Abstract
Secondary ion mass spectrometry (SIMS) applied in the event-by-event bombardment/detection mode is uniquely suited for the characterization of individual nano-objects. In this approach, nano-objects are examined one-by-one, allowing for the detection of variations in composition. The validity of the analysis depends upon the ability to physically isolate the nano-objects on a chemically inert support. This requirement can be realized by deposition of the nano-objects on a Nano-Assisted Laser Desorption/Ionization (NALDI™) plate. The featured nanostructured surface provides a support where nano-objects can be isolated if the deposition is performed at a proper concentration. We demonstrate the characterization of individual nano-objects on a NALDI™ plate for two different types of nanometric bacteriophages: Qβ and M13. Scanning electron microscope (SEM) images verified that the integrity of the phages is preserved on the NALDI™ substrate. Mass spectrometric data show secondary ions from the phages are identified and resolved from those from the underlying substrate.
Keywords: nano-objects, event-by-event SIMS analysis, ToF-SIMS, gold clusters, NALDI™
Introduction
The characterization of individual nano-objects presents new challenges for secondary ion mass spectrometry (SIMS). As their size decreases versus the volume required for full projectile energy dissipation, one reaches confined sample volumes, which can lead to a shift in secondary ion emission.[1] The latter can be further affected by the nature of the substrate supporting the nano-objects. We present here the case of SIMS applied to the characterization of biological nano-objects of ‘subcritical assay dimensions’, specifically two types of nanometric bacteriophages. Our approach uses event-by-event bombardment/detection, involving bombardments with individual 520-keV and identification of the ejecta from each individual impact. Provided individual nano-objects are spatially isolated on the surface, the secondary ions from an impacted object can be collected and resolved from those of other objects or the substrate. Thus, compositional information specific to the objects can be obtained using the coincidence method.[2]
A critical issue in the study of individual nano-objects is to maintain their physical and chemical integrity during examination. Unanticipated chemical reactivity can occur with nano-sized objects. For instance, a mix of individual polystyrene nanospheres and boehmite alumina whiskers can lead to the formation of nanocomposite flakes due to the electrostatic nature of the whiskers.[3] For the present study, we deposited the bacteriophages on a NALDI™ plate, which features a surface made of silicon nanowires of 20 nm in diameter and 100–500 nm in length.[4] The silicon nanowires on the plate provide a suitable platform to deposit individual nano-objects. Moreover, the surface of the nanowires is functionalized with (pentafluorophenyl)propyldimethylsilane,[5] which makes the substrate inert to bio-objects and hence preserves their integrity.
The bacteriophages investigated here are Qβ and M13 phages as models of complex nano-objects. The first phage is spherical, 25nm in diameter, and encapsulates a single-stranded RNA within the protein shell assembly.[6] The second is filamentous, 900nm in length, 7–9 nm in width, and encapsulates a circular single-stranded DNA.[7] In both cases, secondary ions from both the protein shell and the encapsulated ribonucleotides were identified in the conventional (total) mass spectra and resolved from those of the underlying nanowires in the coincidence mass spectra.
Experimental
Sample preparation
M13 phage was purchased from New England Biolabs Inc. (Ipswich, MA). Qβ phage was provided by R. Young (Texas A&M University). NALDI™ plates were provided by Bruker Daltonics Inc. (Billerica, MA). Desalting columns were purchased from Bio-Rad (Hercules, CA). The original buffer in the desalting columns was exchanged by deionized water (18.2 MΩcm, Millipore, Billerica, MA) four times based on the buffer exchange protocol provided by the manufacturer. Each phage sample was diluted by mixing 1 µL of stock phage solution with 49 µL of deionized water. The diluted phage solution was eluted through the desalting column to remove most of the salt and glycerol used to preserve phages. The eluate was collected and drop-casted onto a 1 × 1 cm2 NALDI™ plate.
ToF-SIMS analysis
The samples were analyzed in the event-by-event bombardment/detection mode with a custom-built SIMS instrument comprising an Au-Liquid Metal Ion Source (Au-LMIS 20 kV) coupled to a 100-kV Pegase Platform[8] and a time-of-flight (ToF) mass spectrometer equipped with a two-stage electrostatic mirror. The primary ion projectiles are mass-selected using a Wien filter and focused into the ToF chamber. The experiments were performed under single projectile impacts (rate ~1000 projectiles/s). Impacts on the surface are detected via electron emission from a negatively biased sample surface. Co-emitted secondary ions from single impacts are detected by an eight-anode detector and analyzed by a multi-channel time-to-digital converter. The data are recorded and processed event-by-event with a custom data acquisition system.[9]
Scanning electron microscopy
A Jeol-7500 Cold Field Emission scanning electron microscope (SEM) was used to examine the integrity of the phages on the NALDI™ plates. These plates have a stainless steel backing, and thus are not affected by charging. They were attached to the specimen holders with carbon tape. Electron beam conditions are noted in Fig. 1.
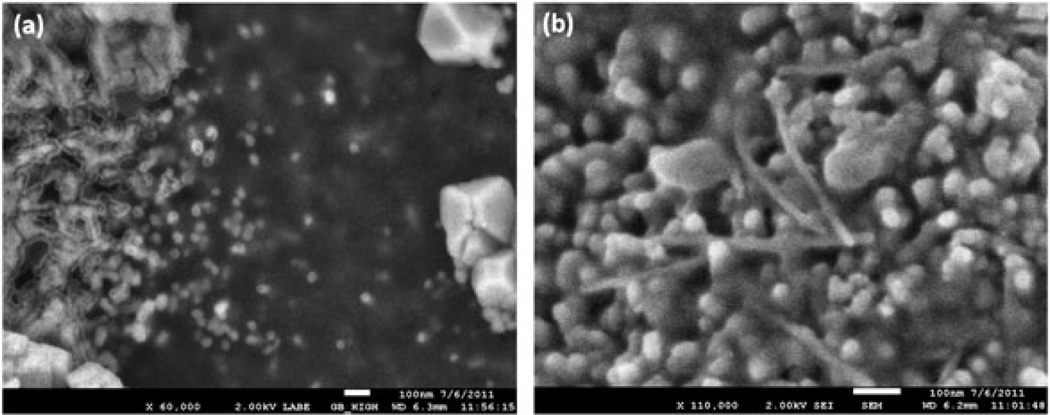
Figure 1.
SEM images of the a) spherical Qβ phages and b) filamentous M13 phages drop-casted onto a NALDI™ plate (scale bar = 100 nm).
Results and discussion
The SEM images of the Qβ and M13 phages on the NALDI™ plates (Fig. 1) show that the phages are spatially dispersed with their integrity preserved. We note negligible agglomeration after deposition onto the nanowire-modified surface. The examination of the Qβ phages with SEM could only be accomplished in the electron backscattering mode. In this mode, we obtained a weak electron signal from the Qβ phages; however, the underlying Si nanowires were invisible. The SEM images were also affected by salt deposit. We base our claim of isolated Qβ phages on the NALDI™ plate on: (i) the fact that we deposited the phages from a very dilute solution (concentration similar to the M13 phage solution); (ii) the SEM image showing (weak) backscattered electron signal from isolated phages. The mass spectral data are from a 500-µm area bombarded stochastically with 1 to 5 million projectiles. A small amount of impacts (84k and 18k for the Qβ and M13 phage, respectively) occurred on an individual bacteriophage. We refer here to impacts where the ion emission met the coincidence/anticoincidence conditions. Data from these selected events were summed for statistics.
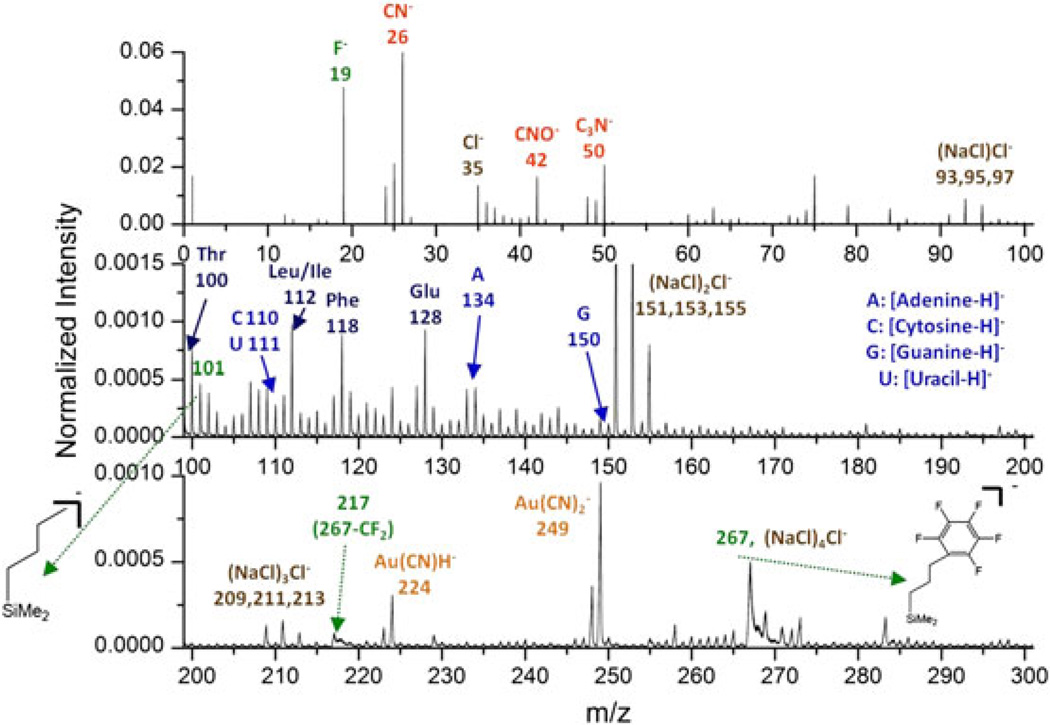
The total mass spectrum of the Qβ phages on the NALDI™ plate is presented in Fig. 2. The spectrum of the M13 phages shares similar features (data not shown). Substrate signals can be identified at m/z 267 as the intact molecular ion of the surface modifier ((pentafluorophenyl)propyldimethylsilane), and m/z 19, 101, 217 as fragments from the modifier. The presence of salt causes an isobaric interference at m/z 267 ((NaCl)4Cl−). Signals relevant to the phages mainly reside in the mass range from m/z 100 to 200, including amino acid residues from the protein shell assembly and nucleobases of the encapsulated ribonucleotides. The formation of Au adducts, e.g. Au(CN)H− and , results from the process where Au atoms are ablated from the massive Au projectile and then recombine with CN- fragments from the objects.[10]
Figure 2.
Negative ion mass spectrum of the Qβ phages on a NALDI™ plate. Intensities are normalized to the number of projectile impacts (~8 × 106).
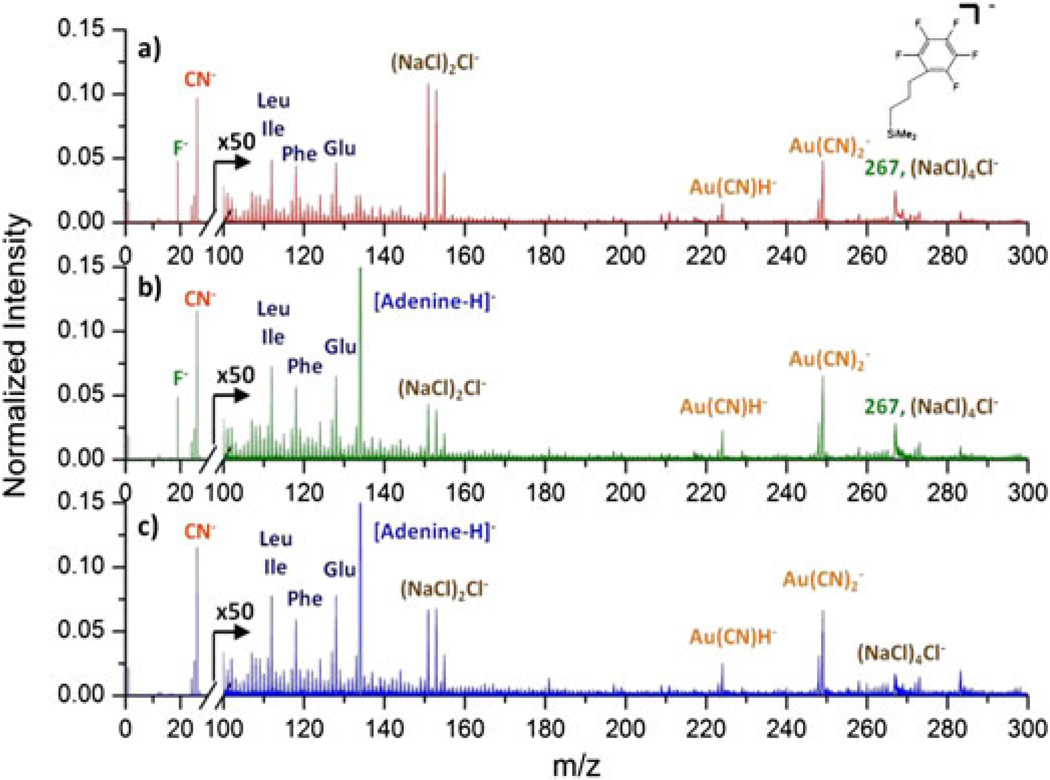
To resolve compositional information of the nano-objects from that of the substrate, coincidence mass spectra can be obtained using the co-emitted secondary ions from those events containing ion emission specific to the objects. The intensity of ions in the coincidence mass spectrum is normalized to the number of the selected events. The coincidence mass spectrum displayed in Fig. 3b shows ions co-emitted with [Adenine-H]- from the Qβ phages. Peaks specific to the phages are enhanced in the coincidence mass spectrum; however, peaks corresponding to the substrate at m/z 19 and 267 are still present. The remaining substrate signals can be attributed to impacts on the regimes where phages and nanowires are in physical contact, i.e. grazing impacts.
Figure 3.
a) Total mass spectrum of the Qβ phages on a NALDI™ plate. b) Coincidence mass spectrum of ions co-emitted with [Adenine-H]− from the phages. c) Coincidence/anticoincidence mass spectrum constituted by events containing [Adenine-H]− but without F−. Coincidence and coincidence/anticoincidence mass spectra are normalized to the corresponding numbers of events.
Compared to the grazing impacts, direct or ‘bulls-eye’ impacts on the Qβ phages are expected to yield specific information to the phage because the resulting ejecta originate only from the inside or the top of the nano-objects. Based on the difference in ejecta from these two types of impacts, a mass spectrum derived only from bulls-eye impacts on the Qβ phages can be extracted from the parent coincidence mass spectrum by removing the events containing substrate specific ions. This additional selection is based on an anticoincidence with the substrate specific ions. The new coincidence/anticoincidence mass spectrum presented in Fig. 3c is constituted by events containing [Adenine-H]−but without F−. The peak at m/z 19 is therefore absent from the spectrum, and the peak at m/z 267 is largely decreased but not totally eliminated. The remaining signal is attributed to the isobaric ion (NaCl)4Cl− and to the events in which F− was not detected.
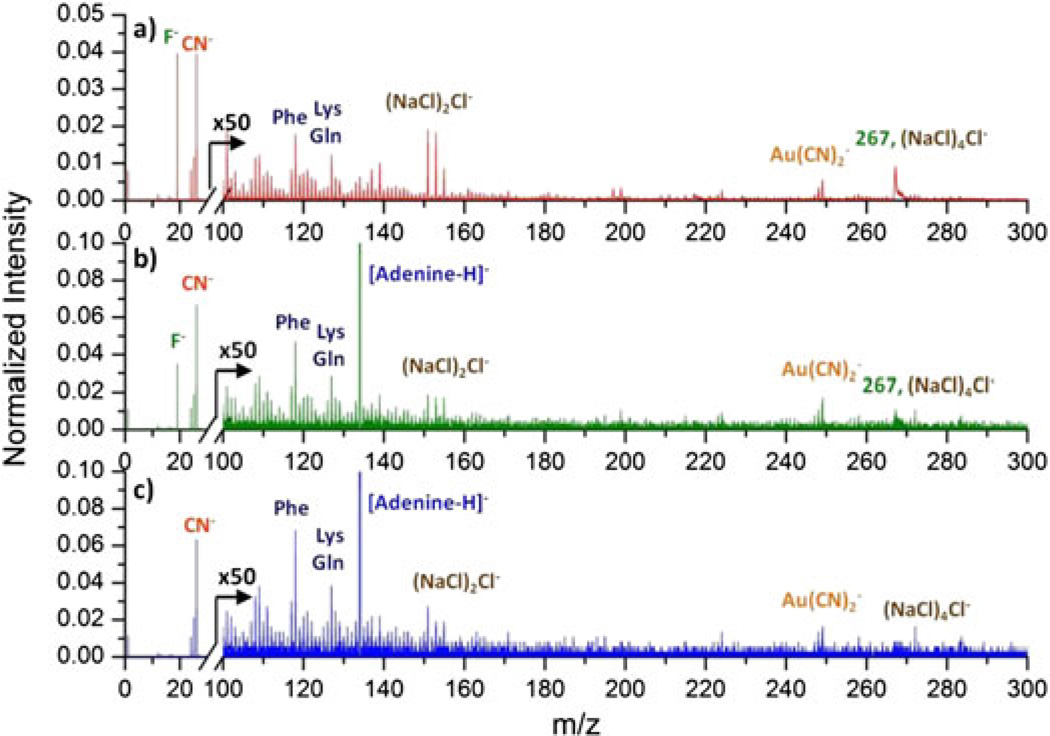
The total spectrum of the M13 phages was processed with the same methodology, and the resulting coincidence and coincidence/anticoincidence mass spectra are presented in Fig. 4b and 4c. It is noteworthy that the decrease of peak at m/z 267 is more prominent in the coincidence mass spectrum compared to that for the Qβ phages. This is due not only to the fact that there is a lower salt content which causes isobaric interference but also to the fact that the long M13 phages provide many more sites for bulls-eye impacts per phage. The ratio of the number of events in the coincidence/anticoincidence mass spectrum to the number of events in the coincidence mass spectrum is larger for the M13 phage than the Qβ phage (57% vs 44%). This ratio represents the proportion of bulls-eye impacts among bulls-eye and grazing impacts. In addition, the presence of phage-related ions in the coincidence/anticoincidence mass spectrum also implies that the depth of emission of the projectile might be close to the width of the M13 phage, which is ~7–9 nm. If the depth of emission is larger than the width, ejecta from bulls-eye impacts would most likely include ions from both the phage and the underlying substrate. Thus, we could not obtain a mass spectrum of the phage shell or core alone.
Figure 4.
a) Total mass spectrum of the M13 phages on a NALDI™ plate. b) Coincidence mass spectrum of ions co-emitted with [Adenine-H]− from the phages. c) Coincidence/anticoincidence mass spectrum constituted by events containing [Adenine-H]− but without F−.
Conclusion
We present a characterization of individual bacteriophages isolated on a nanostructured substrate. This approach preserves the integrity of the bio-objects studied. The event-by-event bombardment/detection methodology allows one to extract chemical information from individually dispersed nano-objects. The coincidence methodology provides primary selection of object-specific information out of the total mass spectrum. The combination of coincidence with anticoincidence demonstrates further resolution between two types of impacts on nanoobjects. The effectiveness of the approach depends on both the size and shape of the nano-objects, which affect the proportion of bulls-eye impacts and grazing impacts. Further resolution may be achieved incorporating additional coincidence or anticoincidence. The scope of coincidence/anticoincidence signal selection in ion co-emission analysis is limited only by the data required for statistical validation.
Acknowledgements
This work was supported by the National Science Foundation (Grant CHE-0750377). We thank Dr. Ry Young for the Qβ phage as a gift.
References
- 1.Pinnick V, Rajagopalachary S, Verkhoturov SV, Kaledin L, Schweikert EA. Anal. Chem. 2008;80:9052. doi: 10.1021/ac8014615. [DOI] [PubMed] [Google Scholar]
- 2.Park MA, Gibson KA, Quinones L, Schweikert EA. Science. 1990;248:988. doi: 10.1126/science.248.4958.988. [DOI] [PubMed] [Google Scholar]
- 3.Pinnick VT, Verkhoturov SV, Kaledin L, Bisrat Y, Schweikert EA. Anal. Chem. 2009;81:7527. doi: 10.1021/ac9014337. [DOI] [PubMed] [Google Scholar]
- 4.Daniels RH, Dikler S, Li E, Stacey C. J. Assoc. Lab. Autom. 2008;13:314. [Google Scholar]
- 5.Go EP, Apon JV, Luo G, Saghatelian A, Daniels RH, Sahi V, Dubrow R, Cravatt BF, Vertes A, Siuzdak G. Anal. Chem. 2005;77:1641. doi: 10.1021/ac048460o. [DOI] [PubMed] [Google Scholar]
- 6.Golmohammadi R, Fridborg K, Bundule M, Valegård K, Liljas L. Structure. 1996;4:543. doi: 10.1016/s0969-2126(96)00060-3. [DOI] [PubMed] [Google Scholar]
- 7.Stopar D, Spruijt RB, Wolfs CJAM, Hemming MA. Biochim. Biophys. Acta. 2003;1611:5. doi: 10.1016/s0005-2736(03)00047-6. [DOI] [PubMed] [Google Scholar]
- 8.Della-Negra S, Arianer J, Depauw J, Verkhoturov SV, Schweikert EA. Surf. Interface Anal. 2011;43:66. [Google Scholar]
- 9.Rickman RD, Verkhoturov SV, Hager GJ, Schweikert EA. Int. J. Mass Spectrom. 2005;241:57. [Google Scholar]
- 10.Guillermier C, Della Negra S, Rickman RD, Hager GJ, Schweikert EA. Int. J. Mass Spectrom. 2007;263:298. [Google Scholar]