Abstract
Recognition of injured mitochondria for degradation by macroautophagy is essential for cellular health, but the mechanisms remain poorly understood. Cardiolipin is an inner mitochondrial membrane phospholipid. We found that rotenone, staurosporine, 6-hydroxydopamine and other pro-mitophagy stimuli caused externalization of cardiolipin to the mitochondrial surface in primary cortical neurons and SH-SY5Y cells. RNAi knockdown of cardiolipin synthase or of phospholipid scramblase-3, which transports cardiolipin to the outer mitochondrial membrane, decreased mitochondrial delivery to autophagosomes. Furthermore, we found that the autophagy protein microtubule-associated-protein-1-light chain-3 (LC3), which mediates both autophagosome formation and cargo recognition, contains cardiolipin-binding sites important for the engulfment of mitochondria by the autophagic system. Mutation of LC3 residues predicted as cardiolipin-interaction sites by computational modeling inhibited its participation in mitophagy. These data indicate that redistribution of cardiolipin serves as an “eat-me” signal for the elimination of damaged mitochondria from neuronal cells.
Mitochondria are indispensable for aerobic life in eukaryotic cells; yet injured or dysfunctional mitochondria generate ROS and release mediators that kill cells. In response, eukaryotic cells developed mechanisms to sequester and degrade mitochondria through macroautophagy. Although once thought of as a non-selective bulk degradation process, the selective recognition of dysfunctional mitochondria for mitochondrial autophagy (mitophagy) has been recently established1-4. Yet, molecular signals underlying recognition of injured mitochondria remain incompletely defined3, 4, particularly in neurons, and mammals lack the adaptor proteins identified in yeast2. Mitochondria, like their bacterial ancestors, contain in their inner membrane the phospholipid cardiolipin (CL)5, which is not found in any other organelle. Here, we provide evidence that the recognition of this ancient phospholipid serves an important defensive function for the elimination of damaged mitochondria in both primary and transformed neuronal cells.
In healthy mitochondria, CL is localized to the inner mitochondrial membrane (IMM), where it interacts with proteins to support cristae, stabilize respiratory chain complexes6, and modulate autophagy and cell death7. The unique structure of CL includes a compact, negatively charged head group, while mammalian LC3 contains basic surface patches8. Just as the collapse of plasma membrane phosphatidylserine asymmetry serves an important signaling function for phagocytosis9, we investigated the hypothesis that CL externalization to the outer mitochondrial membrane (OMM) acts as an elimination signal for mitophagy.
RESULTS
Rotenone elicits increased autophagy and mitophagy
Disruptions in mitophagy are centrally implicated in neurodegenerative diseases and aging10-12. Mitophagy was elicited in primary rat cortical neurons and SH-SY5Y neuroblastoma cells using the complex I inhibitor rotenone (Fig. 1) under sublethal or prelethal conditions (Supplementary Fig. S1a). The ubiquitin-fold domain of LC3 is involved in a lipidation reaction by which LC3 is covalently bound to autophagic membranes, and LC3 puncta are used as specific markers of autophagosomes13. Rotenone elicited increases in GFP-LC3 puncta (Fig. 1a,b; Supplementary Fig. S1b) and in the percentage of GFP-LC3 puncta colocalizing with mitochondria (Fig. 1c,d; Supplementary Fig. S1c), indicating a preferential increase in “mitophagosomes” relative to total autophagosomes.
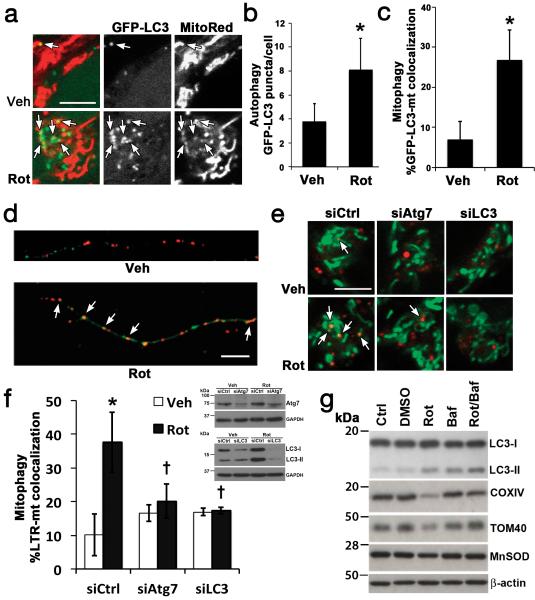
Fig. 1. Rotenone-induced mitophagy.
Rotenone increased GFP-LC3 puncta and colocalization with mitochondria (arrows) in SH-SY5Y cells (a-c, 1 μM) and primary cortical neurons (d, 250 nM), quantified in Fig. 3f and Supplementary Fig. S1b-c. Rotenone increased delivery of MitoTracker Green-stained mitochondria to Lysotracker Red (LTR)-stained lysosomes (e,f), inhibited by siRNA knockdown of Atg7 or LC3 in SH-SY5Y cells. Inset: RNAi knockdown. Rotenone decreased IMM (COXIV), OMM (TOM40) and matrix (MnSOD) protein expression levels in primary neurons (g), reversed by bafilomycin (Baf)-inhibition of autolysosomal degradation, quantified in Supplementary Fig. S1e. Mean +/− s.d. of n=7 independent experiments for b,c, and n=3 independent experiments for f,g (see Statistics Source Data Table); * p < 0.05 vs. vehicle control; † p < 0.05 vs. Rot-siCtrl. Scale = 10 μm. See Supplementary Fig. S7 for uncropped blots.
Rotenone induced a decrease in p62 (SQSMT1), levels, consistent with increased autophagic flux (Supplementary Fig. S1d). Colocalization of mitochondria with LysoTracker Red confirmed maturation and delivery of cargo to lysosomes, which was inhibited by siRNA knockdown of the autophagy proteins Atg7 or LC3 (Fig. 1e,f). The loss of mitochondrial proteins from the IMM, OMM, and matrix subcompartments was reversed by co-treatment with bafilomycin to prevent autophagosome-lysosome fusion13 (Fig. 1g; Supplementary Fig. S1e). Likewise, sublethal staurosporine (STS) treatments elicited mitophagy in primary neurons (Supplementary Fig. S1f-h) and in SH-SY5Y cells, reversed by siRNA knockdown of Atg7 or LC3 (Supplementary Fig. S1i).
CL translocates to the mitochondrial outer surface in response to pro-mitophagy stimuli
To analyze the mitochondrial redistribution of CL during mitophagy, we purified and characterized OMM and IMM fractions (Supplementary Fig. S2a,b) from mitochondria isolated from rotenone- or STS-treated neurons, and measured the levels of CL by quantitative liquid chromatography-mass spectrometry (LC-MS). Rotenone treatment increased both the total content and the numbers of individual molecular species of CL detected in the OMM fraction (Fig. 2a; Table 1; Supplementary Table S1). Under basal conditions, ~0.8% of CL resides in the OMM fraction from cortical neurons. Rotenone elicited an ~10–fold increase in CL content in the OMM, whether the data were normalized to mitochondrial protein (Table 1), or to total mitochondrial lipid phosphorus (Fig. 2b). There were no major changes in the spectra for the other major mitochondrial phospholipids (Supplementary Fig. S2c). Phosphatidic acid (PA) is another anionic phospholipid present in mitochondrial membranes. We found that the OMM content of PA was increased, but to a lesser extent than CL (Table 1). STS-treated neurons showed similar changes (Fig. 2b; Table 1).
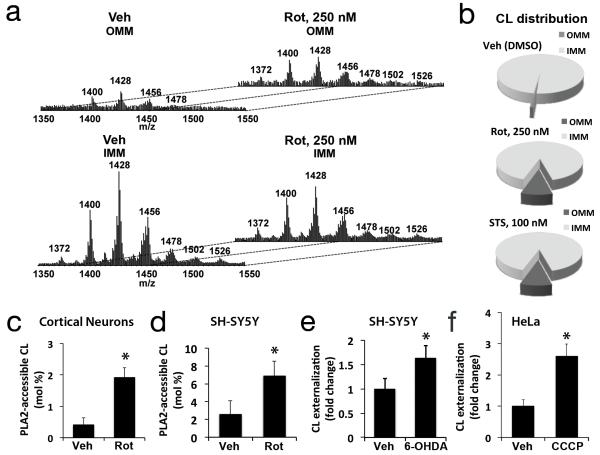
Fig. 2. Analysis of mitochondrial CL distribution and externalization.
IMM and OMM fractions isolated from primary neurons following the indicated treatments were lipid extracted for LC-MS analysis. MS spectra (a) demonstrated increased CL content of the OMM after rotenone treatment, with diversification of the cluster distribution to the 7 clusters exhibited by the IMM. Pie charts showing the CL distribution between IMM and OMM fractions from toxintreated neurons, normalized to mitochondrial lipid Pi (b). Treatment with rotenone caused significant increases in PLA2-hydrolyzable (surface accessible) CL assessed by LC-MS in primary cortical neurons (c, 250 nM) and SH-SY5Y cells (d, 1 μM). Treatment with 6-OHDA and CCCP increased surface exposure of CL probed with Annexin V in SH-SY5Y (e, 120 μM) and Parkin-expressing-HeLa cells (f, 20 μM), respectively. Mean +/− s.d. of n=3 independent experiments for c-f (see Statistics Source Data Table); * p < 0.05 vs. Control.
Table 1. Content of CL and PA in IMM and OMM fractions of rat cortical neurons treated with rotenone or staurosporine.
| Vehicle | Rot (250 nM, 2h) | STS (100 nM, 2h) | ||||
|---|---|---|---|---|---|---|
| PL nmol/mg protein | PL nmol/mg protein | PL nmol/mg protein | ||||
| IMM | OMM | IMM | OMM | IMM | OMM | |
| CL | 48.1 ± 0.9 | 0.4 ± 0.1 (0.82%) |
46.6 ± 0.5 | 5.0 ± 0.1 (9.69%) |
39.5 ± 1.0 | 4.2 ± 0.2 (9.61%) |
| PA | 1.7 ± 0.2 | 0.2 ± 0.1 (10.53%) |
2.0 ± 0.1 | 1.3 ± 0.1 (39.39%) |
3.4 ± 0.2 | 1.2 ± 0.1 (26.09%) |
Primary cortical neurons were isolated from day 16 embryonic rats, pooled and cultured. Primary cortical neurons (DIV 7-9) were treated with rotenone or staurosporine for 2h followed by isolation of IMM and OMM fractions as described in Methods. Lipids were extracted and CL or PA identified and quantitated by LC-MS analysis using ion trap and Q-TOF platforms. Results are expressed as the Mean +/− s.d. of n=5 experiments. The % of the phospholipid present in the OMM fraction is shown in parentheses.
Neuronal CLs are comprised of multiple species resolvable by LC-MS, sharing the same headgroup, but with different fatty acyl chains. Control MS spectra for CL exhibited 7 IMM and 4 OMM CL clusters, while rotenone-treated samples displayed 7 clusters for both IMM and OMM fractions (Fig. 2a). Fatty acyl chain analysis revealed an identical distribution of fatty acids in the most abundant species of each cluster (Supplementary Table S1), suggesting redistribution of pre-existing CL molecular species without fatty acyl chain remodeling. Notably, no increased CL peroxidation products were detected under these early, mitophagy-inducing treatment conditions involving low doses of rotenone or STS, in contrast to the well-documented increases in lipid oxidation that occur during apoptosis14.
To determine if CL was exposed to the outer surface of the OMM, and is not simply enriched due to increased isolation of IMM-OMM contact sites, we employed several techniques. Western blot analysis showed no detectable contamination of OMM fractions by adenine nucleotide translocase, an IMM contact site protein15 (Supplementary Fig. S2b). We also developed two quantitative assays that do not involve biochemical fractionation. Intact mitochondria isolated from control and treated cultures were incubated with anionic phospholipid-selective phospholipase A2 (PLA2), and the CL hydrolysis products monolyso-CL were quantified. Treatment with rotenone caused significant increases in surface-accessible CL in primary cortical neurons and in SH-SY5Y cells (Fig. 2c,d). In contrast, lyso-PA was not detected. Alternatively, we incubated intact, isolated mitochondria with fluorescently labeled Annexin V, which binds CL especially well among anionic phospholipids at μM calcium concentrations (Supplementary Fig. S2d).16 Increased Annexin V binding to isolated, intact mitochondria occurred not only in response to rotenone (Fig. 3a) and STS (Supplementary Fig. S2e), but also in other well-established models of mitophagy: 6-hydroxydopamine (6-OHDA) treatment of SH-SY5Y cells17 and carbonylcyanide-3-chlorophenylhydrazone (CCCP)-treatment of Parkin-expressing HeLa cells3 (Fig. 2e, 2f; Supplementary Fig. S5g). 6-OHDA injury is fully reversible at the early time point studied18. Thus, CL externalization and mitophagy occur prior to commitment to cell death in response to multiple injury stimuli.
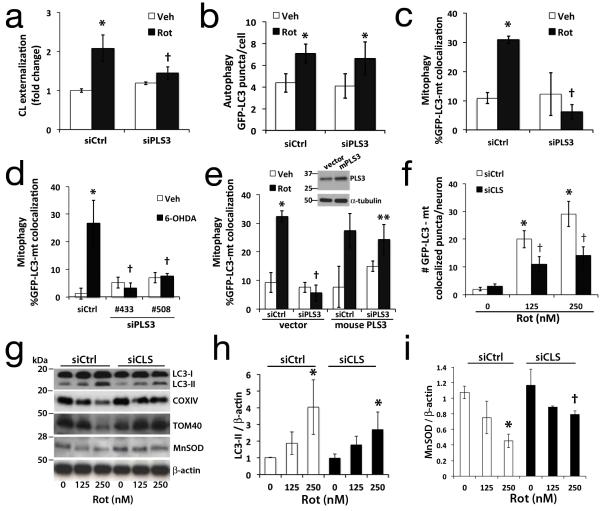
Fig. 3. RNAi knockdown of cardiolipin enzymes attenuate CL exposure and mitophagy.
SH-SY5Y cells expressing control (siCtrl) or scramblase-3 siRNA #508 (siPLS3) × 72h were stained with Mitotracker Green FM, a transmembrane potential-independent dye, and treated with rotenone (1 μM). Anionic phospholipids exposed to the outer surface of isolated mitochondria were quantified by flow cytometry for Alexa 647-annexin V binding (a). PLS3 knockdown had no effect on Rot-induced autophagy (b), but decreased mitophagy in SH-SY5Y cells treated with rotenone (c, 1 μM) or 6-OHDA (d, 120 μM). The effects of siPLS3 were recapitulated using a second siRNA #433 (d, Supplementary Fig. S3b, S3g), and reversed by transfection with an RNAi-resistant mouse PLS3 vector (e). Primary neurons transfected with cardiolipin synthase siRNA (siCLS) or scrambled siCtrl were treated with rotenone (250 nM) 72h later and analyzed for autophagy (Supplementary Fig. S1b) or mitophagy by either colocalization analysis (f), or with mitochondrial protein immunoblot (g) and densitometry (h,i; Supplementary Fig. S4c). Knockdown efficiencies are shown in Supplementary Figs. S3 & S4. Mean +/− s.d. of n=4 independent experiments for b,c, and n=3 independent experiments for a, df, h-i (see Statistics Source Data Table); * p < 0.05 vs. vehicle; † p < 0.05 vs. toxin-treated-siCtrl; ** p < 0.05 vs. Rot-siPLS3. See Supplementary Fig. S7 for uncropped blots.
CL externalization is important for cargo recognition during mitophagy
To determine whether the collapse of CL asymmetry was important for mitophagy, we knocked-down phospholipid scramblase-3 (PLS3) (Supplementary Fig. S3a,b), a mitochondrial enzyme responsible for the translocation of CL from the IMM to the OMM19. This prevented the rotenone-induced increase in CL exposure to the mitochondrial surface (Fig. 3a), inhibited the increase in GFP-LC3 mitochondrial colocalization without affecting autophagy induction (Fig. 3b, 3c), and inhibited the autophagic delivery of mitochondria to lysosomes and loss of mitochondrial proteins (Supplementary Fig. S3c,d). Similar effects were observed for 6-OHDA (Fig. 3d; Supplementary Fig. S3e) and STS (Supplementary Fig. S1i). PLS3 RNAi did not cause significant changes in mitochondrial membrane potential (Supplementary Fig. S3f) in the presence or absence of rotenone. The effects of PLS3 RNAi in human SH-SY5Y cells were recapitulated using a second siRNA (Fig. 3d; Supplementary Fig. S3g), and reversed by overexpression of mouse PLS3 (Fig. 3e).
To further confirm a role for CL in the recognition of injured mitochondria for mitophagy in primary neurons, we utilized RNAi against CL synthase (CLS). Mammalian cells tolerate reductions to ~45% of total CL without adverse effects on bioenergetics, membrane potential, or ROS20, 21. In primary neurons, CLS RNAi reduced CL levels to 57% +/− 1.4 at 72 h post-transfection. CLS knockdown using two distinct siRNAs (Supplementary Fig. S4a, d) diminished rotenone-induced mitophagy (Fig. 3f, 3g, 3i; Supplementary Fig. S1c,e), with little or no effect on general autophagy (Fig. 3h; Supplementary Fig. S1b). CLS knockdown also prevented STS-induced loss of mitochondrial proteins (Supplementary Fig. S1f) and reduced mitochondrial colocalization with GFP-LC3 puncta (Supplementary Fig. S1h), with no significant effects on LC3-II shift (Supplementary Fig. S1f) or the overall number of GFP-LC3 puncta elicited (Supplementary Fig. S1g). While CLS knockdown could affect other aspects of mitochondria or ER biology, there were no significant effects on basal or rotenone-induced mitochondrial membrane potential (Supplementary Fig. S4b,f) or ATP levels, which remained at 96.5% +/− 16 of control levels. Taken together with LC3 mutagenesis studies described below, these data support a role for CL in cargo selection in response to several mitophagy-inducing stimuli.
CL has been reported to bind several proteins, including the C-terminal domain of the autophagy protein Beclin 122,the immunity-related GTPase IRGM7, and the voltage dependent anion channel (VDAC), which is implicated in depolarization-induced mitophagy23, 24. As full length Beclin 1 does not interact with membranes22, we examined whether the Beclin 1 C-terminal domain could be involved. However, cleavage of Beclin 1 to form the C-terminal fragment25 was not observed under the sublethal or prelethal conditions used in this study to elicit mitophagy (Fig. 4a-c). Thus, it is unlikely that this interaction contributes to mitochondrial targeting to the autophagosome in these contexts.
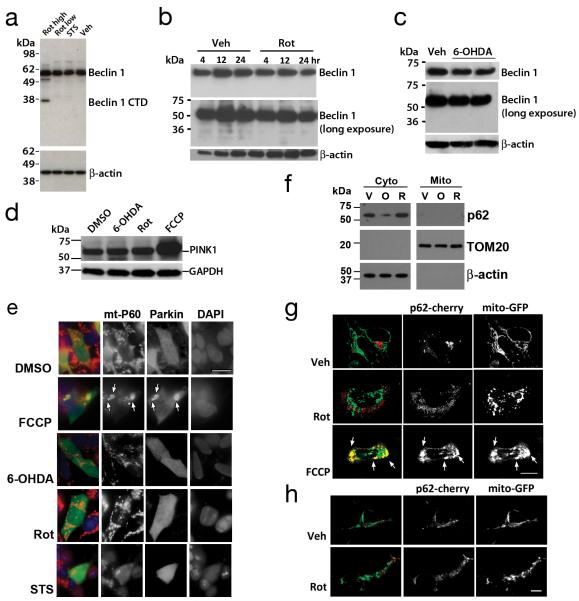
Fig. 4. Effects of Rot, STS or 6-OHDA on Beclin 1 cleavage and distribution of Parkin and p62.
Western blot analysis showed Beclin 1 cleavage products (<49 kDa) were not observed in primary neurons exposed to mitophagy-inducing sublethal rotenone (250 nM × 2h: Rot low) or staurosporine (100 nM × 2h: STS) concentrations (a). However, Beclin-1 was cleaved upon exposure to a lethal dose of rotenone (1 mM × 24h: Rot high). Likewise, SH-SY5Y cells did not show Beclin 1 cleavage upon exposure to rotenone (b) or 6-OHDA (c; duplicate lanes). Data are representative of 2-3 independent experiments per toxin. Cell lysates from stable PINK1-Flag expressing SH-SY5Y cells were treated with 6-OHDA (120 μM), Rot (1 μM), or FCCP (2 μM), and analyzed by Flag immunoblot for PINK1-Flag levels (d). SH-SY5Y cells transfected with HA-Parkin were treated the indicated toxins, immunolabeled for HA (green) and mitochondrial p60 antigen (red) and analyzed for HA-Parkin re-distribution to mitochondria (e). Scale bar: 10μm. FCCP elicited translocation of Parkin to mitochondria (e, arrows), which was not seen with the other mitophagy stimuli. Cytosolic and mitochondrial pellets were prepared from SHSY5Y cells treated with vehicle (V), 6-OHDA (O, 120 μM) or Rot (R, 1 μM). Gels were loaded with cytosolic or mitochondrial proteins, and immunoblotted for p62/SQSMT1, and the indicated fractionation markers (f). Confocal analysis of p62/SQSMT1 distribution in SH-SY5Y cells (g) and in primary cortical neurons (h) that were co-transfected with mCherry-tagged p62/SQSMT1 and mitochondrially targeted GFP. Note recruitment of p62 to large mitochondrial aggregates (g, yellow, arrows) in FCCP-treated cells, which was not observed in control or Rot treated cells. See Supplementary Fig. S7 for uncropped blots.
Parkinsonian toxins initiate mitophagy through distinct mechanisms from FCCP/CCCP
Mitochondrial uncoupling agents such as the protonophores carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) or CCCP trigger depolarization-induced increases in PINK1 accumulation accompanied by Parkin translocation to mitochondria26-28. In contrast, rotenone caused no more than 10-14% decreases of mitochondrial membrane potential in neurons and SH-SY5Y cells (Supplementary Figs. S3f & S4b,f). Moreover, there was no evidence of increased PINK1 levels (Fig. 4d) nor of Parkin translocation to mitochondria in the rotenone or 6-OHDA models (Fig. 4e), in agreement with a previous study contrasting the effects of strong depolarizing agents such as CCCP with the parkinsonian neurotoxins MPP+ and 6-OHDA29. Furthermore, the LC3- and ubiquitin-interacting protein p62 (SQSMT1), which has been variably implicated in Parkin-mediated mitophagy 24, 30, was not recruited to mitochondria in response to rotenone or 6-OHDA (Fig. 4f-h). Thus, the initiation steps elicited by rotenone and 6-OHDA appear to differ from those reported for FCCP or CCCP.
Using FCCP or CCCP as applied to SH-SY5Y cells or Parkin-expressing HeLa cells, we found that CL-modulating RNAi treatments did not prevent depolarization-induced PINK1 stabilization or Parkin translocation to the mitochondria (Supplementary Fig. S5a-c, f). Interestingly, as observed above, CCCP treatment did elicit CL externalization (Fig. 2f, Supplementary Fig. S5g,h), albeit to a lesser extent (~2-fold) than observed in rotenone-treated neurons. Moreover, CL-modulating RNAi treatments partially suppressed mitochondrial colocalization with autophagosomes without affecting overall LC3 puncta formation (Supplementary Fig. S5d,e), and partially inhibited the loss of mitochondrial proteins elicited by CCCP (Supplementary Fig. S5i; See Fig. S5j,k for validation of human CLS knockdown). Overexpression of the fission protein dynamin related protein 1 (Drp1) did not rescue the inhibition of mitophagy by PLS3 knockdown (Supplementary Fig. S5e), suggesting that CL may act downstream of Drp1. These data raise the possibility of pathway convergence downstream of PINK1 or Parkin, and or activation of dual pathways by CCCP.
CL interacts with directly with LC3 for cargo recognition during mitophagy
Although mechanisms of selective autophagy are just beginning to emerge3, 4, LC3 serves as a receptor for cargo adapters that bind protein aggregates31, and its homolog Atg8 functions in an analogous role in yeast2. Examination of the crystal structure of LC3 reveals basic patches at its surface that are not present in the related proteins GABARAP and GATE168. Using a gel retardation assay, we found that recombinant LC3 can directly bind CL liposomes to a much greater extent than to phosphatidylcholine (PC)-only liposomes (Fig. 5a). We also found that LC3 interacts more favorably with CL containing four acyl chains, than with lysoCLs as evidenced by the ratio of phospholipid to LC3 that caused a 50% loss of the LC3 monomeric form entering the gel (IC50) (Fig. 5b). IC50 analysis indicates that LC3 shows a stronger affinity for CL than for phosphatidic acid (PA) or phosphatidylglycerol (PG) (Fig. 5b), supported also by computational molecular dynamics simulations described below (Supplementary Fig. S6d).
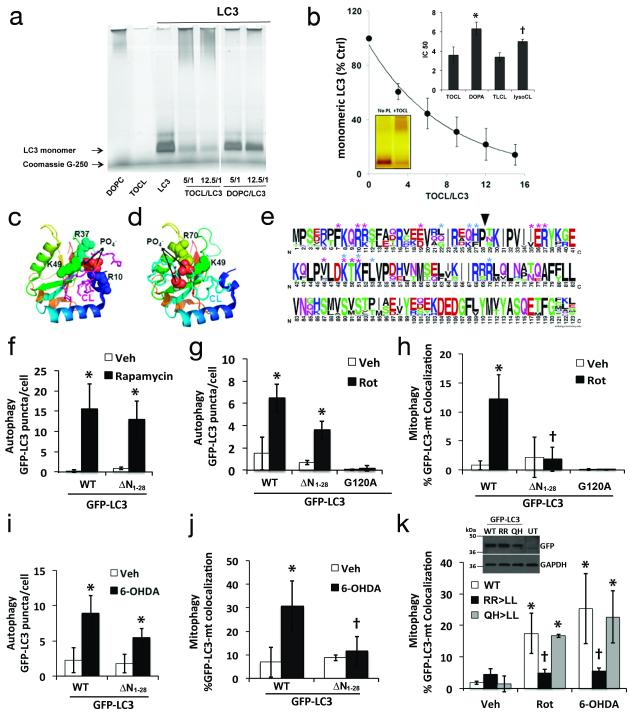
Fig. 5. Mitochondrial recognition for mitophagy requires N-terminal amino acids of LC3.
Recombinant LC3 incubated with tetraoleoylcardiolipin (TOCL) or dioleoylphosphatidylcholine (DOPC) liposomes at the indicated molar ratios were analyzed by Blue Native PAGE (a); liposome binding impaired the gel entry of proteins. Titration of phospholipids (e.g. TOCL) to LC3 was performed to evaluate the ratio that prevents 50% of the LC3 from entering the gels (IC50) as an index of relative affinity (b). Inset: comparison of IC50 values for TOCL vs. dioleoyl-phosphatidic acid (DOPA) and tetralinoleoyl-CL (TLCL) vs. monolyso-trilinoleoyl-CL (lysoCL). The IC50 for DOPG was >15. * p < 0.05 vs. TOCL; † p < 0.05 s. TLCL. Molecular model of interaction sites for CL binding to LC3 by docking analysis. CL is colored magenta in the top-ranked binding site conformation (c), and cyan in the alternate conformation (d), with the phosphates as spheres (oxygen-red, phosphorusorange). Amino acids interacting with CL are represented as spheres along the LC3 ribbon structure, colored blue>orange from N>C terminus. Thirty-nine LC3 family sequences including isoforms A-C were aligned with ClustalW and displayed using WebLogo, with symbol heights corresponding to relative amino acid frequency (e). Asterisks denote residues predicted to contact CL in the favored (magenta) and alternate conformations (cyan). Arrowhead shows position of N-terminal truncation used to create GFP-LC3 deletion mutants, which were analyzed for GFP-LC3 puncta formation (f,g,i) and participation in mitophagy (h.j) in response to the indicated stimuli. Dual point mutations were prepared based on predicted CL contact residues (e), and analyzed for participation in mitophagy elicited by Rot or 6-OHDA (k). Inset: expression of GFP-LC3 plasmids (Uncropped blots in Supplementary Fig. S7). Mean +/− s.d. of n=3 independent experiments for f-h, and n=4 independent experiments for i,j (see Statistics Source Data Table); * p < 0.05 vs. respective Veh. † p < 0.05 vs. toxin-treated-WT.
Analysis of all existing protein crystal structures in complex with CL indicated that side-chain or backbone ‘N’ atoms from K, R or H residues are required for stabilizing the phosphate moieties in CL. Molecular docking analysis predicts CL-binding pockets at the LC3 surface, where Arg and Lys interact with the sterically-constrained head group of CL with two possible conformations (Fig. 5c,d). The top-ranked conformation involves stabilization of both CL phosphate groups (Fig. 5c), resulting in the lowest binding energy. The predicted coordinating amino acids (Fig. 5e), include N-terminal residues of LC3 such as R10 and R11 for the top-ranked conformation or Q26 and H27 for the alternate conformation. Short duration (100 ns) all-atom and longer 1000 ns coarse-grained molecular dynamics simulations also implicate R10 and R11 in the initial interactions of LC3 with CL-containing membranes (Supplementary Fig. S6; SI Note, Supplementary Table S2). The C-terminus of LC3, which becomes cross-linked to autophagosome membranes, is predicted to remain exposed to solvent by coarse-grained modeling (Supplementary Fig. S6e, arrow), compatible with its proposed bridging role.
To test the ability of the N-terminal segment of LC3 to bind CL, we employed custom-synthesized 30-amino acid peptides corresponding to the N-terminal sequence of wild type LC3 or with substitution of leucines at R10,R11. We determined their binding with CL-containing liposomes using the gel retardation assay. After native gel electrophoresis, 69.4±9.7% and 102.4±13.0% (n=8, p<0.001) of wild type and mutant LC3, respectively, were detectable as unbound peptide, indicating that these residues, which are highly conserved among vertebrates (Supplementary Table S3), are important for the ability of LC3 to bind CL.
To establish experimentally key sites of contact between LC3 and CL, we deleted the N-terminal amino acids 1-28 predicted by molecular docking analysis to contain a major portion of the CL binding site (Fig. 5e), and analyzed the ability of this LC3 mutant to participate in autophagy or mitophagy. Consistent with reports that the N-terminal α-helices are not essential for LC3 conjugation during non-selective autophagy32, mutant LC3 participated equivalently to wild-type LC3 in rapamycin-induced autophagy (Fig. 5f), but there was a tendency for reduced participation in response to rotenone or 6-OHDA (Fig. 5g,5i). Strikingly, loss of the N-terminal α-helices abolished the ability of GFP-LC3 to undergo rotenone or 6-OHDA-induced colocalization with mitochondria (Fig. 5h,5j). To test the involvement of two possible CL-binding regions predicted by docking analysis, we produced double point mutants of LC3 in which either R10,R11 or Q26,H27 were mutated. The R10,R11 mutation fully recapitulated the effects of deleting the helices, preventing the injury-induced increase in colocalization with mitochondria (Fig. 5k).
DISCUSSION
Taken together, the data presented in this study support a mechanism by which CL externalized in response to mitochondrial injury interacts with the autophagy protein LC3 to mediate targeted autophagy of mitochondria in primary neurons and transformed neuronal cells (Fig. 6). Interestingly, injured primary neurons exhibited greater fold-increases in externalized mitochondrial CL compared to CCCP-treated HeLa cells. These results suggest that different cell types may show different thresholds for engagement of particular mitophagy pathways, as supported by observations that primary neurons show diminished or delayed Parkin translocation responses to CCCP.33, 34
Fig. 6. Schematic diagram summarizing the proposed role of CL in mitochondrial autophagy.
Mitochondrial injuries induce a PLS3-dependent externalization of CL to the outer surface of mitochondria. Cargo targeting into autophagosomes are facilitated by interactions of CL with the N-terminal domain of LC3. Clearance of CL-exposed mitochondria by mitophagy (green arrows) would be predicted to prevent CL oxidation and accumulation of pro-apoptotic signals (red arrow).
While we cannot exclude the additional contribution of other anionic phospholipids, or of other mitochondrial-autophagosome adaptor proteins, studies of hypoxia-induced adaptors such as NIX and BNIP3 implicate additional signals or receptors for the incorporation of mitochondria into autophagosomes, as mitophagy was only partially blocked in their absence 35, 36. Here, we show several lines of evidence supporting an important role for CL-LC3 binding as a mitophagy signal. In addition to the strongest affinity exhibited by LC3 for CL relative to other phospholipids and the differences predicted by computational modeling, rotenone induced a disproportionate redistribution of CL relative to each of the other major mitochondrial phospholipids (Compare Fig. 2a & Supplementary Fig. S2c). LC-MS analysis demonstrated a significantly lower content of PA than CL in the OMM after pro-mitophageal injury, and the PA that was present was not accessible to PLA2-hydrolysis as an index of exposure to the surface, in agreement with published studies37. Moreover, two independent methods to downregulate OMM CL each suppressed mitochondrial delivery into the autophagolysosomal system. Reciprocally, site-directed mutagenesis of LC3 residues predicted to interact with CL by three computational approaches also suppressed mitophagy. This constellation of findings supports a key role for CL externalization in signaling the engulfment of mitochondria by the autophagic machinery.
Just as clustering or density of interactions is an important component of PS-recognition for phagocytosis38, 39, we hypothesize that a critical density of interactions may be needed to trigger mitophagy. Molecular modeling and simulations predict that clustering of CL around LC3 may serve to stabilize initial electrostatic interactions, with embedding of parts of the protein into the bilayer through hydrophobic interactions (Supplementary Fig. S6d,e). If this is correct, such a clustering mechanism could help ensure that normal transient fluctuations due to lipid transport dynamics do not trigger mitophagy until a critical threshold of LC3-CL interaction density is achieved. It is possible that in some model systems, the threshold may be modulated by concurrent engagement of other cargo-targeting mechanisms.
CL interacts with other mitochondrial proteins such as VDAC, which is also implicated in mitophagy23, 24. Whether or not the effects of CL on VDAC gating functions40 may potentially affect the role of VDAC in mitophagy, however, remains unknown. CL also plays a role in localizing IRGM to the inner membrane-matrix fractions of macrophage mitochondria, allowing IRGM to regulate a cell fate switch between autophagic mycobacterial clearance and apoptosis7. Interestingly, the autophagy-promoting kinase AMPK is able to regulate cardiolipin synthase and nucleoside diphosphate kinase, another mitochondrial protein implicated in regulating the intermembrane distribution of CL41-43. Thus, it is possible that CL distribution is also important for mitophagy elicited downstream of AMPK pathways.
The endosymbiotic theory postulates that eukaryotic cells formed from symbiotic co-evolution with ingested bacteria-like organisms. The advantages of aerobic respiration created new cellular risks, however, as mitochondria are capable of generating ROS and releasing cell death mediators. It appears that yeast and mammals evolved distinct mechanisms to deal with this threat, as yeast proteins that mediate selective mitophagy have no mammalian orthologs2. Notably, the requirement for a specific spatial arrangement of amino acid residues for proteins that bind CL is different from the Ca2+ bridge employed for recognition of phosphatidylserine by phagocytosing cells44, and the particular amino acids forming the CL binding pocket of LC3 are highly conserved in vertebrates (Supplementary Table S3).
Due to their metabolic features and post-mitotic state, neurons are particularly vulnerable to disrupted cellular quality control, and dysregulation of autophagy and cargo recognition are centrally implicated in Parkinson’s disease, Alzheimer’s disease, Huntington’s disease and amyotrophic lateral sclerosis10-12. PINK1 and PARK2, two genes mutated in recessive forms of familial parkinsonism, play important roles in the regulation of CCCP-induced mitophagy3, 24, and phosphorylation of a mitochondrial outer membrane protein regulates mitophagy during hypoxia4. Mitophagy functions as a pro-survival mechanism in PINK1-deficient cells45. Also, CL interaction with IRGM is important for interferon-γ-induced mycobacterial clearance, although the effects were mediated through regulation of mitochondrial fission7. Nevertheless, as other bacteria are targeted intracellularly for autophagy through mechanisms that parallel recognition of protein aggregates46, it is possible that CL may also serve as a molecular pattern recognition signal for xenophagy.
In summary, our data indicate that mammalian neurons have evolved mechanisms to trigger exposure of CL for recognition by the autophagy machinery, contributing another layer to cellular defenses against the malfunctioning mitochondrial endosymbiont. Given that strong apoptotic stimuli result in caspase-mediated cleavage of Beclin 125, and oxidized CL is required for the release of pro-apoptotic factors into the cytosol, we further propose that, if the mitophagic machinery fails to eliminate damaged mitochondria, externalized CLs may then undergo oxidation as a prelude to apoptosis47 (Fig. 6).
Supplementary Material
Acknowledgments
Supported in part by funding from the National Institutes of Health [AG026389 (CTC), NS065789 (CTC), F32AG030821 (RKD), NS061817 (HB), HL70755 (VEK), OH008282 (VEK), U19AIO68021 (HB,VEK), NS076511 (HB,VEK), ES020693 (VEK,YYT)]. We thank many generous scientists listed in the SOM Materials for gifts of reagents, and Daniel Efrain Winnica for assistance with primary neuron cultures.
Footnotes
Author Contributions: J.J., R.K.D. and J.F.J. conducted and analyzed experiments in the primary neuron, SH-SY5Y and HeLa cell systems, respectively. A.A.A., Y.Y.T., V.A.T. collected and analyzed the LC-MS data. K.Z.Q.W. contributed to LC3 mutagenesis and PLS3 knockdown studies. O.O.K performed the liposome studies. D.M., N.Y., J.K.-S., I.H.S. and I.B. performed computational modeling. K.B., G.B. and Z.H. developed and optimized specific assays for the study. C.B. and S.W. performed specialized fluorescence microscopy. J.Z., A.M.G., E.K.S., R.W. and A.C. contributed to the PINK1, Parkin, p62 and flux analyses. C.T.C., H.B., V.E.K. planned and designed the study, analyzed data, developed the conceptual model and wrote the manuscript. All authors discussed the results, and contributed to writing or commenting on the manuscript.
Methods and Supplementary Information
Methods and Supplementary Information are available in the online version of the paper at www.nature.com/naturecellbiology.
Author Information. The authors have no competing financial interests to declare. Reprints and permissions information is available online at www.nature.com/reprints.
References
- 1.Kim I, Lemasters JJ. Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxidants & redox signaling. 2011;14:1919–1928. doi: 10.1089/ars.2010.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of cell biology. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxiainduced mitophagy in mammalian cells. Nature cell biology. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 5.Schlame M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. Journal of lipid research. 2008;49:1607–1620. doi: 10.1194/jlr.R700018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beyer K, Nuscher B. Specific cardiolipin binding interferes with labeling of sulfhydryl residues in the adenosine diphosphate/adenosine triphosphate carrier protein from beef heart mitochondria. Biochemistry. 1996;35:15784–15790. doi: 10.1021/bi9610055. [DOI] [PubMed] [Google Scholar]
- 7.Singh SB, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nature cell biology. 2010;12:1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugawara K, et al. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells. 2004;9:611–618. doi: 10.1111/j.1356-9597.2004.00750.x. [DOI] [PubMed] [Google Scholar]
- 9.Miyanishi M, et al. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 10.Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70:1033–1053. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu JH, Guo F, Shelburne J, Watkins S, Chu CT. Localization of phosphorylated ERK/MAP kinases to mitochondria and autophagosomes in Lewy body diseases. Brain Pathol. 2003;13:473–481. doi: 10.1111/j.1750-3639.2003.tb00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barth S, Glick D, Macleod KF. Autophagy: assays and artifacts. The Journal of pathology. 2010;221:117–124. doi: 10.1002/path.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyurin VA, et al. Oxidative lipidomics of programmed cell death. Methods in enzymology. 2008;442:375–393. doi: 10.1016/S0076-6879(08)01419-5. [DOI] [PubMed] [Google Scholar]
- 15.Crompton M, Barksby E, Johnson N, Capano M. Mitochondrial intermembrane junctional complexes and their involvement in cell death. Biochimie. 2002;84:143–152. doi: 10.1016/s0300-9084(02)01368-8. [DOI] [PubMed] [Google Scholar]
- 16.Andree HA, et al. Binding of vascular anticoagulant alpha (VAC alpha) to planar phospholipid bilayers. The Journal of biological chemistry. 1990;265:4923–4928. [PubMed] [Google Scholar]
- 17.Dagda RK, Zhu J, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson’s disease. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalovich EM, Zhu JH, Caltagarone J, Bowser R, Chu CT. Functional repression of cAMP response element in 6-hydroxydopamine-treated neuronal cells. The Journal of biological chemistry. 2006;281:17870–17881. doi: 10.1074/jbc.M602632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, et al. Role of phospholipid scramblase 3 in the regulation of tumor necrosis factor-alpha-induced apoptosis. Biochemistry. 2008;47:4518–4529. doi: 10.1021/bi701962c. [DOI] [PubMed] [Google Scholar]
- 20.Huang Z, et al. Cardiolipin deficiency leads to decreased cardiolipin peroxidation and increased resistance of cells to apoptosis. Free Radic Biol Med. 2008;44:1935–1944. doi: 10.1016/j.freeradbiomed.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji J, et al. Lipidomics identifies cardiolipin oxidation as a mitochondrial target for redox therapy of brain injury. Nature neuroscience. 2012;15:1407–1413. doi: 10.1038/nn.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W, et al. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell research. 2012;22:473–489. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y, Vashisht AA, Tchieu J, Wohlschlegel JA, Dreier L. Voltage-dependent Anion Channels (VDACs) Recruit Parkin to Defective Mitochondria to Promote Mitochondrial Autophagy. The Journal of biological chemistry. 2012;287:40652–40660. doi: 10.1074/jbc.M112.419721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 25.Wirawan E, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell death & disease. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narendra DP, et al. PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS biology. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondapalli C, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open biology. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okatsu K, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shvets E, Fass E, Scherz-Shouval R, Elazar Z. The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J Cell Sci. 2008;121:2685–2695. doi: 10.1242/jcs.026005. [DOI] [PubMed] [Google Scholar]
- 33.Van Laar VS, et al. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Human molecular genetics. 2011;20:927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai Q, Zakaria HM, Simone A, Sheng ZH. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr Biol. 2012;22:545–552. doi: 10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO reports. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanna RA, et al. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. The Journal of biological chemistry. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Connerth M, et al. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 2012;338:815–818. doi: 10.1126/science.1225625. [DOI] [PubMed] [Google Scholar]
- 38.Borisenko GG, et al. Macrophage recognition of externalized phosphatidylserine and phagocytosis of apoptotic Jurkat cells--existence of a threshold. Archives of biochemistry and biophysics. 2003;413:41–52. doi: 10.1016/s0003-9861(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 39.Ravichandran KS. Find-me and eat-me signals in apoptotic cell clearance: progress and conundrums. The Journal of experimental medicine. 2010;207:1807–1817. doi: 10.1084/jem.20101157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rostovtseva TK, Kazemi N, Weinrich M, Bezrukov SM. Voltage gating of VDAC is regulated by nonlamellar lipids of mitochondrial membranes. The Journal of biological chemistry. 2006;281:37496–37506. doi: 10.1074/jbc.M602548200. [DOI] [PubMed] [Google Scholar]
- 41.Athea Y, et al. AMP-activated protein kinase alpha2 deficiency affects cardiac cardiolipin homeostasis and mitochondrial function. Diabetes. 2007;56:786–794. doi: 10.2337/db06-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onyenwoke RU, et al. AMPK directly inhibits NDPK through a phosphoserine switch to maintain cellular homeostasis. Molecular biology of the cell. 2012;23:381–389. doi: 10.1091/mbc.E11-08-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlattner U, et al. Dual function of mitochondrial Nm23-H4 in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. The Journal of biological chemistry. 2013;288:111–121. doi: 10.1074/jbc.M112.408633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC. Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. The EMBO journal. 1999;18:6329–6338. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dagda RK, et al. Loss of pink1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. The Journal of biological chemistry. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kagan VE, et al. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nature chemical biology. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.