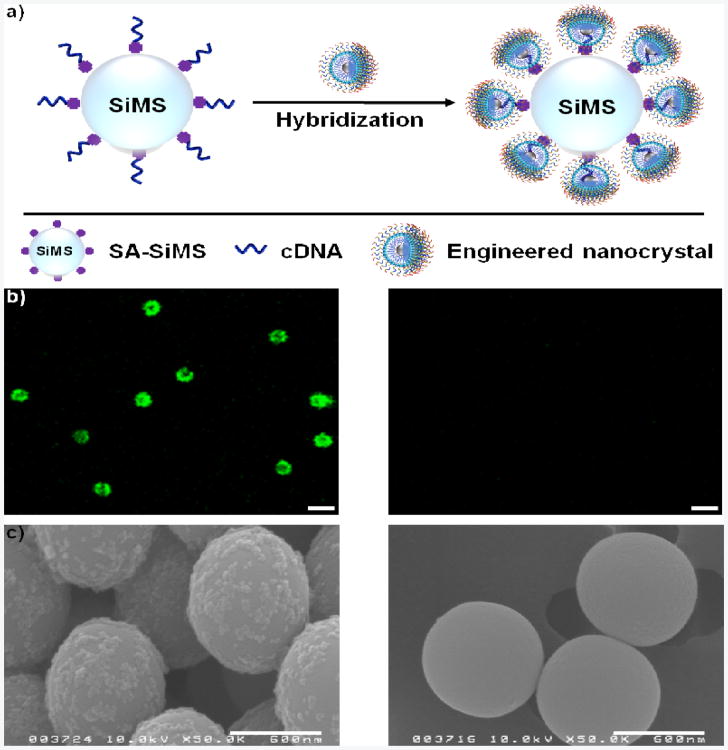
After confirming the stability of functionalized nanocrystals in biological systems, we systematically investigated their ability to recognize desired molecular targets. First of all, the hybridization between modified nanocrystals and their complementary DNA (cDNA) was studied (Figure 3a). Fluorescein isothiocyanate (FITC)-labeled lipid-20 and FITC-labeled lipid-T20 were used as target and control chimeric DNA molecules, respectively. The cDNA here wsa perfectly complementary to lipid-20. The cDNA-conjugated silica microspheres (cDNA-SiMSs) were prepared by immobilizing biotinylated cDNA onto strepavidin-coated SiMSs (SA-SiMSs), and then incubating with engineered Fe-Fe3O4 CSNPs under ambient atmosphere in hybridization buffer (20 mM Tri-HCl, 50 mM NaCl, 5 mM MgCl2) while shaking for 4 hours. For cDNA-SiMSs treated with lipid-20-functionalized Fe-Fe3O4 CSNPs, a strong green fluorescence signal was observed by confocal laser scanning microscopy (CLSM), indicating a high degree of hybridization, while no fluorescence was seen for cDNA-SiMSs mixed with lipid-T20-modified Fe-Fe3O4 CSNPs, suggesting no hybridization (Figure 3b). In addition, coating of lipid-20-functionalized Fe-Fe3O4 CSNPs (instead of lipid-T20-modified Fe-Fe3O4 CSNPs) onto cDNA-SiMSs was viewed by scanning electron microscopy (SEM) (Figure 3c), further showing the selective binding of engineered Fe-Fe3O4 CSNPs to their nucleic acid target. In addition, SEM images also illustrated that the hybridization event did not disrupt the structure integrity of engineered Fe-Fe3O4 CSNPs.
Figure 3.
Hybridization between chimeric DNA molecule-engineered Fe-Fe3O4 CSNPs with their cDNA. a) Schematic illustration of the hybridization reaction. b) CLSM and c) SEM images of cDNA-SiMSs treated with lipid-20 (left) and lipid-T20 (right) functionalized Fe-Fe3O4 CSNPs. Both lipid-20 and lipid-T20 were labeled with FITC. cDNA is perfectly complementary to lipid-20. Scale bars in b): 2 µm, scale bars in c): 600 nm.
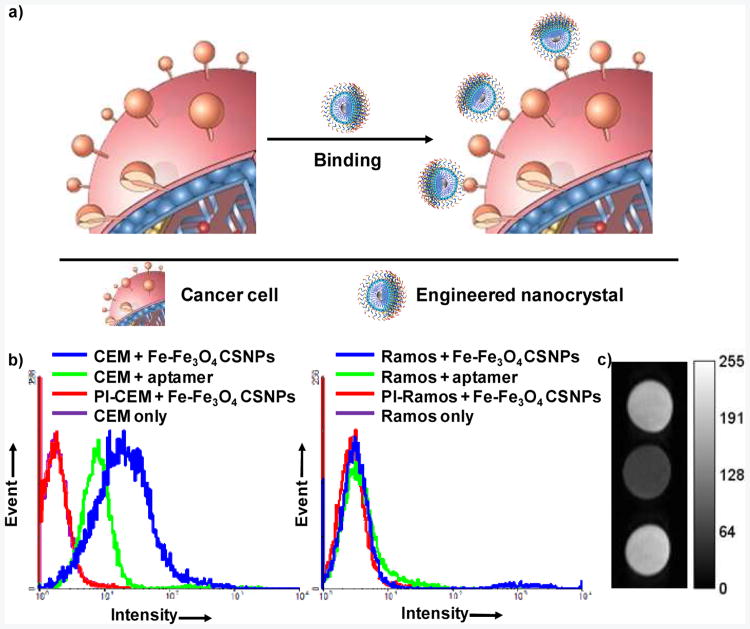
Next, we tested the binding between modified nanocrystals and their target cancer cells, which is of great significance to their use in early cancer diagnosis and efficient cancer therapy. To accomplish this, Fe-Fe3O4 CSNPs were functionalized with a FITC-labeled chimeric aptamer. Aptamers, generated from a process known as SELEX (Systematic Evolution of Ligands by EXponential enrichment), are single-stranded oligonucleotides which can bind to their targets with high affinity and excellent selectivity by folding into distinct secondary or tertiary structures.18 Sgc8 aptamer,19 previously developed to specifically bind to CEM cells (T cell line, human acute lymphoblastic leukemia) using Ramos cells (B cell line, human Burkitt';s lymphoma) as control, was chosen for the study (Figure 4a). According to flow cytometry histograms, an obvious shift (binding) was observed for CEM (target) cells, but only negligible shift was noticed for Ramos (control) cells (Figure 4b). More importantly, even though much fewer engineered Fe-Fe3O4 CSNPs (5 nM, 28 aptamers per functionalized nanoparticle) were used compared to free aptamers (200 nM), a much larger shift was achieved using modified Fe-Fe3O4 CSNPs due to the multivalent effect. The affinity of a ligand to its receptor is highly dependent on its valency (the number of sites available for receptor attachment). Therefore, the presence of multiple aptamers on the surface of functionalized Fe-Fe3O4 CSNPs resulted in greater cooperation, thereby enhancing the binding affinity to target cancer cells. With FITC-labeled chimeric aptamer, the modified Fe-Fe3O4 CSNPs can be used as specific fluorescence imaging agents with excellent sensitivity.
Figure 4.
Specific binding of chimeric DNA molecule engineered-Fe-Fe3O4 CSNPs to target cancer cells. a) Schematic illustration of the binding event, b) Flow cytometry histograms of CEM (target) and Ramos (control) cells incubated with buffer only, aptamer (200 nM) and engineered Fe-Fe3O4 CSNPs (5 nM). PI-CEM and PI-Ramos are CEM and Ramos cells pre-incubated with 1.5 μM unlabeled aptamer. c) T2-weighted MRI image of engineered Fe-Fe3O4 CSNPs only (top), CEM (middle) and Ramos (bottom) cells treated with engineered Fe-Fe3O4 CSNPs. Lipid-PEG-Sgc8 was used as the chimeric DNA molecule here.
Magnetic resonance imaging (MRI) is one of the best noninvasive imaging modalities because of its ability to provide a large amount of spatial and temporal information using various contrast agents, especially iron oxide nanocrystals.20-23 Therefore, we also tested the potential of using engineered Fe-Fe3O4 CSNPs as T2 (transverse relaxation time) contrast agents. Based on a self-amplifying proximity assay,24 when multiple modified Fe-Fe3O4 CSNPs bind to the receptors on their target cancer cells, they act cooperatively to form micrometer-scale clusters, thereby enhancing the net transverse relaxation of neighboring protons and leading to a darker image. T2-weighted MRI was taken for both CEM cells and Ramos cells treated with functionalized Fe-Fe3O4 CSNPs (Figure 4c). Similar darkness was observed for engineered Fe-Fe3O4 CSNPs incubated with buffer only and Ramos cells. However, a significantly darker spot was obtained for modified Fe-Fe3O4 CSNPs mixed with CEM cells, as a result of their highly specific interaction, demonstrating that functionalized Fe-Fe3O4 CSNPs can also serve as selective contrast agents with high performance.
Compared to traditional polymer systems used to produce biocompatible nanocrystals from hydrophobic ones, like polyethylene glycol (PEG), the new method demonstrated here provides a more time- and labor-efficient single-step surface engineering approach: the DNA corona not only renders the functionalized nano-particles with excellent water solubility, but also furnishes them with tunable specific targeting. However, multiple steps are needed to accomplish the phase transfer of hydrophobic nanocrystals using traditional polymer systems, and additional procedures are further required for conjugating targeting ligands. In addition, without the necessity of complicated organic synthesis, all these chimeric DNA molecules can be synthesized on a fully automated DNA/RNA synthesizer. If desired, different modifiers, including commericially-available organic dyes, functional groups, therapeutic agents, or even short polymers (e.g., PEG) can be introduced at any location of the sequence during synthesis.
To summarize, we have developed a one-step facile surface engineering approach for hydrophobic nanocrystals using chimeric DNA molecules. This method is simple, efficient, and can be readily adapted to a broad range of nanocrystals with different size, composition and morphology. Engineered nanocrystals possess excellent dispersity in biological fluids with minimal aggregation and long-term stability. In particular, this novel surface engineering approach equips modified nanocrystals with designer molecular recognition to various molecular addresses, varying from nucleic acids to cancer cells. Based on all these superior features, we believe that this newly developed surface engineering approach will greatly facilitate the use of nanocrystals in many biomedical applications.
Supplementary Material
Acknowledgments
The authors are grateful to Karen Kelly (Interdisciplinary Center for Biotechnology Research, University of Florida) for her technical assistance on transmission electron microscopy and scanning electron microscopy and Hao Liu (Department of Chemistry, University of Florida) for her technical support on Fourier transform infrared spectroscopy. MRI data were obtained at the Advanced Magnetic Resonance Imaging and Spectroscopy (AMRIS) facility in the McKnight Brain Institute of the University of Florida. The authors also sincerely appreciate Dr. Kathryn Williams for her critical comments during the preparation of this manuscript.
Footnotes
Notes: The authors declare no competing financial interest.
Supporting Information. Detailed synthesis and characterization of hydrophobic nanocrystals, chimeric DNA molecules, functionalized hydrophobic nanocrystals, and detailed experimental procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 2.L. Villaraza AJ, Bumb A, Brechbiel MW. Chem Rev. 2010;110:2921–2959. doi: 10.1021/cr900232t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee H, Sun E, Ham D, Weissleder R. Nat Med. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee H, Yoon TJ, Figueiredo JL, Swirski FK, Weissleder R. PNAS. 2009;106:12459–12464. doi: 10.1073/pnas.0902365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Namiki Y, Namiki T, Yoshida H, Ishii Y, Tsubota A, Koido S, Nariai K, Mitsunaga M, Yanagisawa S, Kashiwagi H, Mabashi Y, Yumoto Y, Hoshina S, Fujise K, Tada N. Nat Nanotechnol. 2009;4:598–606. doi: 10.1038/nnano.2009.202. [DOI] [PubMed] [Google Scholar]
- 6.Goya GF, Grazu V, Ibarra MR. Curr Nanosci. 2008;4:1–16. [Google Scholar]
- 7.Brigger I, Dubernet C, Couvreur P. Adv Drug Delivery Rev. 2002;54:631–651. doi: 10.1016/s0169-409x(02)00044-3. [DOI] [PubMed] [Google Scholar]
- 8.Samia ACS, Chen X, Burda C. J Am Chem Soc. 2003;125:15736–15737. doi: 10.1021/ja0386905. [DOI] [PubMed] [Google Scholar]
- 9.Murray CB, Norris DJ, Bawendi MG. J Am Chem Soc. 1993;115:8706–8715. [Google Scholar]
- 10.Sun S, Zeng H, Robinson DB, Raoux S, Rice PM, Wang SX, Li G. J Am Chem Soc. 2004;126:273–279. doi: 10.1021/ja0380852. [DOI] [PubMed] [Google Scholar]
- 11.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sperling RA, Parak WJ. Philos Trans R Soc A. 2010;368:1333–1383. doi: 10.1098/rsta.2009.0273. [DOI] [PubMed] [Google Scholar]
- 13.Lin CAJ, Sperling RA, Li JK, Yang TY, Li PY, Zanella M, Chang WH, Parak WJ. Small. 2008;4:334–341. doi: 10.1002/smll.200700654. [DOI] [PubMed] [Google Scholar]
- 14.Tong S, Hou S, Ren B, Zheng Z, Bao G. Nano Lett. 2011;11:3720–3726. doi: 10.1021/nl201978c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blokzijl W, Engberts JBFN. Angew Chem Int Ed. 1993;32:1545–1579. [Google Scholar]
- 16.Liu H, Zhu Z, Kang H, Wu Y, Sefan K, Tan W. Chem – Eur J. 2010;16:3791–3797. doi: 10.1002/chem.200901546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Sefah K, Liu H, Wang R, Tan W. PNAS. 2010;107:5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sefah K, Shangguan D, Xiong X, O'Donoghue MB, Tan W. Nat Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 19.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. PNAS. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin J, Laurent S, Jo YS, Roch A, Mikhaylova M, Bhujwalla ZM, Muller RN, Muhammed M. Adv Mater. 2007;19:1874–1878. [Google Scholar]
- 21.Babes L, Denizot B, amp x, Tanguy G, Le Jeune JJ, Jallet P. J Colloid Interface Sci. 1999;212:474–482. doi: 10.1006/jcis.1998.6053. [DOI] [PubMed] [Google Scholar]
- 22.Jun Yw, Huh YM, Choi Js, Lee JH, Song HT, KimKim Yoon S, Kim KS, Shin JS, Suh JS, Cheon J. J Am Chem Soc. 2005;127:5732–5733. doi: 10.1021/ja0422155. [DOI] [PubMed] [Google Scholar]
- 23.Jendelová P, Herynek V, Urdzíková L, Glogarová K, Kroupová J, Andersson B, Bryja V, Burian M, Hájek M, Syková E. J Neurosci Res. 2004;76:232–243. doi: 10.1002/jnr.20041. [DOI] [PubMed] [Google Scholar]
- 24.Perez JM, Josephson L, O'Loughlin T, Hogemann D, Weissleder R. Nat Biotechnol. 2002;20:816–820. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.