Abstract
Background: Low level laser irradiation (LLLI) stimulates bone regeneration. However, the molecular mechanisms leading to this is not yet understood. The stepwise subtractive cDNA cloning technology has been developed, coupled with DNA homology searched in DNA database is useful to identify the certain gene.
Aim: In order to understand the mechanism, we attempted to identify genes whose expressions are enhanced by LLLI. MC3T3-E1 osteoblastic cells were irradiated with an 830 nm Ga-Al-As diode laser, and a cDNA library was constructed using subtractive gene cloning.
Material and methods: The cDNA library of osteoblasts which was treated by LLLI was constructed. Nucleotide sequences were analyzed and homology searched in a DNA database using BLASTN program to identify the gene with altered expression. Altered mRNA levels by LLLI were confirmed by reverse transcription polymerase chain reaction (RT-PCR) and real-time PCR.
Results: The DNA sequence of a subtracted gene clone MCL129 indicated high homology (99%) with the microtubule-associated protein 1A (MAP1A) gene. Increase in MAP-1A mRNA level by LLLI was successfully confirmed by RT-PCR and real-time PCR.
Discussion: MAP1A has been shown to promote microtubule assembly and its functional expression. Microtubules play an important role in cell division, cell shape and polarity, cell movement, intracellular transport, signal transduction, and synthesis and secretion of collagen. Thus, enhancement of MAP1A gene expression by LLLI may be one of the molecular mechanisms involved in accelerating bone formation by LLLI.
Conclusion: LLLI irradiation enhances MAP1A gene expression and modulates microtubule assembly and the functional structure of microtubules, in turn, stimulates osteoblastic proliferation and differentiation.
Keywords: low level laser, osteoblasts, microtubule associated protein, gene expression
Introduction
Low level laser irradiation (LLLI) stimulates bone regeneration.1,2) In bone metabolism, in vivo experimental models using biochemical and quantitative histomorphometrical experiments revealed that LLLI stimulates alkaline phosphatase activity, calcium accumulation, and rapid accumulation of reparative new bone in injuries in rat tibia.3,4) In osteoblast cell cultures, we demonstrated that LLLI stimulated bone nodule formation.5) However, the molecular mechanisms leading to these observations are not yet understood. We previously constructed a cDNA library, which were enhanced gene expression in the pre-osteoblastic cell line, MC3T3-E1, using stepwise subtraction.6) In the present study, we further characterized a gene clone, MCL129, by partial nucleotide sequencing and BLASTN homology search using the NCBI DNA database.
Material and methods
Cell culture and LLLI
MC3T3-E1 cells, established from newborn mouse calvaria by Kodama et al.,7) were cultured in minimal essential medium (alpha-MEM; GIBCO BRL) containing 10% fetal calf serum and antibiotics comprising 100 μg/ml penicillin G (Sigma Chemical Co.) and 50 μg/ml gentamicin sulphate (Sigma) in multiwell plates.
A Ga-Al-As diode laser device (Panasonic Inc., Osaka, Japan model: Panalas¯ 1000) was used for LLLI. The technical specification of this laser device was as follows: wavelength: 830 nm and output power: 100–700 mW. Laser irradiation was performed at a distance of 550 mm (area of spot size: 78.5 cm2) from the probe to the cell layer for 20 minutes (power density: 7.64 J/cm2).
DNA sequencing and homology search
Dideoxy-chain termination sequencing8) was performed with fluorescent dye-labelled T7 universal primers (Aloka, Tokyo, Japan) and SequiTherm™ Long-Read™ cycle sequencing kits for Li-Cor¯ Sequencing (Epicentre Technologies, USA). The reaction products were analysed by a 4000LS Long ReadIR™ DNA sequencing system (LI-COR, USA).
Homology for the DNA sequence of MCL129 was searched using the nucleotide blast search (search a nucleotide database using a nucleotide query; BLASTN) with the NCBI DNA database (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
RT-PCR and real time PCR analysis
RT-PCR and real-time PCR reactions were carried out using a DNA thermal analyzer (RFN-Gene™ 6000; Corbett Life Science, Sidney, Australia). Amplification by PCR was started with an initial incubation at 95°C for 15 seconds to activate the Taq DNA polymerase, and then performed at 95°C for 5 seconds and 56°C for 15 seconds by adequate cycles. RT-PCR products were electrophoresed on 1.5% agarose gel, followed by staining with ethidium bromide to examine the size of PCR products. Real-time PCR was carried out with SYBR Premix Ex Taq™ (Perfect Real-Time PCR, Takara, Japan) and a Green PCR kit (Qiagen). To calculate gene expression fold changes, the initial template concentration was derived from the cycle number at which the fluorescent signal crossed the threshold in the exponential phase of the real-time PCR reaction. The mRNA copy unit was given by the cycle threshold value from the fluorescent signal of all the samples, including the standard curve and target genes, following the method provided by Corbett Life Science Company using RFN-Gene™ 6000 software. Details were described in an operation manual, version 1.7.40, 2006. Each assay was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels. The DNA primer sequences were 5′- aaagaccaccaccactcctg–3′ (the forward primer for MAP1A gene); 5′- tgttgctgtggttgggaata-3′ (the reverse primer for MAP1A gene) (predicted size=206 bp); 5′- atcaccatcttccaggag-3′ (the forward primer for GAPDH); and 5′-atcgactgtggtcatgag- 3′ (the reverse primer for GAPDH gene) (predicted size=318 bp). Values were calculated as mean±standard deviation (SD). Comparisons were made between two groups using a Student's t-test.
Results
Fig. 1 shows a partial DNA sequence of the inserted DNA from MCL129.
Figure 1:
The nucleotide sequence of the cDNA inserted in the MCL129 clone.
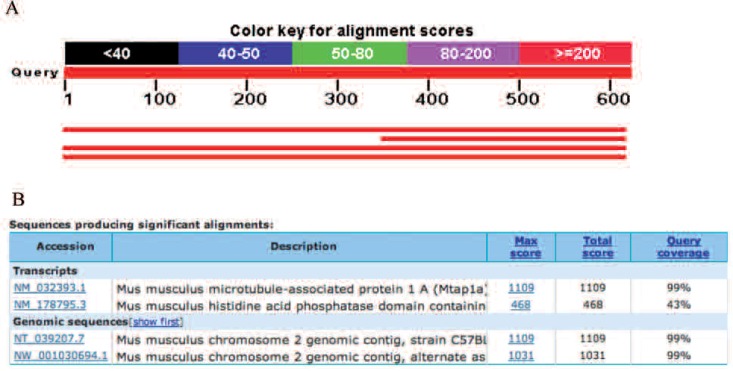
We searched the homology of the DNA sequence (619-bp) of MCL129 with NCBI DNA database using the BLASTN program. We found high homology exhibited by the MAP1A gene shown in Fig. 2.
Figure 2:
Blast hits on the query sequence in the NCBI DNA database. Three blast hits were found with high homology.
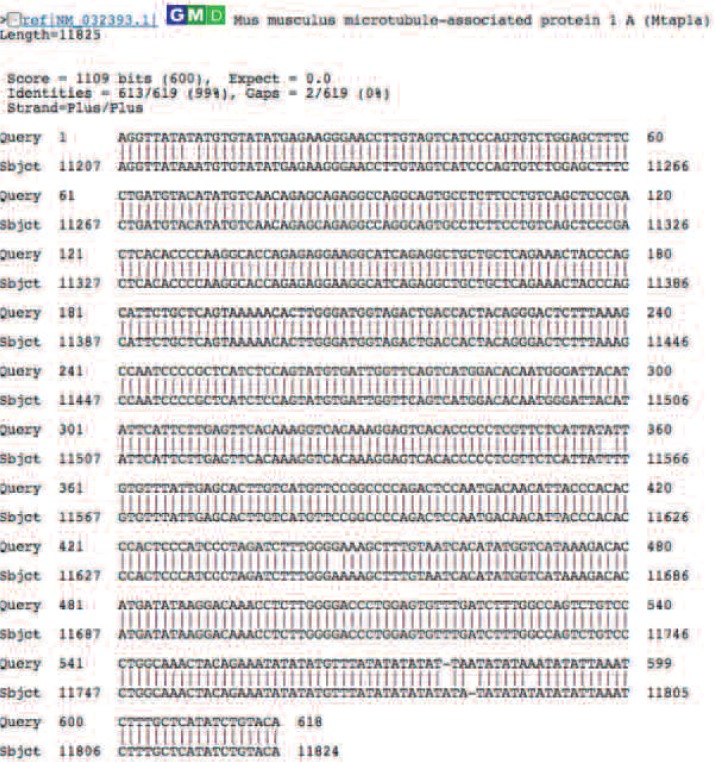
Fig. 3 shows the alignment of the MCL129 DNA sequence with MAP1A (gene ID=NM-032393.1) in the DNA database with 99% homology.
Figure 3:
Identification of the three blast hit gene as MAP1A. Sequence similarity was shown with 99% match in the 517 bp insert of mouse MAP1A.
Since the MCL129 DNA sequence has high homology with MAP1A, we designed DNA primers for PCR amplification of the MAP1A gene from the DNA database and examined the elevated mRNA level of MAP1A by RT-PCR. As shown in Fig. 4, the MAP1A mRNA level in LLLI-MC-3T3-E1 cells was higher than that in non-irradiated cells used as control up to 12-hr after LLLI. In contrast, mRNA levels of GAPDH, the housekeeping gene used as control, showed no difference between LLLI and control MC3T3-E1 cells.
Figure 4:
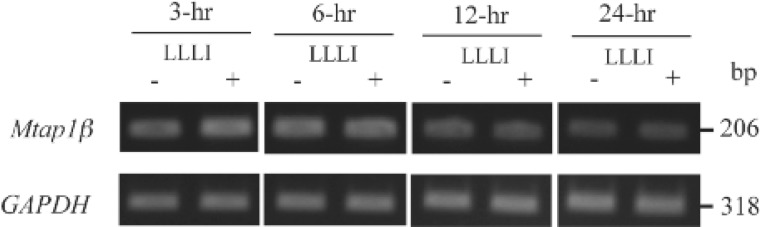
RT-PCR analysis of MAP1A mRNA levels. Amplified each DNA band of MAP1A was greater in LLLI-MC3T3-E1 cells than in control cells. Each GAPDH was used as an internal control.
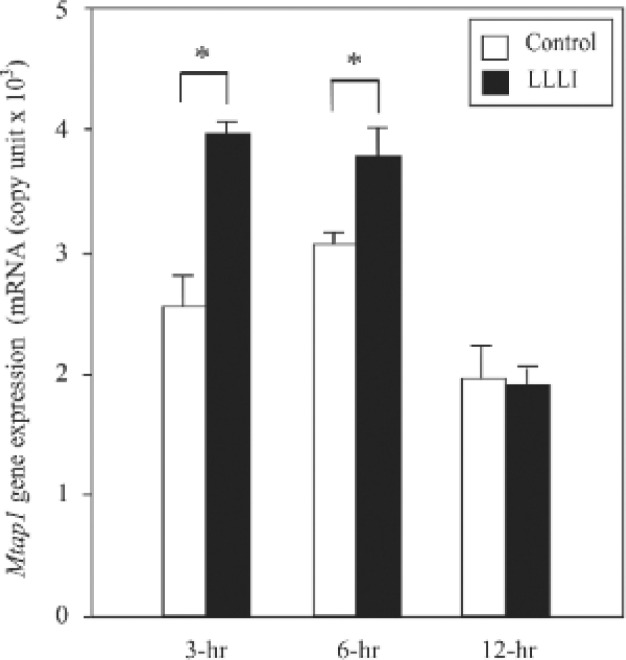
To determine the significant difference of enhancement of MAP1A mRNA level, real-time PCR was performed and results were converted to mRNA copy units. As shown in Fig. 5, the increase in MAP-1A gene expression was significantly different between LLLI and control MC3T3-E1 cells up to 13-hrs after LLLI.
Figure 5:
Real time PCR analysis of MAP1A mRNA levels. The MAP1A mRNA copy unit in LLLI-MC3T3-E1 cells was significantly greater than that in control. Each GAPDH was used as an internal control. (∗p < 0.05, n = 3).
Discussion
Microtubules are composed of alpha-tubulin and betatubulin heterodimers that associate end to end to form protofilaments.9) They elongate to the periphery of the cell according to the inherent dynamic instable properties of microtubules.10) Microtubules play a critical role in various cellular functions such as cell division, cell shape and polarity, cell movement, and intracellular transport.11,12) Microtubules may also be uniquely equipped to transmit signals originating from ligated receptors because they fill the cytoplasm and usually span the distance from the plasma membrane to the nucleus13) MAP1A binds to tubulin via electrostatic interactions and modulate the assembly and the stability of microtubules, mediating their interactions with other intracellular components.14,15)
In this study, we analyzed the DNA sequence of a MCL129 subtracted gene clone and carried out a homology search with the NCBI DNA database. As a result, we discovered that the cDNA insert of MCL129 shared a high homology with the mouse microtubule-associated protein (MAP1A) gene with 99% homology. This paper could be the first report on MAP1A expression in osteoblastic cells. Next, we designed PCR DNA primers of the MAP1A gene and analyzed mRNA levels by RT-PCR, successfully confirming enhanced MAP1A mRNA levels by LLLI. We also quantitatively measured MAP1A mRNA levels and demonstrated the statistical difference in MAP1A mRNA levels between LLI.I and control osteoblasts. These findings clearly show that LLLI significantly enhanced MAP1A gene expression in osteoblasts.
Regarding MAP1A function, structural MAPs interact with microtubules to regulate microtubule dynamics. The microtubule-binding site of MAP1A appears to be present near the amino terminus of the protein and is characterized by 11 repeats of the Lys-Lys-Glu sequence.16) MAP1A has been shown to promote microtubule assembly.17,18) In vitro experiments have demonstrated an increase in microtubule assembly and rates compared with unbound microtubules while n vivo studies have also demonstrated the growth-promoting ability of MAP1A.19) It is worth noting that anti-microtubular agents inhibited pro-collagen secretion and conversion to collagen and specifically inhibited collagen synthesis in chick cranial bone. Disruption of microtubules may interfere with the movement of Golgi-derived vesicles and the resulting accumulation of collagen precursors in the Golgi complex may lead secondarily to an inhibition of synthesis.20) Thus, MAP1A may play important roles in bone formation through collagen synthesis and secretion. Taken together, the enhancement of MAP1A expression by LLLI may be one of the molecular mechanisms involved in accelerating bone formation by LLLI.
Acknowledgments
Supported in part by the “Academic Frontier” Project for Private Universities: a matching fund subsidy from Ministry of Education, Culture, Sports, Science and Technology, 2007–2011, and by Grant-in-Aid for Scientific Research from Japan Society for the Promotion of Science (This study was supported in part by the “Academic Frontier” Project for Private Universities: a matching fund subsidy from Ministry of Education, Culture, Sports, Science and Technology, 2007–2011 and by Grant-in-Aid for Scientific Research from Japan Society for the Promotion of Science (B 20390530, B21390497).
References
- Trelles MA, Mayayo E. (1987): Bone fracture consolidates faster with low-power laser. Laser Surgery Medicine, 7:36–45 [DOI] [PubMed] [Google Scholar]
- Nagasawa A, Kato K, Negishi A. (1991): Bone regeneration effect of low level lasers including argon laser. Laser Therapy, 3:59–62 [Google Scholar]
- Barushka O, Yaakobi T, Oron U. (1995): Effect of low-energy laser (He-Ne) irradiation on the process of bone repair in the rat tibia. Bone, 16:47–55 [DOI] [PubMed] [Google Scholar]
- Yaakobi T, Maltz L, Oron U. (1996): Promotion of bone repair in the cortical bone of the tibia in rats by low energy laser (He-Ne) irradiation. Calcified Tissue International, 59:297–300 [DOI] [PubMed] [Google Scholar]
- Ozawa Y, Shimizu N, Kariya G, Abiko Y. (1998): Low-energy laser irradiation stimulates bone nodule formation at early stages of cell culture in rat calvarial cells. Bone, 22:347–354 [DOI] [PubMed] [Google Scholar]
- Hosoya S, Tamura K, Nomura K, Abiko Y. (1997): Construction of subtracted osteoblast cDNA librery with laser-irradiation-enhanced transcription. Laser Therapy, 9:107–114 [Google Scholar]
- Kodama H, Amagi Y, Sudo H, Kasai S, Yamamoto S. (1981): Establishment of Clonal Osteogenic Cell Line from Newborn Mouse Calvaria. Japanese Journal of Oral Biology, 23: 899–901 [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. (1977): DNA sequencing with chain-terminating inhibitors. Proceeding of National Academy of Science, USA, 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow E, Mandelkow EM. (1995): Microtubules and microtubuleassociated proteins. Current Opinion in Cell Biology, 7:72–81 [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. (1984): Dynamic instability of microtubule growth. Nature, 312:237–242 [DOI] [PubMed] [Google Scholar]
- Downing KH. (2000): Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annul Review Cell and Developmental Biology, 16:89–111 [DOI] [PubMed] [Google Scholar]
- Gelfand VI, Bershadsky AD. (1991): Microtubule dynamics: mechanism, regulation, function. Annul Review Cell and Developmental Biology, 7:93–116 [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Cook TA. (1999): Microtubules and signal transduction. Current Opinion in Cell Biology, 11:81–94 [DOI] [PubMed] [Google Scholar]
- Olmsted JB. (1986): Microtubule-associated proteins. Annul Review Cell and Developmental Biology 2:421–457 [DOI] [PubMed] [Google Scholar]
- Wolff J. (1998): Promotion of microtubule assembly by oligocations: cooperativity between charged groups. Biochemistry 37:10722–10729 [DOI] [PubMed] [Google Scholar]
- Langkopf A, Hammarback JA, Muller R, Valee RB, Garner CC. (1992): Microtubule associated proteins 1a and LC2. Journal of Biological Chemistry, 267:16561–16566 [PubMed] [Google Scholar]
- Pedrotti B, Soffientini A, islsm K. (1993): Sulphonate buffers affect the recovery of microtubule-associated proteins MAP1 and MAP2: evidence that MAP1A promotes microtubule assembly. Cell motility and the cytoskeleton, 25:234–242 [DOI] [PubMed] [Google Scholar]
- Pedrotti B, islsm K. (1994): Purified native microtubule associated protein MAP1a: kinetics of microtubule assembly and MAP1a/tubulin stoichiometry. Biochemistry, 33:12463–12470 [DOI] [PubMed] [Google Scholar]
- Faller EM, Brown DL. (2009): Modulation of microtubule dynamics by MAP. Journal of Neuroscience Research, 87:1080–1089 [DOI] [PubMed] [Google Scholar]
- Ehrlich HP, Ross R, Bornstein P. (1974): Effects of antimicrotubular agents on the secretion of collagen. A biochemical and morphological study. Journal of Cell Biology, 62:390–405 [DOI] [PMC free article] [PubMed] [Google Scholar]