Abstract
Arbuscular mycorrhizal fungi (AMF) form symbioses with most plant species. They are ecologically important determinants of plant growth and diversity. Considerable genetic variation occurs in AMF populations. Thus, plants are exposed to AMF of varying relatedness to each other. Very little is known about either the effects of coexisting AMF on plant growth or which factors influence intraspecific AMF coexistence within roots. No studies have addressed whether the genetics of coexisting AMF, and more specifically their relatedness, influences plant growth and AMF coexistence. Relatedness is expected to influence coexistence between individuals, and it has been suggested that decreasing ability of symbionts to coexist can have negative effects on the growth of the host. We tested the effect of a gradient of AMF genetic relatedness on the growth of two plant species. Increasing relatedness between AMFs lead to markedly greater plant growth (27% biomass increase with closely related compared to distantly related AMF). In one plant species, closely related AMF coexisted in fairly equal proportions but decreasing relatedness lead to a very strong disequilibrium between AMF in roots, indicating much stronger competition. Given the strength of the effects with such a shallow relatedness gradient and the fact that in the field plants are exposed to a steeper gradient, we consider that AMF relatedness can have a strong role in plant growth and the ability of AMF to coexist. We conclude that AMF relatedness is a driver of plant growth and that relatedness is also a strong driver of intraspecific coexistence of these ecologically important symbionts.
Keywords: Rhizophagus irregularis, intraspecific competition, coexistence, symbiosis, mycorrhiza
Introduction
Arbuscular mycorrhizal fungi (AMF) form mutualistic symbioses with 80% of the terrestrial plant species. Through this symbiosis, the fungal partner enhances plant nutrient acquisition, especially phosphate. In return, the plant provides carbohydrates to the fungus (Smith and Read, 2008). Numerous studies have pointed out that AMF have a considerable impact on plant growth and on plant ecology (van der Heijden and Sanders, 2002). AMF affect plant coexistence (Hart et al., 2003). In some cases, this has been shown to be the case by determining the outcome of plant competition (Scheublin et al., 2007). The presence of AMF and the diversity of AMF species, have been shown to be strong determinants of plant community structure, diversity and productivity (Grime et al., 1987; van der Heijden et al., 1998). The role of AMF species diversity is emphasized by the fact that phylogenetically distinct AMF species are also functionally different and their hyphae are spatially partitioned outside the root in the area where they exploit the soil volume (Maherali and Klironomos, 2007). This may improve the number of possible coexisting plant species and their development by enhancing the number of niches (Powell et al., 2009).
In natural ecosystems, individual plants are usually colonized by many different AMF species (Helgason et al., 2002; Opik et al., 2009). Despite this, almost nothing is known about the factors that determine coexistence and competition among AMF in plant roots. Furthermore, many experiments on AMF effects on plant growth are conducted with one plant and one AMF isolate. Only a few studies have explored the effect on plant growth of coexisting AMF species inside plant roots but no consensus has emerged from these studies. In some cases, dual colonization by two AMF species was beneficial for plant growth (Gustafson and Casper, 2006; Jansa et al., 2008) and could be explained by functional complementarity between the AMF species (Koide, 2000). But cases of plant growth depression due to dual fungal inoculation have also been reported (Violi et al., 2007; Janouskova et al., 2009).
Most research on the effects of different AMF on plant growth has focused on different AMF species. One aspect that has been largely overlooked is the effect on plant growth of several coexisting AMF from a population of the same species. This could be important ecologically because some AMF species such as Rhizophagus irregularis exhibit a high genetic diversity even at a small spatial scale (Koch et al., 2004; Croll et al., 2008; Börstler et al., 2010) and vary in their effects on plant growth (Munkvold et al., 2004; Koch et al., 2006). This means that several AMF of the same species, differing in their genetic relatedness and their effects on plant growth, have the opportunity to coexist in the same plant.
So far, explanations of how coexisting AMF influence plant growth have concentrated on explaining the combined growth effects of the fungi based on functional complementarity and functional differences. For example, the extraradical hyphae of different AMF species clearly exploit different parts of the soil compartment around roots, thus, they do not spatially exploit the same resources (Jakobsen et al., 1992; Smith et al., 2000). However, competition among different coexisting symbionts has also been proposed as a likely factor affecting host benefit, with the prediction that increasing competition among symbionts will decrease host benefit (Pearson et al., 1993, 1994; Douglas, 1998). However, competition among AMF has been largely ignored when trying to explain the effects of several AMF species on plant growth and, to our knowledge, the effects of coexistence and competition between AMF individuals of one AMF species have never been studied.
Given that genetic variation is high in AMF populations (Koch et al., 2004; Croll et al., 2008; Börstler et al., 2010), we assume that a plant root must encounter AMF individuals that differ in their relatedness. AMF are thought to be clonal haploid organisms and their genetics is poorly understood (Sanders and Croll, 2010). Hence, in this study, we define relatedness in a broad sense as the degree to which pairs of different AMF in a population share common alleles.
Genetic relatedness in some bacterial populations is known to strongly influence the outcome of competition, even if its influence may be reduced in case of very local competition (Griffin et al., 2004). AMF obtain their nutrition directly from the plant root. Thus, several coexisting AMF probably compete for the same space and resources inside the roots meaning that local competition could be high. Following a kin selection model (Hamilton, 1964), competition is expected to be weaker between closely related individuals than between more distantly related individuals. One scenario is that as two AMF compete more with each other, they have to invest more into competition and that this will reduce how much they can invest in providing resources to the host. If increased competition between symbionts leads to reduced host benefit, then in this scenario, closely related AMF would be expected to increase plant growth more than distantly related AMF. On the other hand, closely related AMF would likely be phenotypically more similar than distantly related AMF and, thus, would be competing for the same space and resources. Thus, in this opposite scenario, local competition could be higher between closely related AMF reducing their positive effect on plant growth. The effects of genetic relatedness, among AMF from a population, on either plant growth or coexistence among the fungi have also never been addressed.
There are three main reasons for why such experiments have not previously been possible. First, it is very labor intensive and difficult to obtain many single-spore isolates of one AMF species from the same field site where they naturally co-occur. Thus, very few appropriate collections exist for conducting such work. Second, it is almost impossible to reliably distinguish among different isolates of the same AMF species within roots using conventional microscopy and morphological characteristics. Third, until recently, no molecular markers were available that could distinguish among different AMF isolates of the same species, that could be used to measure relatedness among AMF isolates and that could be used to quantify the relative abundance of the different AMF isolates within roots.
Recent developments now make it possible to study the role of genetic relatedness in AMF on plant growth and its effects on competition between AMF within roots. First, a set of appropriate markers now exists for distinguishing between AMF individuals and for measuring their relatedness (Croll et al., 2008). Second, quantitative techniques employing such markers can be used to measure relative frequencies of isolate-specific alleles in samples containing AMF DNA (Angelard et al., 2010). We conducted a greenhouse experiment, in which we jointly inoculated plants with genetically different isolates of the AMF R. irregularis, following a gradient of relatedness. The fungi are known to naturally coexist at the same field site. We employed quantitative molecular techniques in one plant species to observe AMF competition. Because such studies have not previously been conducted, we tested the null hypotheses: (1) that relatedness between pairs of AMF does not influence their effect on plant growth and (2) that relatedness between pairs of AMF does not influence the outcome of competition between the AMF. We conclude that relatedness among AMF strongly influences AMF competition in roots and can have an important role in plant ecology through its influence on plant growth.
Materials and methods
Plant species, fungal isolates and inoculation treatments
We used two plant species: Allium porrum and Plantago lanceolata. Many studies have demonstrated positive growth effects of AMF on these two plant species (Kahiluoto and Vestberg, 1998; van der Heijden and Sanders, 2002; Scheublin et al., 2007). Germinated seeds were transplanted into vermiculite for 1 week. Seedlings were transplanted into pots (diameter 12 cm × height 7 cm for P. lanceolata and 32 cm × 10 cm for A. porrum) with autoclaved mixed loam with low P concentration (Orflor, Inc., Geneva, Switzerland; horticol number 1) and sand (ratio 1:1, v/v for P. lanceolata and ratio 2:3 for A. porrum). At the time of transplantation, we inoculated plants with 500 μl of water containing 300 spores of the AMF isolates directly on the roots. In dual inoculation treatments, spores of the two AMF isolates were mixed in equal quantities and then 300 spores of this mixture was applied to the plant roots. Controls were inoculated with 500 μl of sterilized water. The experiment was conducted in a greenhouse under controlled conditions (temperature 22–25 °C, humidity 40%–60% and natural lighting).
We used isolates of R. irregularis. These isolates were previously ascribed to the species Glomus intraradices and then to G. irregulare (Stockinger et al., 2009). Recently, G. irregulare was renamed as R. irregularis(Krüger et al., 2012). All isolates are single-spore cultures originating from one field located at Hausweid, Tänikon, Switzerland (Anken et al., 2004). All the AMF isolates used in this study were produced in an in vitro culture system containing root inducing T-DNA-transformed Daucus carota roots in a sterile medium (Bécard and Fortin, 1988). Genetic and phenotypic differences among the isolates have already been shown (Koch et al., 2004, 2006). The genetic distance among these isolates was already determined (Croll et al., 2008). We chose six isolates known as A1, A5, B2, B12, C2 and C5. Figure 1 shows the genetic distance between these isolates. We generated 16 different treatments: six single isolate treatments, three double isolate treatments between closely related isolates (A1+B2, A5+B12 and C2+C5), three double isolate treatments with a moderate genetic distance between the two isolates (B2+A5, A1+B12 and B2+B12) and three double isolate treatments with more distant isolates (A5+C5, B2+C2 and B12+C2). The relatedness between each pair of isolates was determined by the number of identical alleles at 13 loci. This was represented as a proportion, where 1 equals a different allele at all 13 loci and 0 represents identical alleles at all 13 loci. The loci are described in (Croll et al., 2008). There was one control treatment without AMF. Pairs of isolates within the classes ‘closely related' and ‘distant' each had an equal genetic distance. The three treatments within the ‘moderately distant' class displayed different measured genetic distances (see Figure 1 for distances between each pair). This also gave us a gradient of genetic relatedness among the pairs of isolates that could be later used in regression analysis. Each of the 16 treatments was replicated 12 times with each plant species, leading to a total of 392 experimental units.
Figure 1.
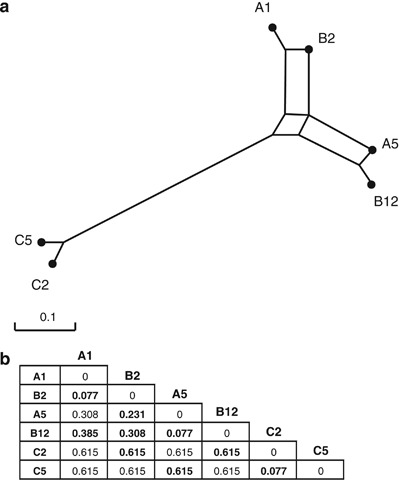
(a) Minimum spanning network of the six R. irregularis isolates based on allele size for 13 microsatellites loci; and (b) the related distance matrix (modified from Croll et al., 2008). Numbers in bold represent genetic distance for pairs of isolates used in this study.
Measurements of plant traits
The experiment was conducted for 16 weeks. Non-destructive plant growth measurements were made after 4, 8 and 12 weeks. To estimate plant growth at those time points, we made non-destructive measurements on plant height (cm), leaf thickness (mm) and chlorophyll content of 3 different leaves of each plant (using a Konica Minolta Inc., Tokyo, Japan, SPAD-501 chlorophyll meter). Leaf thickness was not measured in A. porrum as the device used to measure leaf thickness damaged the leaves of this plant. The same non-destructive measurements were also made at 16 weeks. In addition, after 16 weeks, we collected shoots and dried them at 60 °C for 48 h in order to measure the aboveground plant dry mass.
There were significant correlations between shoot dry weight at the final harvest and all non-destructive measurements (Supplementary Table S1). A composite variable called ‘phenotype' was calculated using values from each of the non-destructive measurements. To make the composite variable ‘phenotype' in each plant species, each of the variables from non-destructive measurements were divided by their highest value. This is known as a fixed weight composite variable (Grace and Bollen, 2008). Hence, all non-destructive variables ranged from 0–1, with the highest value of the variable equal to 1. All the modified variables were summed for each plant species and the mean of this new variable was called ‘phenotype'. The composite variable ‘phenotype' also showed a strong correlation with plant dry weight (Supplementary Table S1; Supplementary Figure S1). We used the composite variable ‘phenotype' for further statistical analysis for the time points 4, 8 and 12 weeks because it was the best predictor of plant dry mass (Supplementary Table S1; Supplementary Figure S1).
We then collected and washed part of the root system of each plant for molecular analyses. P. lanceolata roots were dried for two days at 60 °C. After drying and DNA extraction, we did not consider the DNA of P. lanceolata to be of sufficient quality for quantitative molecular tests. However, it was of sufficient quality to use in standard PCR to reveal that the inoculated roots were colonized by the expected AMF and that the uninoculated plants were not colonized (see Supplementary Methods). Hence, all the quantitative molecular analyses were only performed with DNA from A. porrum roots.
Measurement of total fungal colonization using quantitative PCR
We used quantitative PCR (q-PCR) to estimate the total colonization by AMF in the roots of A. porrum. This method has previously been used by (Gamper et al., 2008). However, because we used a different locus, namely the RAD15 gene, for amplification we also had to test that it was reliable and verify that the gene is in the same copy number per AMF isolate (Corradi et al., 2007). We obtained a strong correlation between the amount of AMF DNA used for q-PCR and how much DNA was amplified, showing that this was a reliable method (Supplementary Figure S2). See Supplementary Information for full details of the methods for testing the q-PCR.
To estimate the level of AMF colonization in A. porrum, roots were crushed using liquid nitrogen and total DNA was extracted using the DNeasy plant mini Kit (Qiagen, Inc., Gaithersburg, MD, USA). Total AMF colonization was assessed by measuring the quantity of AMF DNA in a given sample using q-PCR amplification of the RAD15 gene. No DNA was amplified from uninoculated roots using the same primers. q-PCRs were run on the DNA extracted from root samples, after standardization of the total DNA concentration among samples at 2 ng μ−1. The q-PCR mix contained: 0.8 ng of genomic DNA, 19.2 μl of 2X Power Sybr green (Applied Biosystems, Foster city, CA, USA), 3.8 μl of each of the two primers at concentration of 3 μM and 3.8 μl of water in a total volume of 31 μl. q-PCR reactions were carried out on a Prism AB7900 q-PCR machine (Applied Biosystems). Thermal cycling conditions included 2 min at 50 °C, 10 min at 95 °C, followed by 45 cycles of 15 s at 95 °C and 60 s at 60 °C, then one cycle of 15 s at 95 °C and 15 s at 60 °C. Ct values were calculated with the SDS2.4 software (Applied Biosystems).
The resulting Ct values from the q-PCR were used to estimate the amount of AMF inside the roots. The Ct values generated in q-PCR were transformed into 2−ΔCt, following Livak and Schmittgen (2001). Because q-PCR was made on DNA, there was reference gene. Thus, in the transformation, we set the highest observed Ct value as a reference. The highest Ct value corresponded to the sample with the lowest total fungal colonization (sample from treatment: A5+C5; replicate 12; where Ct=32.841). All Ct values in the dataset were then transformed as follows ΔCt=Ct (sample) −32.841.
Measurement of the relative proportion of different AMF isolates in roots
The presence and relative abundance of each R. irregularis isolate was measured using specific marker amplification (see Supplementary Information for an explanation of why we used this molecular approach). To discriminate between the six isolates, four different markers were used: Bg196, Bg273, Bg276 and Nrint (Croll et al., 2008). The same amplification protocol was used for all markers. The PCR mix contained 10 ng of genomic DNA, 1 μl of 10 × PCR buffer including 15 mM MgCl2 (QBiogene, Morgan Irvine, CA, USA), 0.19 mM dNTPs, 0.25 μM of each of the two primers and 0.15 U Taq DNA polymerase (Qbiogene) in a total volume of 10 μl. Thermal cycling conditions included 3 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at 53 °C and 45 s at 72 °C and a final elongation of 7 min at 72 °C. Forward primers were either hexachlorofluorescein phosphoramidite or fluorescein fluorescence-labelled to allow capillary fragment length visualization on the ABI-3100 Genetic Analyser (Applied Biosystems ). A mixture of 1 μl of PCR product, 0.3 μl of Hi-Di formamide was prepared for loading. Data from the ABI-3100 Genetic Analyser were manually scored using GENEMAPPER 4.0 (Applied Biosystems).
Relative abundance of the different AMF isolates in the dual inoculation treatments was calculated using relative peak heights of each replicate, following (Angelard et al., 2010). We made two separate DNA extractions of each replicate, and we made two independent PCRs on each DNA extraction, giving four values for each replicate. The four values were pooled for each replicate. We used the marker Nrint for measuring the relative frequency of alleles between the isolates A1 and B2 (272 bp and 270 bp, respectively); A5 and C5 (270 bp and 265 bp, respectively); B2 and C2 (270 bp and 265 bp, respectively) and B12 and C2 (270 bp and 265 bp, respectively). We used the marker Bg196 for measuring the relative frequency of alleles between the isolates A1 and B12 (301 bp and 312 bp, respectively); B2 and B12 (301 bp and 312 bp, respectively) and A5 and B12 (301 bp and 312 bp, respectively). We used the genetic marker Bg273 to distinguish between C2 and C5 frequency (248 bp and 247 bp, respectively), and to distinguish between B2 and A5 frequency, we used the marker Bg276 (307 bp and 278 bp, respectively).
From the data on the proportion of each AMF isolate we also calculated a variable called ‘frequency of the most abundant isolate.' This was calculated by looking at the proportion of colonization represented by the most abundant isolate in each plant within a treatment, irrespective of which isolate was the most abundant. This gave a value of how much the more dominant fungus colonized the roots compared to the less dominant fungus irrespective of which particular fungal isolate was most often the dominant fungus in replicate plants of the same treatment.
Statistical analyses
To test for differences in plant growth effects among the different AMF treatments, one-way analyses of variance were performed on the dry mass of both species at week 16 with the factor AMF treatment (with 16 levels). We performed a pairwise mean comparisons between all treatments using the Tukey–Kramer honest significant difference test.
We tested if the composite variable ‘phenotype' (at 4, 8 and 12 weeks), plant dry mass (at 16 weeks) and frequency of the most abundant isolate were explained by the relatedness between the coexisting fungal isolates, using a linear regression model. This was performed on a subset of the data, including only treatments with two fungal isolates. We also tested if the frequency of the most abundant isolate was correlated with leek dry mass, using a Pearson correlation test.
To investigate if there were differences in overall AMF colonization among AMF treatments, we performed a Kruskal–Wallis non-parametric test on the 2−ΔCt values. We did this because the 2−ΔCt values did not exhibit a normal distribution. Pairwise comparisons were tested with a pairwise Wilcoxon test with Bonferroni corrections.
Finally, we made statistical analyses on the three relatedness classes of the mycorrhizal treatments (closely related, moderately distant and distant). We tested if there were differences in the frequency of the most abundant isolate among the three relatedness classes. A one-way analysis of variance was performed with the factor relatedness class (three levels). Means comparisons were made by performing pairwise Tukey tests. All the statistical tests were computed in the statistical software R 2.7.2 (R Development Core Team, 2008).
Results
Effect of AMF relatedness on plant growth
Plant growth, as measured with the composite variable ‘phenotype', was significantly influenced by the genetic distance between the pairs of AMF isolates. This was true in both plant species at weeks 4 and 8. At week 12, the regression was not significant in A. porrum, but marginally significant in P. lanceolata (Figures 2a and b). At the final harvest, shoot dry mass was significantly affected by relatedness in both plant species (Figures 2a and b). In all cases, significant linear regressions between the genetic distance of the coexisting fungal isolates and plant growth were negative (Figures 2a and b). Thus, dual inoculation with closely related fungi provided greater benefits for the plant than inoculation with two more distantly related fungi.
Figure 2.
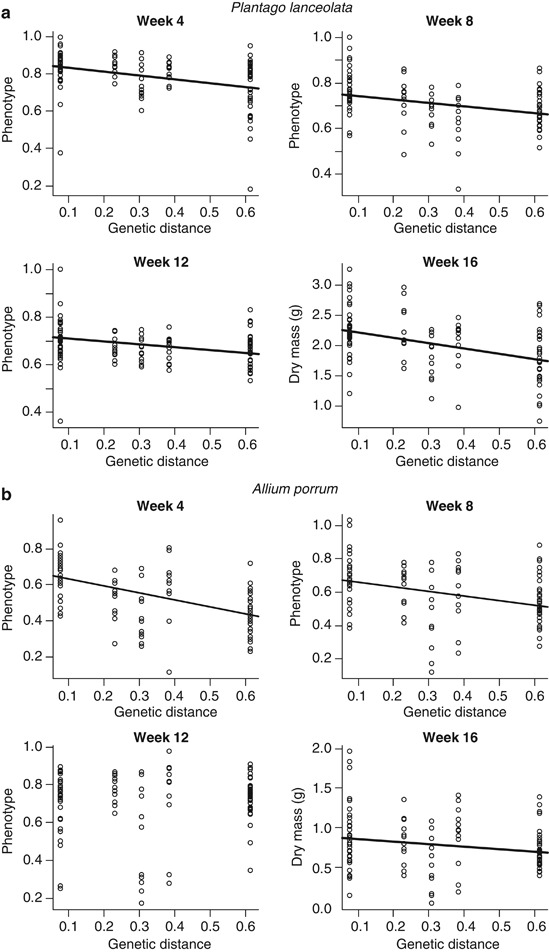
Relationship between (a) P. lanceolata and (b) A. porrum growth and the genetic distance between coexisting AMF. The line represents the linear regression (lm model). P. lanceolata, week 4, F1,102=17.41, R2=0.142; week 8, F1,102=11.57, R2=0.102; week 12, F1,102=3.72, R2=0.04, and week 16, F1,102=20.86, R2=0.17. A. porrum, week 4, F1,105=42.4, R2=0.288; week 8, F1,105=14.67, R2=0.124, and week 16, F1,105=4.227, R2=0.039. All regressions P⩽0.001, except P. lanceolata, week 12 (P=0.057) and A. porrum dry mass at week 16 (P=0.042).
The dry mass of P. lanceolata and A. porrum differed significantly among AMF treatments (F ratios of the analysis of variances, F15,168=4.76, P⩽0.001 and F15,175=3.98, P⩽0.001, respectively). Tukey–Kramer honest significant difference tests showed that most of the time double inoculation treatments did not provide a significantly larger growth than single inoculation treatments (Supplementary Figures S3a and S3b). In P. lanceolata, plants inoculated with B12 and C2 alone were significantly larger than plants inoculated with the dual inoculation treatment with B12+C2 (Supplementary Figure S3a). We observed the same trend in A. porrum, where the single treatments B2 and B12 provided the highest mean plant dry mass, whereas the double inoculation treatment B2+B12 provided the smallest plant dry mass (Supplementary Figure S3b). Thus, in several cases, being colonized by two fungi was not beneficial and sometimes even provided less benefit than not being colonized by mycorrhizal fungi. For example, P. lanceolata in the treatment B12+C2 had significantly lower dry mass than uninoculated plants.
Total AMF colonization
The amount of fungal DNA inside leek roots was measured in all treatments. There was no amplification of RAD15 in uninoculated plants. Colonization differed significantly among the different mycorrhizal treatments (Kruskal–Wallis λ2=51.3279, d.f.=14, P⩽0.001), meaning that the level of AMF colonization was dependent on the identity of the R. irregularis isolates. One single AMF treatment (C5) showed a significantly higher amount of fungal colonization than the treatments B2, A1, A5+B12, B2+A5 and A1+B12 (Supplementary Figure S4). Plants with the treatment B12+C2 showed the highest mean colonization but the variance within this treatment was very large and thus did not lead to significant differences as compared to the other treatments (Supplementary Figure S4). The variance in total fungal colonization (as measured by fungal DNA concentration) was much higher in some dual inoculation treatments than in single inoculation treatments, especially in treatments A5+C5 and B12+C2 (Supplementary Figure S4).
Effect of AMF relatedness on the intensity of competition between AMF isolates
We investigated fungal competition by measuring the frequency of each fungus inside the leek roots of dual inoculation treatments. From this data, we were then able to calculate the proportion of the most abundant fungus in each dual treatment as a measure of the strength of competition between fungal pairs. There was a strong significant effect of AMF relatedness on the strength of competition between the pairs. We observed that when the fungi were closely related, they were able to coexist in almost equal proportions (Figure 3). As the genetic distance increased between pairs, competition between the fungi increased. Between the distant pairs, one isolate almost always outcompeted the other isolate, but usually not completely (Figure 3). On average, one fungus dominated the other in a proportion of ∼95% to 5% (median value; mean value 90% to 10%). However, among replicates of the same treatment, it was not always the same fungal isolate of a given pair that outcompeted the other. This effect was highly significant if the data were grouped into relatedness classes and tested with an analysis of variance (Figure 3) or analyzed as a linear regression without grouping the treatments into three different relatedness classes (Supplementary Figure S5).
Figure 3.
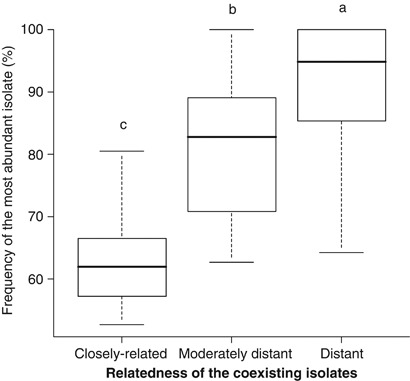
Box plot showing the strength of fungal competition, as measured by the frequency of the most abundant fungal isolate, between pairs of closely related, moderately distant and distant fungal isolates in A. porrum roots. Horizontal lines within the boxes represent the median. Bars represent the range of variation including 90% of the values. The F ratio for the analysis of variance (ANOVA) on frequency of the most abundant isolate in the three relatedness classes was F(2,101)=60.32; P⩽0.001. Different letters above boxes show significant differences according to Tukey pairwise tests (P⩽0.0036).
The frequency of the most abundant isolate was also correlated with leek shoot dry mass at the final harvest (Figure 4). Although this effect was not very strong, it indicates that increasing fungal competition potentially influences the benefit of the fungi on plant growth.
Figure 4.
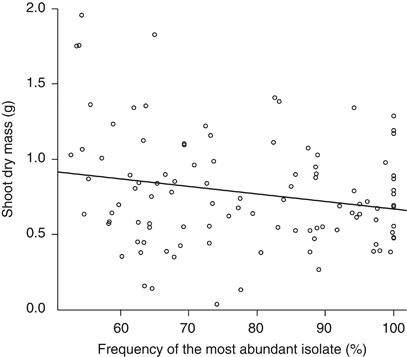
Correlation between strength of fungal competition, as measured by frequency of the most abundant isolate, and the shoot dry mass of A. porrum. Correlation coefficient=−0.209 (P=0.033).
Discussion
The results clearly allowed us to reject the two null hypotheses as relatedness between AMF significantly influenced the symbiotic effects of AMF on the growth of both plant species and strongly influenced the outcome of competition between the fungi in A. porrum.
Effects of AMF relatedness on plant growth
The effect of the fungi on plant growth, in both plant species, was greater when the AMF were more closely related. This is congruent with some reports showing that dual colonization by different AMF species results in reduced plant growth (Violi et al., 2007; Janouskova et al., 2009). However, this is the first study that has specifically tested the effect of a gradient of genetic relatedness in AMF on their symbiotic effect.
The magnitude of the effect of AMF relatedness plant growth was similar in both plant species. The range of genetic relatedness caused a 27.34% and 27.67% increase in biomass from the lowest to the highest values of dry weight in P. lanceolata and A. porrum, respectively. We view this as a surprisingly large change in plant growth given that the plants were only offered pairs of AMF of the same species with a relatively shallow gradient of relatedness. Furthermore, the magnitude of growth difference was very similar to that found in another study where Plantago was inoculated with communities of different AMF families or different species of the same AMF genera (Maherali and Klironomos, 2007). The fungi were chosen out of a relatively small pool of available isolates, all of which coexist at the same field site (Croll et al., 2008). This made the testable gradient of genetic relatedness relatively shallow. However, subsequent work at the same field site (Börstler et al., 2010), that was not restricted to isolates already in culture, revealed much higher levels of genetic variation in this AMF species than that observed by (Croll et al., 2008). Thus, the gradient of AMF relatedness of this AMF species would be much steeper in the field than that tested in this study. Furthermore, the plants were only offered two different AMF. At the field site, a large number of genetically different R. irregularis individuals could co-colonize the plants. With the markers available at present, it would not be possible to scale up the study to investigate the co-colonization of more than two of those AMF to measure how they coexist in the roots.
Communities of AMF, composed of different AMF families, have previously been shown to positively influence Plantago growth compared to AMF of the same genera (Maherali and Klironomos, 2007). Our results on plant growth are of a similar magnitude to that study but in an opposite direction and within AMF species. This could indicate that when plants are colonized by different AMF families, their improved growth could potentially be offset by genetic diversity in the population of individual AMF species.
An alternative explanation of the effects could be that plants inoculated with distantly related AMF were not well colonized and this resulted in low plant growth. However, dual inoculated A. porrum were well colonized (Supplementary Figure S4), thus ruling out this possibility in one plant species and we know that Plantago was colonized by all the fungal isolates used in this study. Furthermore, there was no correlation between the total colonization levels in A. porrum and plant dry mass or the genetic distance of the coexisting fungi (data not shown). Thus, total colonization levels do not explain the effect of AMF relatedness on plant growth in A. porrum.
Genetic relatedness influences fungal coexistence
The effect of relatedness on coexistence of the fungal pairs in A. porrum was highly significant and large. The strength of competition was high between distantly related AMF, with one isolate almost completely outcompeting the other. By inoculating A. porrum plants singly with each AMF, we were able to show that all of the fungi were readily able to colonize the roots. Therefore, the poor colonization by some fungi in the distantly related treatments could not have been due to inability to colonize the roots, but must have been due to the genetic identity of the other fungal partner. One possibility is that one of the AMF managed to colonize the roots first and then prevent the other from colonizing. There is some evidence for this among replicate pairs of distantly related AMF. Although one fungus almost always dominated the other in a mean proportion of 90% to 10%, it was not always the same isolate of the pair that dominated. The other possibility is that both isolates colonized the roots but then competition became intense, rather than initial prevention. However, this seems less likely from our data as one AMF would be expected to consistently outcompete the other, which was not the case.
Direct versus plant mediation of fungal competition
A previous study has shown that competition between AMF can be plant mediated (Pearson et al., 1993). Plants are able to sanction less beneficial AMF and allocate resources to a more beneficial fungus (Bever et al., 2009; Kiers et al., 2011). The results of this study cannot easily be explained by a plant-mediated effect. First, in such a scenario, the plant should always sanction the same fungus, which was not the case. Second, it should sanction the fungus that is least beneficial. This was not the case in these experiments. For example, the fungus A5 provided significantly less benefit to A. porrum than B12 (Supplementary Figure S3) but these two closely related fungi coexisted together in fairly equal proportions, with A5 sometimes occupying more of the roots than B12. The other possibility is that direct competition between pairs of fungi occurred. While there is no obvious evidence supporting plant-mediated competition or direct competition, it is impossible to say from these experiments which occurred. Furthermore, they would not have to be mutually exclusive.
Fusion of isolates and genetic competition
The above interpretations consider direct competition between pairs of fungi. Another scenario involving genetic compatibility is conceivable. Fusion between some of these AMF isolates has previously been shown, resulting in AMF that subsequently contain DNA from both parents (Croll et al., 2009). Thus, the apparent proportion of each fungal isolate in the roots, as measured by DNA amplification, could represent the proportion of compatible genetic material from the two fungi that coexist following fusion. In this case, we still consider the proportions observed to represent the degree of competition or compatibility between the fungi but at the genetic level rather than the individual level. We cannot say from this data whether our results represent direct competition between individual fungi or competition between genetically incompatible nuclei inside fused AMF.
Why does AMF relatedness influence plant growth?
We suggest two potential mechanisms why increased relatedness between AMF should promote more plant growth than distantly related AMF. First, distantly related fungi may have to invest more into competition than closely related pairs. To do this, we assume that the fungus would have to either produce molecules that are inhibitory to the other fungus, but these would have to be regulated by highly specific recognition system. Alternatively, the fungi could alter their efficiency of taking up carbohydrates from the host in the presence of a competitor. This could result in greater carbohydrate requirements from the plant, thus reducing the amount of carbohydrates available for plant growth. Costs of fungal competition could also result in less investment by the fungus into external hyphae, resulting in the fungus exploiting a smaller soil volume and supplying fewer nutrients to the plant. While we cannot say if the strength of competition between AMF directly influenced plant growth, these two variables were weakly correlated in leek (Figure 4). Second, if fusion between AMF occurred, the fused AMF may have had a lower symbiotic efficiency than the parents. It is known that fusion between AMF can result in AMF that invest differently into production of hyphae outside the roots (Croll et al., 2009). Moreover, previous studies show that AMF fusion can result in fungi with reduced growth effects on plants compared to the parents (Angelard et al., 2010). However, it is currently unknown whether the symbiotic effect of the AMF resulting from fusion is influenced by the relatedness of the two parents.
Conclusions
We conclude that genetic relatedness between AMF is a strong determinant of plant growth. Relatedness among AMF in populations is potentially a powerful driver of plant growth in plant communities, given the magnitude of the effect on plant growth with such a narrow range of relatedness, and that the range of genetic relatedness in AMF populations is much greater than that tested in this study. Furthermore, while it is known that plants are normally colonized by many different AMF almost nothing is known about the factors that determine AMF coexistence and competition. Our study points to a strong role of AMF genetics and relatedness in determining AMF intraspecific coexistence inside plant roots. Until now, the role of intraspecific AMF relatedness for both these important aspects of ecology has been overlooked.
Acknowledgments
We thank the Swiss National Science Foundation with a grant to IRS (310030B_144079/1). We also thank Daniel Croll, Jeremy Bonvin and Nicolas Salamin for their help in the choice of genetic markers, lab work and statistical analyses, respectively.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Angelard C, Colard A, Niculita-Hirzel H, Croll D, Sanders IR. Segregation in a mycorrhizal fungus alters rice growth and symbiosis-specific gene transcription. Curr Biol. 2010;20:1216–1221. doi: 10.1016/j.cub.2010.05.031. [DOI] [PubMed] [Google Scholar]
- Anken T, Weisskopf P, Zihlmann U, Forrer H, Jansa J, Perhacova K. Long-term tillage system effects under moist cool conditions in Switzerland. Soil Till Res. 2004;78:171–183. [Google Scholar]
- Bécard G, Fortin JA. Early events of vesicular arbuscular mycorrhiza formation on Ri T-DNA trnasformed roots. New Phytol. 1988;108:211–218. doi: 10.1111/j.1469-8137.1988.tb03698.x. [DOI] [PubMed] [Google Scholar]
- Bever JD, Richardson SC, Lawrence BM, Holmes J, Watson M. Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecol Lett. 2009;12:13–21. doi: 10.1111/j.1461-0248.2008.01254.x. [DOI] [PubMed] [Google Scholar]
- Börstler B, Thiery O, Sykorova Z, Berner A, Redecker D. Diversity of mitochondrial large subunit rDNA haplotypes of Glomus intraradices in two agricultural field experiments and two semi-natural grasslands. Mol Ecol. 2010;19:1497–1511. doi: 10.1111/j.1365-294X.2010.04590.x. [DOI] [PubMed] [Google Scholar]
- Corradi N, Croll D, Colard A, Kuhn G, Ehinger M, Sanders IR. Gene copy number polymorphisms in an arbuscular mycorrhizal fungal population. Appl Environ Microbiol. 2007;73:366–369. doi: 10.1128/AEM.01574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, Giovannetti M, Koch AM, Sbrana C, Ehinger M, Lammers PJ, et al. Nonself vegetative fusion and genetic exchange in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 2009;181:924–937. doi: 10.1111/j.1469-8137.2008.02726.x. [DOI] [PubMed] [Google Scholar]
- Croll D, Wille L, Gamper HA, Mathimaran N, Lammers PJ, Corradi N, et al. Genetic diversity and host plant preferences revealed by simple sequence repeat and mitochondrial markers in a population of the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 2008;178:672–687. doi: 10.1111/j.1469-8137.2008.02381.x. [DOI] [PubMed] [Google Scholar]
- Douglas AE. Host benefit and the evolution of specialization in symbiosis. Heredity. 1998;81:599–603. [Google Scholar]
- Gamper HA, Young JPW, Jones DL, Hodge A. Real-time PCR and microscopy: are the two methods measuring the same unit of arbuscular mycorrhizal fungal abundance. Fungal Genet Biol. 2008;45:581–596. doi: 10.1016/j.fgb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Grace JB, Bollen KA. Representing general theoretical concepts in structural equation models: the role of composite variables. Environ Ecol Stat. 2008;15:191–213. [Google Scholar]
- Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- Grime JP, Mackey JML, Hillier SH, Read DJ. Floristic diversity in a model system using experimental microcosms. Nature. 1987;328:420–422. [Google Scholar]
- Gustafson DJ, Casper BB. Differential host plant performance as a function of soil arbuscular mycorrhizal fungal communities: experimentally manipulating co-occurring Glomus species. Plant Ecol. 2006;183:257–263. [Google Scholar]
- Hamilton WD. Genetical evolution of social behaviour I. J Theor Biol. 1964;7:1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- Hart MM, Reader RJ, Klironomos JN. Plant coexistence mediated by arbuscular mycorrhizal fungi. Trends Ecol Evol. 2003;18:418–423. [Google Scholar]
- Helgason T, Merryweather JW, Denison J, Wilson P, Young JPW, Fitter AH. Selectivity and functional diversity in arbuscular mycorrhizas of co-occurring fungi and plants from a temperate deciduous woodland. J Ecol. 2002;90:371–384. [Google Scholar]
- Jakobsen I, Abbott LK, Robson AD. External hyphae of vesicular-arbuscular mycorrhizal fungi associated with trifolium-subterraneum l.1. Spread of hyphae and phosphorus inflow into roots. New Phytol. 1992;120:371–380. [Google Scholar]
- Janouskova M, Seddas P, Mrnka L, van Tuinen D, Dvorackova A, Tollot M, et al. Development and activity of Glomus intraradices as affected by co-existence with Glomus claroideum in one root system. Mycorrhiza. 2009;19:393–402. doi: 10.1007/s00572-009-0243-4. [DOI] [PubMed] [Google Scholar]
- Jansa J, Smith FA, Smith SE. Are there benefits of simultaneous root colonization by different arbuscular mycorrhizal fungi. New Phytol. 2008;177:779–789. doi: 10.1111/j.1469-8137.2007.02294.x. [DOI] [PubMed] [Google Scholar]
- Kahiluoto H, Vestberg M. The effect of arbuscular mycorrhiza on biomass production and phosphorus uptake from sparingly soluble sources by leek (Allium porrum L.) in Finnish field soils. Biol Agric Hortic. 1998;16:65–85. [Google Scholar]
- Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science. 2011;333:880–882. doi: 10.1126/science.1208473. [DOI] [PubMed] [Google Scholar]
- Koch AM, Croll D, Sanders IR. Genetic variability in a population of arbuscular mycorrhizal fungi causes variation in plant growth. Ecol Lett. 2006;9:103–110. doi: 10.1111/j.1461-0248.2005.00853.x. [DOI] [PubMed] [Google Scholar]
- Koch AM, Kuhn G, Fontanillas P, Fumagalli L, Goudet J, Sanders IR. High genetic variability and low local diversity in a population of arbuscular mycorrhizal fungi. Proc Natl Acad Sci USA. 2004;101:2369–2374. doi: 10.1073/pnas.0306441101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide RT. Functional complementarity in the arbuscular mycorrhizal symbiosis. New Phytol. 2000;147:233–235. [Google Scholar]
- Krüger M, Krüger C, Walker C, Stockinger H, Schussler A. Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol. 2012;193:970–984. doi: 10.1111/j.1469-8137.2011.03962.x. [DOI] [PubMed] [Google Scholar]
- Livak KL, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maherali H, Klironomos JN. Influence of Phylogeny on fungal community assembly and ecosystem functioning. Science. 2007;316:1746–1748. doi: 10.1126/science.1143082. [DOI] [PubMed] [Google Scholar]
- Munkvold L, Kjoller R, Vestberg M, Rosendahl S, Jakobsen I. High functional diversity within species of arbuscular mycorrhizal fungi. New Phytol. 2004;164:357–364. doi: 10.1111/j.1469-8137.2004.01169.x. [DOI] [PubMed] [Google Scholar]
- Opik M, Metsis M, Daniell TJ, Zobel M, Moora M. Large-scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol. 2009;184:424–437. doi: 10.1111/j.1469-8137.2009.02920.x. [DOI] [PubMed] [Google Scholar]
- Pearson JN, Abbott LK, Jasper DA. Mediation of competition between 2 colonizing Va mycorrhizal fungi by the host plant. New Phytol. 1993;123:93–98. doi: 10.1111/j.1469-8137.1994.tb04263.x. [DOI] [PubMed] [Google Scholar]
- Pearson JN, Abbott LK, Jasper DA. Phosphorus, soluble carbohydrates and the competition between 2 arbuscular mycorrhizal fungi colonizing subterranean clover. New Phytol. 1994;127:101–106. doi: 10.1111/j.1469-8137.1994.tb04263.x. [DOI] [PubMed] [Google Scholar]
- Powell JR, Parrent JL, Hart MM, Klironomos JN, Rillig MC, Maherali H. Phylogenetic trait conservatism and the evolution of functional trade-offs in arbuscular mycorrhizal fungi. Proc Biol Sci. 2009;276:4237–4245. doi: 10.1098/rspb.2009.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2008R: A language and environment for statistical computing R Foundation for Statistical Computing: Vienna, Austria; ISBN 3-900051-07-0, Available at http://www.Rproject.org . [Google Scholar]
- Sanders IR, Croll C. Arbuscular mycorrhiza: the challenge to understand the genetics of the fungal partner. Annu Rev Genet. 2010;44:271–292. doi: 10.1146/annurev-genet-102108-134239. [DOI] [PubMed] [Google Scholar]
- Scheublin TR, Van Logtestijn RSP, Van der Heijden MGA. Presence and identity of arbuscular mycorrhizal fungi influence competitive interactions between plant species. J Ecol. 2007;95:631–638. [Google Scholar]
- Smith FA, Jakobsen I, Smith SE. Spatial differences in acquisition of soil phosphate between two arbuscular mycorrhizal fungi in symbiosis with Medicago truncatula. New Phytol. 2000;147:357–366. [Google Scholar]
- Smith SE, Read DJ.2008Mycorrhizal Symbiosis3rd ednAcademic Press: San Diego, CA, USA; p787 [Google Scholar]
- Stockinger H, Walker C, Schussler A. 'Glomus intraradices DAOM197198', a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol. 2009;183:1176–1187. doi: 10.1111/j.1469-8137.2009.02874.x. [DOI] [PubMed] [Google Scholar]
- van der Heijden MGA, Boller T, Wiemken A, Sanders IR. Different arbuscular mycorrhizal fungal species are potential determinants of plant community structure. Ecology. 1998;79:2082–2091. [Google Scholar]
- van der Heijden MGA, Sanders IR. Mycorrhizal Ecology. Springer: Berlin, Germany; 2002. [Google Scholar]
- Violi HA, Treseder KK, Menge JA, Wright SF, Lovatt CJ. Density dependence and interspecific interactions between arbuscular mycorrhizal fungi mediated plant growth, glomalin production, and sporulation. Can J Bot-Rev Can Bot. 2007;85:63–75. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


