Abstract
Microbes exist in a range of metabolic states (for example, dormant, active and growing) and analysis of ribosomal RNA (rRNA) is frequently employed to identify the ‘active' fraction of microbes in environmental samples. While rRNA analyses are no longer commonly used to quantify a population's growth rate in mixed communities, due to rRNA concentration not scaling linearly with growth rate uniformly across taxa, rRNA analyses are still frequently used toward the more conservative goal of identifying populations that are currently active in a mixed community. Yet, evidence indicates that the general use of rRNA as a reliable indicator of metabolic state in microbial assemblages has serious limitations. This report highlights the complex and often contradictory relationships between rRNA, growth and activity. Potential mechanisms for confounding rRNA patterns are discussed, including differences in life histories, life strategies and non-growth activities. Ways in which rRNA data can be used for useful characterization of microbial assemblages are presented, along with questions to be addressed in future studies.
Keywords: community rRNA, microbial activity, microbial growth, ribosomes, environmental samples, ecosystem processes
Introduction
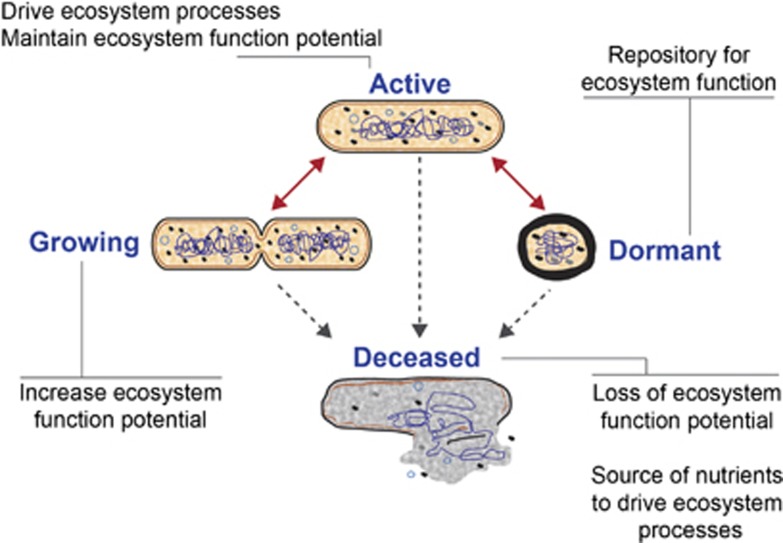
Microorganisms have essential roles in shaping and controlling virtually all ecosystems including the atmosphere, oceans, soils and plant- and animal-associated biomes. Microbes exist in different metabolic states in these systems: growing, active, dormant and recently deceased (Figure 1). These metabolic states correspond to different degrees of influence that microbes can have on their environment. Therefore, to understand the relationships between microbial community structure and ecosystem functions, it is important to accurately associate microbial identity with concurrent metabolic state. Simultaneous identification of microbes and their metabolic states has been a longstanding goal in microbial ecology, and methods to achieve this have recently been accumulating in our molecular toolboxes.
Figure 1.
Microorganism metabolic states and their contribution to ecosystem functioning. Viable microorganisms exist in one of three general metabolic states that are all subject to mortality. Definitions of terms: Growing—cells are actively dividing, Active—cells are measurably metabolizing (catabolic and/or anabolic processes) but are not necessarily dividing, Dormant—cells are not measurably dividing or metabolizing, Deceased—cells are not metabolically active or capable of becoming metabolically active in the future, but intact macromolecules may persist.
Nucleic-acid analysis has proven to be effective for characterizing the phylogenetic, taxonomic and functional structure of microbial assemblages, but this approach has limitations when attempting to assess current metabolic state. Ribosomal RNA genes (rRNA genes) are frequently used to identify microorganisms present in environmental samples regardless of metabolic state, while ribosomal RNA (rRNA) has been widely applied to characterize the growing or active microbes. We found >100 studies that used rRNA for these purposes, including recent studies using rRNA to identify currently active microbes (for example, Muttray and Mohn, 2000; Duineveld et al., 2001; Mills et al., 2005; Schippers et al., 2005; Gentile et al., 2006; DeAngelis et al., 2010; Jones and Lennon, 2010; Brettar et al., 2011; Egert et al., 2011; Gaidos et al., 2011; Wüst et al., 2011; Mannisto et al., 2012; Hunt et al., 2013). However, conflicting patterns between rRNA content and growth rate indicate that rRNA is not a reliable metric for growth or activity and in some cases may be grossly misleading. Virtually all molecular characterization methods are imperfect, but we suggest that using rRNA analyses to evaluate microbial assemblages requires that limitations and underlying assumptions be clearly identified and understood. Here, we explore critical limitations and potential causes of inconsistent rRNA/activity relationships. We then suggest employing rRNA abundance data as an index of potential activity and propose a framework for future application. The reader should note that RNA extraction methods are important in interpreting the validity of any downstream RNA-based results. Often in the literature, purification and analytical methods for RNA differ and are not shown to be reproducible and quantitative. As techniques advance, methods are continuously improved and new experimental results are presented. From a technical point of view, it is extremely arduous to re-interpret older results based on new methodological improvements and is beyond the scope of this review. However, from an epistemological point of view, it is important to keep in mind potential methodological biases to ensure that the assumptions of the relationship between RNA and activity are clearly articulated, and to recognize the specific limitations of applying a broad generalization for RNA content to environmental samples. With this in mind, we discuss studies that utilized several different experimental approaches; thus, observed discrepancies between rRNA abundance and activity are very likely to be at least in part biological in origin and not simply methodological artifact. We focus on bacteria, which have been extensively studied, but many of the limitations discussed here are likely relevant for other microbes, including archaea, fungi and algae.
rRNA and its use in microbial ecology
The cell's total RNA pool is mainly composed of rRNA (82–90%) (Tissieres and Watson, 1958; Neidhardt and Magasanik, 1960; Neidhardt, 1987). As an integral structural component of ribosomes, rRNA is a fundamental constituent of all known microorganisms and most rRNA found in a cell is ribosome associated (Lindahl, 1975; Nomura et al., 1984). Total RNA concentration is generally proportional to rRNA concentration and to the number of ribosomes in the cell, and has often been employed as a proxy for both (Kerkhof and Ward, 1993; Poulsen et al., 1993; Bremer and Dennis, 1996). In pure-culture experiments, cell counts can be done to determine RNA or ribosome concentration per cell. In mixed communities, other methods of normalization are necessary. Commonly, RNA or rRNA concentration is normalized to the number of cells using DNA concentration to calculate the RNA:DNA or an rRNA:rRNA gene ratio (for example, Kemp et al., 1993; Kerkhof and Ward, 1993; Poulsen et al., 1993; Muttray et al., 2001), since DNA concentration per cell is generally more stable than RNA concentration. Note, however, that while cell genome content commonly varies less than RNA content, genome abundance per cell can vary significantly and therefore could influence RNA:DNA measurements (Schaechter et al., 1958; Cooper and Helmstetter, 1968; Sukenik et al., 2012), but this issue will not be addressed here.
Historically, rRNA analyses have been used to quantify populations' growth rates in mixed microbial communities (for example, Poulsen et al., 1993; Muttray et al., 2001), but recent application has shifted toward the more qualitative approach using rRNA to identify currently active microbial populations in a mixed community (for example, Jones and Lennon, 2010; Kamke et al., 2010; Campbell et al., 2011; DeAngelis et al., 2011; Gaidos et al., 2011; Reid et al., 2011; Baldrian et al., 2012; Mannisto et al., 2012; Mattila et al., 2012; Simister et al., 2012; Campbell and Kirchman, 2013; Hunt et al., 2013; Yarwood et al., 2013). Two principal lines of evidence used to support rRNA as an indicator of current activity originate from earlier studies testing how rRNA scales with growth rate. First, total RNA and rRNA content correlate well with growth rate for a handful of microbes in pure culture, over a wide range of growth rates under balanced growth conditions (that is, growing in an unchanging environment) (Schaechter et al., 1958; Neidhardt and Magasanik, 1960; Rosset et al., 1966; Koch, 1970; Kemp et al., 1993; Kerkhof and Ward, 1993; Poulsen et al., 1993; Wagner, 1994; Bremer and Dennis, 1996; Ramos et al., 2000). Second, decreased rRNA content is associated with decreased growth rate for some organisms growing under specific nutrient-limiting conditions (Mandelstam and Halvorson, 1960; Davis et al., 1986; Kramer and Singleton, 1992; Tolker-Nielsen et al., 1997). Note that the relationship between rRNA concentration and growth rate is frequently coupled with the assumption that activity and growth are synonymous. Here, we distinguish growth from activity; while all growing organisms are active, not all active organisms are growing (Figure 1). Experimental evidence demonstrates numerous limitations to use rRNA to quantify population growth rates in mixed communities, many of which have been addressed in methodological reviews (for example, Molin and Givskov, 1999). However, while most of these limitations are also pertinent when attempting to identify which microbes are currently active in a community, these limitations are frequently overlooked or ignored in practice. Here, we provide a summary of limitations (Box 1) that pertain to the relationship between rRNA and current activity, and discuss relevant examples to assess the information that rRNA data can actually provide.
Box 1: Limitations of rRNA as an indicator of current microbial activity (References include the seminal studies that were later often overly generalized to support rRNA–activity relationship).
Concentration of rRNA and growth rate are not always simply correlated; therefore, the relationship between rRNA and activity is not likely consistent (Schaechter et al., 1958; Mandelstam and Halvorson, 1960; Flärdh et al., 1992, Kemp et al., 1993; Tolker-Nielsen et al., 1997; Binder and Liu, 1998; Lepp and Schmidt, 1998; McKillip et al., 1998; Kerkhof and Kemp, 1999; Morgenroth et al., 2000; Oda et al., 2000; Schmid et al., 2001; Worden and Binder, 2003).
The relationship between rRNA concentration and growth rate can differ significantly among taxa; therefore, relative rRNA abundance will likely not provide robust information regarding which taxa are relatively more active in a community (Mandelstam and Halvorson, 1960; Wade and Robinson, 1965; Rosset et al., 1966; Kemp et al., 1993; Pang and Winkler, 1994; Oda et al., 2000; Binnerup et al., 2001; Worden and Binder, 2003).
Dormant cells can contain high numbers of ribosomes; therefore, in environments that could likely contain dormant cells, employing rRNA to identify current activity is highly problematic (Chaloupecky, 1964; Bishop and Doi, 1966; Chambon et al., 1968; Filion et al., 2009; Sukenik et al., 2012).
The relationship between non-growth activities and concentration of rRNA has not yet been investigated.
Critical analysis of rRNA as an indicator of current activity
Concentration of rRNA and growth rate are not always simply correlated
The first line of evidence that has been used to support a predictable relationship between the presence of rRNA and current activity is based on pure-culture studies assessing growth under balanced growth conditions. However, even under constrained conditions (balanced growth) the correlation between growth rate and rRNA concentration is commonly not straightforward and in some cases breaks down altogether. For example, the relationship between growth rate and rRNA content is not linear or consistent across all measured growth rates. Under balanced growth conditions, Synechococcus and Prochlorococcus strains can have a three-phase relationship between growth and rRNA concentration: (1) at low growth rates, rRNA concentration remains constant, (2) at intermediate growth rates, rRNA concentration increases linearly with growth rate and (3) at higher growth rates, rRNA content decreases as growth rate increases (Binder and Liu, 1998; Worden and Binder, 2003). For these organisms, rRNA concentration is not a robust proxy for growth rate. We argue that rRNA will also not be a robust measure of current activity, since changes in growth-associated activity must impact total activity. Additionally, balanced growth conditions are unlikely in most environments. Little work has characterized how rRNA concentration varies with growth rate under more environmentally realistic non-steady state conditions. Kerkhof and Kemp (1999) identified three different relationship patterns between rRNA concentration and growth rate for Proteobacteria strains under non-steady state conditions: a direct linear relationship, an indirect relationship in which cell growth rate consistently lagged behind rRNA concentration or no discernible relationship. The latter was observed in Vibrio fischeri, and included periods during which growth rate decreased while rRNA content increased. Again, since growth activity likely accounts for much of total activity, these results indicate that using rRNA concentration to assess current activity or changes in activity over time is problematic. Further evidence showing potential for misleading environmental interpretations includes significant increase in cellular rRNA in Aphanizomenon ovalisporum cells transitioning from vegetative to dormant state (Sukenik et al., 2012). These results indicate that a measurable increase in rRNA abundance does not necessarily indicate an increase in activity.
A second line of evidence cited to support rRNA as an indicator of current activity arises from studies on RNA stability under different growth-limiting conditions (for example, carbon or nutrient limitations). Several studies have reported that exponentially growing cells subjected to nutrient starvation degrade much of their rRNA in a relatively short time. However, the dynamics of cellular rRNA may be strongly tied to previous growth conditions. For example, Azotobacter agilis was grown on different substrates, then starved for 72 h (Sobek et al., 1966). When grown on glucose, RNA did not decrease during the starvation period, but O2 consumption dramatically dropped, indicating that cell activity (that is, respiration) declined. In another study, Rhodopseudomonas palustris cells were grown at different growth rates, then carbon-starved (Oda et al., 2000). The rRNA concentration of R. palustris cells grown at maximum growth rate decreased by ∼50% within a week of starvation; however, cells grown at lower rates before the starvation period were able to maintain near pre-starvation rRNA concentrations for more than a week of starvation. These results indicate that measurable rRNA concentration can be influenced not only by current conditions, but also by life history (that is, the sequence of events that impacted an organism up to a given time point, and the resulting physiological response to these events).
The relationship between rRNA concentration and growth rate can differ significantly among taxa
Relating rRNA concentration and growth rate becomes even more problematic when considering microbial assemblages. rRNA concentration may correlate well with growth rate in some strains of bacteria, but correlations can differ significantly between strains (Wade and Robinson, 1965; Kemp et al., 1993; Pang and Winkler, 1994; Binnerup et al., 2001; Worden and Binder, 2003). Even at the ‘species' level of bacteria, the relationship between rRNA and growth rate can differ significantly between subpopulations (Rosset et al., 1966; Licht et al., 1999). Hence, using rRNA to compare relative activity or changes in activity between taxa will likely provide misleading information.
Dormant cells can contain high numbers of ribosomes
Dormant organisms contain measurable amounts of rRNA (Chambon et al., 1968) and in some cases can contain significantly more rRNA in dormancy than in a vegetative state (Sukenik et al., 2012). Detectability of rRNA in dormant cells can be affected more by methodology (due to changes in cell structure) than by low levels of rRNA (Filion et al., 2009). The issue of dormant cells containing measurable rRNA concentrations can be especially problematic when using rRNA data to identify currently active organisms in environments likely to contain many dormant organisms such as soil, deep subsurface, frozen environments or the atmosphere. One approach to discounting rRNA in dormant cells is to estimate the rRNA concentration per cell for specific taxa by calculating rRNA:rRNA gene ratios, then defining a minimum cutoff value for activity (for example, DeAngelis et al., 2011; Jones and Lennon, 2010). However, rRNA:rRNA gene ratios have been characterized for very few bacteria in dormant state. The limited available evidence demonstrates the difficulties in establishing a suitable universal cutoff value for rRNA:rRNA gene ratio. For example, an RNA:DNA ratio of around 5 was found both in dormant Bacillus megaterium (Chambon et al., 1968) and in bacteria growing at the rapid pace of ∼0.5 h−1 (Kerkhof and Ward, 1993).
The relationship between non-growth activities and concentration of rRNA has not been investigated
Finally, the relationship between rRNA concentration and growth rate is commonly considered to be equivalent to that between rRNA and activity. However, many microbial activities are not necessarily related to growth, including those associated with maintenance, such as cell motility, osmoregulation, defense against oxidative stress, communication, exopolysaccharide production or conjugation (van Bodegom, 2007). To our knowledge, no published work has investigated the relationship between non-growth activities and rRNA concentration. It has been hypothesized that under certain stress conditions, microbes can dramatically increase the portion of metabolism geared toward non-growth maintenance activities (Schimel et al., 2007), indicating that, under appropriate conditions, non-growth activities may contribute significantly to ecosystem processes.
Relationship between rRNA, growth and activity: physiological links
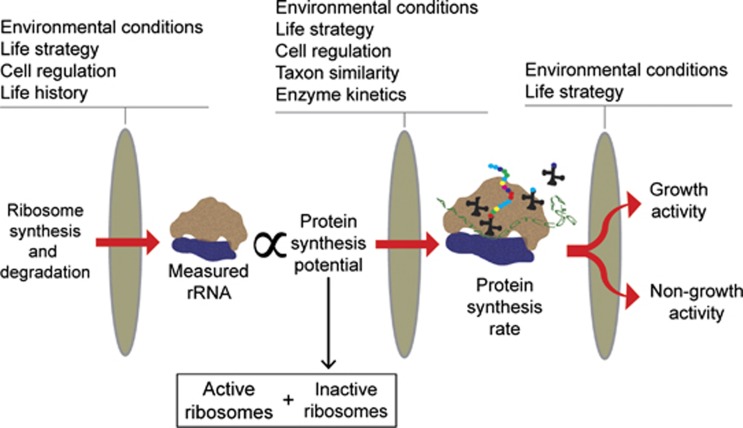
The multi-level modulation and regulation of most cell functions may easily invalidate simple correlations between current metabolic state and rRNA abundance. For example, the relationship between microbial activity and measurable rRNA can be influenced by heterogeneity of cell physiology within a population (Licht et al., 1999), changes in the ratio of non-growth to growth-specific metabolic activity, life history (Oda et al., 2000), life strategy (Flärdh et al., 1992; Lepp and Schmidt, 1998; Mitchell et al., 2009; Sukenik et al., 2012), sample heterogeneity, changing environmental conditions and of course fundamental enzyme kinetics (that is, substrate concentration). Additionally, the concentration of rRNA in a cell at a given point in time is the net result of rRNA synthesis (that is, transcription) and degradation rates (Gausing, 1977), each of which may be under distinct controls. All of these factors can affect the relationship between ribosome turnover and microbial activity at multiple levels (Figure 2) and should be considered when analyzing rRNA data from environmental samples.
Figure 2.
From ribosome cycling to microbial activity: factors that affect ribosome quantities and their relationship to microbial activities. Life strategy is defined here as the potential physiological responses, hardwired via DNA into an organism or population, toward two competing goals: surviving and reproducing.
rRNA analyses in community ecology
rRNA-based measurements can provide meaningful insight into microbial community dynamics. rRNA directly relates to a population's potential to catalyze the specific function of protein synthesis, and can therefore document the relative expression of this function. rRNA-based measurements provide a specific piece of information in the spectrum of molecular approaches (including metagenomics, metatranscriptomics, metaproteomics and community proteogenomics) that are increasingly applied to study microbial communities. Metagenomic data provide information about the functional potential of a sample, without providing insight into current metabolic state. Metatranscriptomic data come one step closer to current metabolic state, without providing direct evidence of translation or enzyme activity. Metaproteomic data come an additional step closer to current metabolic state, by identifying enzymes expressed in a community, but without providing direct evidence of enzyme activity. While rRNA is a product of transcription, community rRNA data are more analogous to metaproteomic than to metatranscriptomic (mRNA) data; rRNA is generally much more stable than mRNA (Snyder and Champness, 2007), and is not translated to protein but instead acts as a structural component of housekeeping catalysts (ribosomes). Therefore, rRNA data can provide evidence of the relative expression of an enzyme, with the explicit function of protein synthesis, for different populations in a community. Analogously, in metaproteomics, environmental proteins are characterized to provide information about specific enzymatic functions that are expressed (Wilmes and Bond, 2004). Taking this analogy one step further, the community proteogenomics approach can be used to map the expressed function of a community (metaproteomic data) onto the available template of potential functions (metagenomic data) (Verberkmoes et al., 2009), to provide valuable information about how environmental changes correspond to changes in community expression in the context of community composition. Similarly, rRNA data can be mapped onto rRNA gene data to illuminate relative ribosomal expression of the total community. However, it is important to recognize that while enzyme/protein data come closer than gene and transcript data to identifying real-time activity, the presence of an enzyme does not unequivocally denote current activity for a given function, because many factors control enzymatic activity in vivo (Nannipieri et al., 2002). Similarly, the presence of rRNA is indicative of protein synthesis potential, not of realized protein synthesis (Figure 2). The number of ribosomes present at a given time limits the maximum protein synthesis activity for a population, but does not directly inform about realized protein synthesis activity. The distinction between actual activity and potential activity is critical when attempting to identify and characterize the dynamics of organisms that drive ecosystem functions (Figure 1).
Applications in microbial ecology: future directions
What does measuring ‘protein synthesis potential' tell us about microbial populations? The relationship between the number of ribosomes and the ability to synthesize proteins links the quantity of rRNA in a population with its potential for growth and acclimation (that is, to upregulate or change currently expressed metabolic functions). rRNA can represent potential future activity, in addition to reflecting historical activity and conditions (as discussed above). For example, some microorganisms increase ribosome concentration as they enter a dormant state, a life strategy that provides them with higher protein synthesis potential, and therefore potentially higher fitness, as they return to a vegetative state when environmental conditions improve (Sukenik et al., 2012). Similarly, non-dormant populations maintaining ribosome levels above current protein synthesis demands likely have the ability to rapidly shift metabolic functions to adapt to changing conditions, thereby becoming better competitors (Koch, 1971; Alton and Koch, 1974; Flärdh et al., 1992).
Recognizing that rRNA concentration reflects past, current and future activities in addition to different life strategies restricts its utility as a metric of real-time activity, but provides the basis for generating and testing important hypotheses. Several studies show that under repeated temporal patterns of changing environmental conditions, microbes may develop an anticipatory life strategy, enduring one phase of the cycle while preparing for a more favorable phase that regularly follows. Further, accumulating or maintaining rRNA during periods of low metabolic activity may confer a competitive advantage during a favorable phase of the cycle. In Synechococcus sp. incubated under light and dark diurnal cycles, rRNA content increased during dark periods compared with light periods; in contrast, growth occurred during the light periods and ceased during the dark periods (Lepp and Schmidt, 1998). Similar results were found for a strain of Prochlorococcus in which expression of ribosomal genes was higher during a dark cycle than during a light cycle (Zinser et al., 2009). Further evidence for anticipatory behavior in bacteria was found in E. coli manifesting a Pavlovian-type response to a primary stimulus by preemptively modifying genetic expression for a secondary stimulus before it occurred (Tagkopoulos et al., 2008; Mitchell et al., 2009). Anticipatory strategies may also take place on a seasonal scale: at the end of a summer dry-down period, Mediterranean soil communities showed almost no measurable microbial activity (based on CO2 production), yet total extractable bacterial 16S rRNA was similar to that found after the microbes become activated by the first wet-up event (Placella et al., 2012), which could reflect anticipation for the upcoming annual rainy season (Barnard et al., 2013). If anticipatory life strategies reflected in rRNA concentrations are common in microbial populations experiencing repeated cyclic patterns, then can this information be meaningfully applied to predict future changes in ecosystem function?
To utilize rRNA data to characterize microbial assemblages, we need to better our understanding of how these data relate to environmental conditions and community interactions; this understanding could be furthered by several experimental approaches:
Coupling direct measurements of metabolic activity to rRNA data.
Explicitly testing the relationship between non-growth activities and rRNA concentrations.
Characterizing ribosome turnover under different environmental conditions.
Conclusion
A number of pure-culture studies have shown a correlation between growth rate and rRNA concentration. This relationship makes intuitive and biological sense, since rRNA is a critical component of ribosomes, and ribosomes are necessary to synthesize protein. However, the correlation between real-time activity and rRNA in environmental samples is inconsistent due to differences in life histories, life strategies and non-growth activities. Using rRNA analysis as a general indicator of currently active microbes in environmental samples is not valid under many circumstances, and may actually hinder progress connecting microbial activities to ecosystem functions. Considering rRNA measurements as indicators of protein synthesis potential provides microbial ecologists with a robust framework, facilitating a more prudent yet comprehensive understanding of the complex dynamics at play in microbial communities.
Acknowledgments
We thank Jim Prosser, Josh Schimel, Eoin Brodie and Laurent Philippot for constructive comments. SJB was supported by a National Science Foundation Graduate Research Fellowship. RLB was funded by the European Community's Seventh Framework Programme under grant agreement PIOF-GA-2008-219357. DOE Genomic Science Program grant (FOA DE-PS02-09ER09-25 award #0016377) to MKF.
The authors declare no conflict of interest.
References
- Alton TH, Koch AL. Unused protein synthetic capacity of Escherichia coli grown in phosphate-limited chemostats. J Mol Biol. 1974;86:1–9. doi: 10.1016/s0022-2836(74)80002-1. [DOI] [PubMed] [Google Scholar]
- Baldrian P, Kolarík M, Štursová M, Kopecky J, Valášková V, Vētrovsky T, et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012;6:248–258. doi: 10.1038/ismej.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard RL, Osborne CA, Firestone MK.2013Responses of soil bacterial and fungal communities to extreme desiccation and rewetting ISME Je-pub ahead of print 4 July 2013; doi: 10.1038/ismej.2013.104 . [DOI] [PMC free article] [PubMed]
- Binder BJ, Liu YC. Growth rate regulation of rRNA content of a marine Synechococcus (cyanobacterium) strain. Appl Environ Microbiol. 1998;64:3346–3351. doi: 10.1128/aem.64.9.3346-3351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnerup S, Bloem J, Hansen B, Wolters W, Veninga M, Hansen M. Ribosomal RNA content in microcolony forming soil bacteria measured by quantitative 16S rRNA hybridization and image analysis. FEMS Microbiol Ecol. 2001;37:231–237. [Google Scholar]
- Bishop H, Doi R. Isolation and characterization of ribosomes from Bacillus subtilis spores. J Bacteriol. 1966;91:695–701. doi: 10.1128/jb.91.2.695-701.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer H, Dennis PP.1996Modulation of Chemical Composition and Other Parameters of the Cell by Growth Rate2nd edn.ASM Press: Washington, DC; [DOI] [PubMed] [Google Scholar]
- Brettar I, Christen R, HöFle MG. Analysis of bacterial core communities in the central Baltic by comparative RNA–DNA-based fingerprinting provides links to structure–function relationships. ISME J. 2011;6:195–212. doi: 10.1038/ismej.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA. 2011;108:12776–12781. doi: 10.1073/pnas.1101405108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Kirchman DL. Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J. 2013;7:210–220. doi: 10.1038/ismej.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaloupecky V. Ribosomes in growing and non-growing bacterial cells. Folia Microbiol. 1964;9:232–237. doi: 10.1007/BF02875842. [DOI] [PubMed] [Google Scholar]
- Chambon P, DuPraw EJ, Kornberg A. Biochemical studies of bacterial sporulation and germination. J Biol Chem. 1968;243:5101–5109. [PubMed] [Google Scholar]
- Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol. 1968;31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- Davis BD, Luger SM, Tai PC. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis KM, Silver WL, Thompson AW, Firestone MK. Microbial communities acclimate to recurring changes in soil redox potential status. Environ Microbiol. 2010;12:3137–3149. doi: 10.1111/j.1462-2920.2010.02286.x. [DOI] [PubMed] [Google Scholar]
- DeAngelis KM, Wu CH, Beller HR, Brodie EL, Chakraborty R, DeSantis TZ, et al. PCR Amplification-independent methods for detection of microbial communities by the high-density microarray PhyloChip. Appl Environ Microbiol. 2011;77:6313–6322. doi: 10.1128/AEM.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duineveld BM, Kowalchuk GA, Keijzer A, Van Elsas JD, Van Veen JA. Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl Environ Microbiol. 2001;67:172. doi: 10.1128/AEM.67.1.172-178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egert M, Schmidt I, Höhne H-M, Lachnit T, Schmitz RA, Breves R. Ribosomal RNA-based profiling of bacteria in the axilla of healthy males suggests right-left asymmetry in bacterial activity. FEMS Microbiol Ecol. 2011;77:146–153. doi: 10.1111/j.1574-6941.2011.01097.x. [DOI] [PubMed] [Google Scholar]
- Filion G, Laflamme C, Turgeon N, Ho J, Duchaine C. Permeabilization and hybridization protocols for rapid detection of Bacillus spores using fluorescence in situ hybridization. J Microbiol Methods. 2009;77:29–36. doi: 10.1016/j.mimet.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Flärdh K, Cohen P, Kjelleberg S. Ribosomes exist in large excess over the apparent demand for protein-synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J Bacteriol. 1992;174:6780–6788. doi: 10.1128/jb.174.21.6780-6788.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidos E, Rusch A, Ilardo M. Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: community spatial structure, rare members and nitrogen-cycling guilds. Environ Microbiol. 2011;13:1138–1152. doi: 10.1111/j.1462-2920.2010.02392.x. [DOI] [PubMed] [Google Scholar]
- Gausing K. Regulation of ribosome production in Escherichia coli: synthesis and stability of ribosomal RNA and of ribosomal protein messenger RNA at different growth rates. J Mol Biol. 1977;115:335–354. doi: 10.1016/0022-2836(77)90158-9. [DOI] [PubMed] [Google Scholar]
- Gentile G, Giuliano L, D'auria G, Smedile F, Azzaro M, De Domenico M, et al. Study of bacterial communities in Antarctic coastal waters by a combination of 16S rRNA and 16S rDNA sequencing. Environ Microbiol. 2006;8:2150–2161. doi: 10.1111/j.1462-2920.2006.01097.x. [DOI] [PubMed] [Google Scholar]
- Hunt DE, Lin Y, Church MJ, Karl DM, Tringe SG, Izzo LK, et al. Relationship between abundance and specific activity of bacterioplankton in open ocean surface waters. Appl Environ Microbiol. 2013;79:177–184. doi: 10.1128/AEM.02155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA. 2010;107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamke J, Taylor MW, Schmitt S. Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. ISME J. 2010;4:498–508. doi: 10.1038/ismej.2009.143. [DOI] [PubMed] [Google Scholar]
- Kemp P, Lee S, LaRoche J. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol. 1993;59:2594. doi: 10.1128/aem.59.8.2594-2601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhof L, Ward B. Comparison of nucleic acid hybridization and fluorometry for measurement of the relationship between RNA/DNA ratio and growth rate in a marine bacterium. Appl Environ Microbiol. 1993;59:1303. doi: 10.1128/aem.59.5.1303-1309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhof L, Kemp P. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol Ecol. 1999;30:253–260. doi: 10.1111/j.1574-6941.1999.tb00653.x. [DOI] [PubMed] [Google Scholar]
- Koch AL. Overall controls on the biosynthesis of ribosomes in growing bacteria. J Theor Biol. 1970;28:203–231. doi: 10.1016/0022-5193(70)90053-6. [DOI] [PubMed] [Google Scholar]
- Koch AL. The adaptive responses of Escherichia coli to a feast and famine existence. Adv Microbiol Physiol. 1971;6:147–217. doi: 10.1016/s0065-2911(08)60069-7. [DOI] [PubMed] [Google Scholar]
- Kramer JG, Singleton FL. Variations in rRNA content of marine Vibrio spp. during starvation-survival and recovery. Appl Environ Microbiol. 1992;58:201. doi: 10.1128/aem.58.1.201-207.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepp P, Schmidt T. Nucleic acid content of Synechococcus spp. during growth in continuous light and light/dark cycles. Arch Microbiol. 1998;170:201–207. doi: 10.1007/s002030050634. [DOI] [PubMed] [Google Scholar]
- Licht TR, Tolker-Nielsen T, Holmstrøm K, Krogfelt KA, Molin S. Inhibition of Escherichia coli precursor-16S rRNA processing by mouse intestinal contents. Environ Microbiol. 1999;1:23–32. doi: 10.1046/j.1462-2920.1999.00001.x. [DOI] [PubMed] [Google Scholar]
- Lindahl L. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J Mol Biol. 1975;92:15–37. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- Mandelstam J, Halvorson H. Turnover of protein and nucleic acid in soluble and ribosome fractions of non-growing Escherichia coli. Biochim Biophys Acta. 1960;40:43–49. doi: 10.1016/0006-3002(60)91313-5. [DOI] [PubMed] [Google Scholar]
- Mannisto MK, Kurhela E, Tiirola M, Haggblom MM. Acidobacteria dominate the active bacterial communities of Arctic tundra with widely divergent winter-time snow accumulation and soil temperatures. FEMS Microbiol Ecol. 2012;84:47–59. doi: 10.1111/1574-6941.12035. [DOI] [PubMed] [Google Scholar]
- Mattila HR, Rios D, Walker-Sperling VE, Roeselers G, Newton ILG. Characterization of the active microbiotas associated with honey bees reveals healthier and broader communities when colonies are genetically diverse. PLoS One. 2012;7:e32962. doi: 10.1371/journal.pone.0032962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKillip JL, Jaykus LA, Drake M. rRNA stability in heat-killed and UV-irradiated enterotoxigenic Staphylococcus aureus and Escherichia coli O157:H7. Appl Environ Microbiol. 1998;64:4264–4268. doi: 10.1128/aem.64.11.4264-4268.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills HJ, Martinez RJ, Story S, Sobecky PA. Characterization of microbial community structure in Gulf of Mexico gas hydrates: comparative analysis of DNA-and RNA-derived clone libraries. Appl Environ Microbiol. 2005;71:3235. doi: 10.1128/AEM.71.6.3235-3247.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, et al. Adaptive prediction of environmental changes by microorganisms. Nature. 2009;460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- Molin S, Givskov M. Application of molecular tools for in situ monitoring of bacterial growth activity. Environ Microbiol. 1999;1:383–391. doi: 10.1046/j.1462-2920.1999.00056.x. [DOI] [PubMed] [Google Scholar]
- Morgenroth E, Obermayer A, Arnold E, Brühl A, Wagner M, Wilderer P. Effect of long-term idle periods on the performance of sequencing batch reactors. Water Sci Technol. 2000;41:105–113. [Google Scholar]
- Muttray AF, Mohn WW. Quantitation of the population size and metabolic activity of a resin acid degrading bacterium in activated sludge using slot-blot hybridization to measure the rRNA:rDNA ratio. Microb Ecol. 2000;38:348–357. doi: 10.1007/s002489901005. [DOI] [PubMed] [Google Scholar]
- Muttray AF, Yu Z, Mohn WW. Population dynamics and metabolic activity of Pseudomonas abietaniphila BKME-9 within pulp mill wastewater microbial communities assayed by competitive PCR and RT-PCR. FEMS Microbiol Ecol. 2001;38:21–31. [Google Scholar]
- Nannipieri P, Kandeler E, Ruggiero P.2002Enzyme activities and microbiological and biochemical processes in soilIn: Burns RG, Dick RP (eds)Enzymes in the Environment: Activity, Ecology, and Applications Marcel Dekker, Inc.: New York, NY; 1–33. [Google Scholar]
- Neidhardt FC, Magasanik B. Studies on the role of ribonucleic acid in the growth of bacteria. Biochim Biophys Acta. 1960;42:99–116. doi: 10.1016/0006-3002(60)90757-5. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC. Chemical Composition of Escherichia coli. vol. 1. American Society for Microbiology: Washington, DC; 1987. [Google Scholar]
- Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Oda Y, Slagman S-J, Meijer WG, Forney LJ, Gottschal JC. Influence of growth rate and starvation on fluorescent in situ hybridization of Rhodopseudomonas palustris. FEMS Microbiol Ecol. 2000;32:205–213. doi: 10.1111/j.1574-6941.2000.tb00713.x. [DOI] [PubMed] [Google Scholar]
- Pang H, Winkler HH. The concentrations of stable RNA and ribosomes in Rickettsia prowazekii. Mol Microbiol. 1994;12:115–120. doi: 10.1111/j.1365-2958.1994.tb01000.x. [DOI] [PubMed] [Google Scholar]
- Placella SA, Brodie EL, Firestone MK. Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc Natl Acad Sci USA. 2012;109:10931–10936. doi: 10.1073/pnas.1204306109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen L, Ballard G, Stahl D. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos C, Molbak L, Molin S. Bacterial activity in the rhizosphere analyzed at the single-cell level by monitoring ribosome contents and synthesis rates. Appl Environ Microbiol. 2000;66:801. doi: 10.1128/aem.66.2.801-809.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid NM, Addison SL, Macdonald LJ, Lloyd-Jones G. Biodiversity of active and inactive bacteria in the gut flora of wood-feeding huhu beetle larvae (Prionoplus reticularis) Appl Environ Microbiol. 2011;77:7000–7006. doi: 10.1128/AEM.05609-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosset R, Julien J, Monier R. Ribonucleic acid composition of bacteria as a function of growth rate. J Mol Biol. 1966;18:308–320. doi: 10.1016/s0022-2836(66)80248-6. [DOI] [PubMed] [Google Scholar]
- Schaechter M, Maaloe O, Kjeldgaard N. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. Microbiology. 1958;19:592. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Schimel J, Balser T, Wallenstein M. Microbial stress-response physiology and its implications for ecosystem function. Ecology. 2007;88:1386–1394. doi: 10.1890/06-0219. [DOI] [PubMed] [Google Scholar]
- Schippers A, Neretin LN, Kallmeyer J, Ferdelman TG, Cragg BA, Parkes RJ, et al. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature. 2005;433:861–864. doi: 10.1038/nature03302. [DOI] [PubMed] [Google Scholar]
- Schmid M, Schmitz-Esser S, Jetten M, Wagner M. 16S-23S rDNA intergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: implications for phylogeny and in situ detection. Environ Microbiol. 2001;3:450–459. doi: 10.1046/j.1462-2920.2001.00211.x. [DOI] [PubMed] [Google Scholar]
- Simister R, Taylor MW, Tsai P, Fan L, Bruxner TJ, Crowe ML, et al. Thermal stress responses in the bacterial biosphere of the Great Barrier Reef sponge, Rhopaloeides odorabile. Environ Microbiol. 2012;14:3232–3246. doi: 10.1111/1462-2920.12010. [DOI] [PubMed] [Google Scholar]
- Snyder L, Champness W.2007Molecular Genetics of Bacteria3rd ednASM Press: Washington, DC [Google Scholar]
- Sobek J, Charba J, Foust W. Endogenous metabolism of Azotobacter agilis. J Bacteriol. 1966;92:687–695. doi: 10.1128/jb.92.3.687-695.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukenik A, Kaplan-Levy RN, Welch JM, Post AF. Massive multiplication of genome and ribosomes in dormant cells (akinetes) of Aphanizomenon ovalisporum (Cyanobacteria) ISME J. 2012;6:670–679. doi: 10.1038/ismej.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagkopoulos I, Liu Y-C, Tavazoie S. Predictive behavior within microbial genetic networks. Science. 2008;320:1313–1317. doi: 10.1126/science.1154456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissieres A, Watson JD. Ribonucleoprotein particles from Escherichia coli. Nature. 1958;182:778–780. doi: 10.1038/182778b0. [DOI] [PubMed] [Google Scholar]
- Tolker-Nielsen T, Larsen MH, Kyed H, Molin S. Effects of stress treatments on the detection of Salmonella typhimurium by in situ hybridization. Int J Food Microbiol. 1997;35:251–258. doi: 10.1016/s0168-1605(97)01242-7. [DOI] [PubMed] [Google Scholar]
- van Bodegom P. Microbial maintenance: a critical review on its quantification. Microb Ecol. 2007;53:513–523. doi: 10.1007/s00248-006-9049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberkmoes NC, Denef VJ, Hettich RL, Banfield JF. Systems biology: functional analysis of natural microbial consortia using community proteomics. Nat Rev Microbiol. 2009;7:196–205. doi: 10.1038/nrmicro2080. [DOI] [PubMed] [Google Scholar]
- Wade HE, Robinson HK. The distribution of ribosomal ribonucleic acids among subcellular fractions from bacteria and the adverse effect of the membrane fraction on the stability of ribosomes. Biochem J. 1965;96:753–765. doi: 10.1042/bj0960753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. The regulation of ribosomal RNA synthesis and bacterial cell growth. Arch Microbiol. 1994;161:100–109. doi: 10.1007/BF00276469. [DOI] [PubMed] [Google Scholar]
- Wilmes P, Bond PL. The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ Microbiol. 2004;6:911–920. doi: 10.1111/j.1462-2920.2004.00687.x. [DOI] [PubMed] [Google Scholar]
- Worden AZ, Binder BJ. Growth regulation of rRNA content in Prochlorococcus and Synechococcus (marine cyanobacteria) measured by whole-cell hybridization of rRNA-targeted peptide nucleic acids. J Phycol. 2003;39:527–534. [Google Scholar]
- Wüst PK, Horn MA, Drake HL. Clostridiaceae and Enterobacteriaceae as active fermenters in earthworm gut content. ISME J. 2011;5:92–106. doi: 10.1038/ismej.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood S, Brewer E, Yarwood R, Lajtha K, Myrold D. Soil microbe active community composition and capability of responding to litter addition after 12 yeears of no inputs. Appl Environ Microbiol. 2013;79:1385–1392. doi: 10.1128/AEM.03181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser ER, Lindell D, Johnson ZI, Futschik ME, Steglich C, Coleman ML, et al. Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS ONE. 2009;4:e5135. doi: 10.1371/journal.pone.0005135. [DOI] [PMC free article] [PubMed] [Google Scholar]