Abstract
Nitrogen (N) is an essential nutrient in the sea and its distribution is controlled by microorganisms. Within the N cycle, nitrite (NO2−) has a central role because its intermediate redox state allows both oxidation and reduction, and so it may be used by several coupled and/or competing microbial processes. In the upper water column and oxygen minimum zone (OMZ) of the eastern tropical North Pacific Ocean (ETNP), we investigated aerobic NO2− oxidation, and its relationship to ammonia (NH3) oxidation, using rate measurements, quantification of NO2−-oxidizing bacteria via quantitative PCR (QPCR), and pyrosequencing. 15NO2− oxidation rates typically exhibited two subsurface maxima at six stations sampled: one located below the euphotic zone and beneath NH3 oxidation rate maxima, and another within the OMZ. 15NO2− oxidation rates were highest where dissolved oxygen concentrations were <5 μM, where NO2− accumulated, and when nitrate (NO3−) reductase genes were expressed; they are likely sustained by NO3− reduction at these depths. QPCR and pyrosequencing data were strongly correlated (r2=0.79), and indicated that Nitrospina bacteria numbered up to 9.25% of bacterial communities. Different Nitrospina groups were distributed across different depth ranges, suggesting significant ecological diversity within Nitrospina as a whole. Across the data set, 15NO2− oxidation rates were decoupled from 15NH4+ oxidation rates, but correlated with Nitrospina (r2=0.246, P<0.05) and NO2− concentrations (r2=0.276, P<0.05). Our findings suggest that Nitrospina have a quantitatively important role in NO2− oxidation and N cycling in the ETNP, and provide new insight into their ecology and interactions with other N-cycling processes in this biogeochemically important region of the ocean.
Keywords: nitrification, nitrite oxidation, nitrospina, oxygen minimum zone, pyrosequencing
Introduction
Nitrogen (N) is a required nutrient for all organisms and limits primary production across large areas of the ocean (Mills et al., 2004). Although N is present in many different chemical forms in the sea, dissolved nitrite (NO2−) alone may be used by microorganisms either as a reductant (under aerobic conditions) or oxidant (under anaerobic conditions) owing to its intermediate redox state. In the aerobic ocean, NO2− is oxidized to nitrate (NO3−) by NO2−-oxidizing bacteria (NOB); this constitutes the second step of nitrification and is expected to be tightly coupled to the first step, oxidation of ammonia (NH3) by NH3-oxidizing bacteria and NH3-oxidizing Archaea. No single organism is known to carry out both steps of nitrification. Costa et al. (2006) argue that this reflects optimization of pathway length to maximize energy production, and that because NO2− oxidation is energetically less favorable, it must occur more rapidly than NH3 oxidation and consume NO2− nearly instantaneously. However the coupling between these groups is fundamentally unstable: Graham et al. (2007) demonstrated that perturbation of NH3 oxidation in bioreactors lead to chaotic behavior among NOB, and evidence for such temporal decoupling in the ocean was observed off southern California (Beman et al., 2010). NH3 and NO2− oxidation can also be decoupled spatially; light is thought to directly or indirectly inhibit NH3-oxidizing Archaea, NH3-oxidizing bacteria and NOB to varying degrees, with NOB being more sensitive (reviewed by Lomas and Lipschultz (2006). This may contribute to the accumulation of NO2− at the base of the euphotic zone—a widespread feature known as the primary NO2− maximum (PNM). Although prevailing evidence indicates that PNM primarily results from incomplete phytoplankton reduction of NO3− (Lomas and Lipschultz, 2006, Beman et al., 2012), this remains uncertain because direct quantitative comparisons between NH3 and NO2− oxidation, and the microorganisms involved, are rare (Ward et al., 1989a, Lipschulz et al., 1990).
Of particular relevance is the recent discovery that marine Thaumarchaeota (formerly marine group 1 Crenarchaeota) oxidize NH3 (reviewed by Francis et al. (2007); these organisms number up to 1028 cells in the ocean (Karner et al., 2001) and even modest per cell rates of archaeal NH3 oxidation imply substantial global N turnover. In fact, recent studies indicate that NH3 oxidation occurs more rapidly and over a broader depth range than previously appreciated (Yool et al., 2007, Beman et al., 2008, Lam et al., 2009, Santoro et al., 2010, Beman et al., 2012). A NO2− rarely accumulates in the ocean (discussed above), this necessitates a corresponding sink for NO2−. Mincer et al. (2007) proposed that correlation between NH3-oxidizing Archaea and relatives of known NO2−-oxidizing Nitrospina bacteria is evidence of metabolic coupling; these groups showed similar distributions in both Monterey Bay and the North Pacific Subtropical Gyre. More recently, Füssel et al. (2012) reported that Nitrospina numbered up to 5.4% of microbial cells and were detected where NO2− was oxidized in the oxygen minimum zone (OMZ) along the Namibian coast. This included depths were dissolved oxygen (DO) concentrations were extremely low, indicating that NO2− oxidation persists even under low DO. In fact, NO2−-based metabolism is central in the world's OMZs (Francis et al., 2007; Wright et al., 2012), where DO concentrations fall below 20 μM because of microbial respiration (Paulmier and Ruiz-Pino, 2009). The accumulation of NO2− within OMZs is a diagnostic of anaerobic N and sulfur cycling (Ulloa et al., 2012), and among the processes that use NO2− as an oxidant under these conditions, anammox and denitrification lead to gaseous N loss from the ocean (Lam and Kuypers, 2011). However, the source of NO2− that fuels these processes is poorly constrained. NH3 oxidation may contribute ca. 50% of NO2− supply to anammox in the Black Sea (Lam et al., 2007), whereas NO3− reduction coupled to anammox was most important (at least 67% of NO2− demand) in the eastern tropical South Pacific (ETSP) OMZ (Lam et al., 2009), and Ward et al. (2009) demonstrated that conventional denitrification was dominant in the Arabian Sea.
The eastern tropical North Pacific Ocean (ETNP) is the largest oceanic OMZ (Paulmier and Ruiz-Pino, 2009), and so has an important role in oceanic—and global—N cycling. Annually, ∼26 Tg of dissolved N is converted to gaseous forms in the ETNP (DeVries et al., 2012) via microbial processes that require oxidized N. Our work and that of others has also identified relatively high rates of NH3 oxidation in the ETNP (Ward and Zafiriou, 1988, Sutka et al., 2004, Beman et al., 2008, 2012); yet NO2− oxidation has not been previously measured in this region. As NO2− oxidation can persist under <1 μM DO, NOB may directly compete with other organisms for NO2− in OMZs (Lipschultz et al., 1990, Füssel et al., 2012); in the ETNP OMZ, the accumulation of NO2− and its isotopic composition suggest that NO3− may be continually reduced and reoxidized through dynamic N and NO2− cycling (Casciotti and McIlvin, 2007). Adding to this complexity, OMZs are expanding as a consequence of climate change (Stramma et al., 2008, Keeling et al., 2010), potentially altering the rates and distribution of N transformations in the water column (Gilly et al., 2013). In the ETNP, N loss rates have varied several-fold over the past few decades in response to variations in the DO (Deutsch et al., 2011). Placing quantitative bounds on microbially driven biogeochemical processes occurring in OMZs—and specifically the ETNP—is therefore essential.
We quantified NO2− oxidation rates and NOB distributions across an oceanographic transect through northern portions of the ETNP and the southern Gulf of California (GOC). Using an in situ floating array and stable isotope tracer, 15NO2− oxidation rates were measured at high resolution with depth; NOB were quantified using real-time quantitative PCR (QPCR) and next-generation pyrosequencing of bacterial communities in corresponding DNA samples. This parallels our work on NH3 oxidation for the same stations and depths (Beman et al., 2012). Dual study of these processes—which presumably are biogeochemically coupled, and together constitute nitrification—are rare in the literature. We compared 15NO2− oxidation rate profiles with 15N-labeled ammonium (15NH4+) oxidation rates, depth profiles of NOB and key environmental factors such as NO2− concentrations and DO concentrations.
Materials and methods
Sampling and biogeochemical measurements
Samples were collected in July and August of 2008 aboard the R/V New Horizon; seven stations were occupied for several days each. A Seabird Electronics (Bellevue, WA, USA) SBE 9 conductivity-temperature-depth sensor package equipped with additional sensors was used to measure conductivity, temperature, depth, DO concentrations, chlorophyll fluorescence, light transmission and photosynthetically active radiation. Water samples were collected using 10 l polyvinyl chloride bottles deployed on the conductivity-temperature-depth rosette; samples for nutrient measurements and Winkler titrations were typically collected on the first cast and analyzed within hours of collection to characterize the chemical structure of the water column. We selected depths for rate measurements and collection of DNA samples based primarily on nutrient concentrations: 1–3 depths above the primary NO2− and NH4+ maxima were sampled, followed by 5–7 depths spaced every 5 m and several additional depths spaced every 10–20 m. 15NO2− oxidation rates were measured at each depth on a free-floating array (see below), and DNA samples were collected at the same depths (see below). Water samples for rate measurements and DNA samples were collected on the same cast at all stations.
Details of biogeochemical measurements are provided in Beman et al. (2012). Briefly, oxygen concentrations were measured using a SBE oxygen sensor and corrected on the basis of Winkler titrations (r2=0.997 for 187 samples for the cruise). Winkler-corrected SBE oxygen data are shown for the casts where rate and DNA samples were collected. NH4+ concentrations were measured using the fluorometric method of Holmes et al. (1999), and NO2− and NO3− concentrations were measured using standard colorimetric techniques (Strickland and Parsons, 1972); NO3− was reduced to NO2− with cadmium for measurement. r2 for standard curves were: 0.996–0.999 (oxygen measurements), 0.959–0.999 (NH4+ measurements) and 0.993–0.999 (NO2− and NO3− measurements).
In situ free-floating array
We deployed a free-floating array to mimic in situ conditions for 15NO2− oxidation rate measurements (Beman et al., 2012). 15NO2− oxidation was measured via addition of 15N-labeled NO2− to sample bottles suspended at multiple depths in the water column; water samples were collected into 250 ml polycarbonate bottles in the dark using silicone tubing, samples were overfilled by three volumes to avoid oxygen contamination, chilled 15N-labeled NaNO2 was added and bottles were sealed. Bottles were attached to the array immediately before deployment at dawn, and the array was deployed for ca. 24 h under in situ temperature and light conditions. Our deepest array-based incubations were placed at 160 m depth, and samples collected from depths >160 m were incubated onboard the ship at 12 °C in the dark within temperature-controlled incubators; this includes 18 samples reported here. Following array recovery, samples were filtered through 0.2 μm syringe filters, collected in 50 ml high-density polyethylene bottles, residual 15NO2− was removed (see below) and samples frozen at −20 °C until analysis at the University of Hawaii.
15NO2− oxidation rate measurements
15NO2− oxidation rates were measured by adding 98 atom percent (atom%) 15NO2− to a final concentration of 48–192 nmol l−1 (depending on in situ NO2− concentrations) and measuring the accumulation of 15N label in the NO3− pool after incubation for ∼24 h; excess 15NO2− was removed using methods modified from Granger and Sigman (2009). Upon recovery of samples from the in situ array, sulfamic acid (∼8–10 μl ml−1) was added, the samples were shaken and allowed to sit for >5 min, and then samples were neutralized by adding NaOH (4 M, ∼11 μl ml−1) before storing frozen (−20 °C) until analysis. The efficiency of 15NO2− removal using this method was tested in the laboratory and our results indicated a minor amount of 15NO3− was present after the sulfamic acid treatment, most likely owing to a small quantity of 15NO3− contamination in K15NO2 (see Olson, 1981; Ward, 1987; Lipschultz et al., 1990). However, the amount of 15N label remaining was not significantly higher at concentrations of 15NO2− <200 nmol l−1, and so does affect our rate calculations.
The δ15N value of N2O produced from NO3− using the denitrifier method (Sigman et al., 2001) was measured using methods described in Popp et al. (1995) and Dore et al. (1998). Briefly, N2O produced from NO3− was transferred from a reaction vial, cryofocused, separated from other gases using a 0.32-mm inner diameter 25 m × 5 μm CP-PoraBOND Q (Agilent Technologies, Santa Clara, CA, USA) capillary column at room temperature and introduced into the ion source of a MAT252 mass spectrometer through a modified GC-C I interface (Thermo Scientific, Bremen, Germany). Isotopic reference materials (United States Geological Survey-32, National Institute of Standards and Technology-3 and University of Hawaii NaNO3) bracketed every nine samples and δ15N values measured online were linearly correlated (r2=0.979) with accepted reference material δ15N values. Accuracy and precision were further evaluated by multiple analyses of a sodium NO3− solution for which the δ15N value of the solid NaNO3 (that is, University of Hawaii NaNO3) had been previously determined using an online carbon-N analyzer coupled with an isotope ratio mass spectrometer (ConFlo II Delta-Plus; Thermo Scientific), and were found to be <±0.5‰ (±0.00018 atom% 15N) for samples containing >2.5 nmol of NO3−.
Initial atom% enrichment of the substrate at the beginning of the experiment (noNO2−, see Equation (1)) was calculated by isotope mass balance based on NO2− concentrations, assuming that the 15N activity of unlabeled NO2− was 0.3663 atom% 15N. Rates of 15NO2− oxidation (15Rox) were calculated using Equation (1) from Ward et al. (1989b):
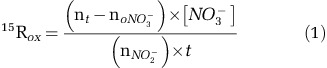 |
where nt is the atom% 15N in NO3− measured at time t, noNO3− is the measured atom% 15N of unlabeled NO3−, [NO3−] is the concentration of the NO3− pool and nNO2− is the exponential average atom% of NO2− over time t. nNO2− was calculated based on ambient NO2− concentrations and atom% (assumed to be 0.3663), added 15N-labeled NO2−, and measured 15NH4+ oxidation rates (producing unlabeled NO2−) reported by Beman et al. (2012).
DNA and RNA extraction
At each depth, 4 l of seawater was collected from the conductivity-temperature-depth rosette and divided into two sets of 2-l samples, which were then filtered through separate 25 mm diameter 0.2 μm Suppor filters (Pall Corporation, Port Washington, NY, USA) using a peristaltic pump. Filters were flash-frozen in liquid N and stored at −80 °C until DNA and RNA extraction, with one set of filters dedicated to each. Details of DNA extraction were reported in Beman et al. (2012). Filters for RNA extraction were frozen in RLT buffer with beta-mercaptoethanol added (Qiagen, Valencia, CA, USA), and RNA was extracted using the Qiagen RNeasy kit following Church et al. (2005) but with the following modifications: tubes containing filters, glass beads and buffer were first agitated for 80 s on a FastPrep machine (MP Biomedicals, Solon, OH, USA) at setting 5.5; each sample and an equal volume of 70% ethanol solution was then bound to the RNeasy spin column, and purified and eluted following the manufacturer's instructions. We used Turbo DNase (Ambion, Life Technologies Corporation, Carlsbad, CA, USA) to remove carry-over DNA, and samples were treated with 10% DNase buffer and 5% DNase for 20 min at 37 °C.
Complementary DNA was generated from extracted RNA using the Invitrogen SuperScript III Reverse Transcriptase kit (Life Technologies Corporation), following the manufacturer's instructions. In brief, ten microliters of RNA extract, 1 μl each of random hexamers and 1 μl 10 mM dNTPs were incubated at 65 °C for 5 min, and then placed on ice for 1 min; samples were then incubated with 10 μl of complementary DNA synthesis mix (2 μl 10 × reverse transcriptase buffer, 4 μl 25 mM MgCl2, 2 μl 0.1 M dithiothreitol, 1 μl RNase OUT (40 U−μl−1) and 1 μl SuperScript III Reverse Transcriptase) at 25 °C for 10 min, then 55 °C for 50 min, and the reverse transcriptase reaction was terminated at 85 °C for 5 min. Remaining RNA was removed through incubation with RNase H at 37 °C for 20 min.
Quantitative PCR
QPCR assays were performed on a Stratagene MX3005P (Agilent Technologies), using the following reaction chemistry: 12.5 μl SYBR Premix F (Epicentre Biotechnologies, Madison, WI, USA), 2 mM MgCl2, 0.4 μM of each primer, 1.25 units AmpliTaq polymerase (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA), 40 ng μl−1 bovine serum albumin and 1 ng DNA in a final volume of 25 μl. 16S rRNA genes from Nitrospina were amplified in triplicate using the primers NitSSU_130 F (5′-GGGTGAGTAACACGTGAATAA-3′) and NitSSU_282 R (5′-TCAGGCCGGCTAAMCA-3′ Mincer et al., 2007). Coefficient of variations of triplicate QPCR reactions averaged 5%. Cycling conditions were modified from Mincer et al. (2007): the total number of cycles was reduced from 50 to 30, and the length of the detection step was increased from 1 to 7 s. The partial 16S rRNA sequence from the uncultured marine Nitrospinaceae bacterium recovered on BAC EB080L20_F04 was used as a QPCR standard; this sequence was synthesized at Blue Heron Biotechnologies (Bothell, WA, USA) and used in tenfold dilutions from 107 to 102 16S rRNA genes per μl to generate standard curves (r2=0.987–0.994). narG and napA assays followed Lam et al. (2009) and were used to screen select complementary DNA samples (see text); negative controls did not exhibit amplification.
Pyrosequencing
DNA samples collected at all depths at stations 1–6 were sequenced using Titanium chemistry on the Roche 454 (Branford, CT, USA) FLX platform at Research and Testing Laboratories. No samples from station 7 were sequenced. Bacteria-specific primers 27F (5′-GAGTTTGATCNTGGCTCAG-3′) and 519R (5′-GWNTTACNGCGGCKGCTG-3′ Engelbrektson et al., 2010) modified with additional degeneracies (Ns in primer sequence) were used to amplify a portion of the bacterial 16S rRNA gene. Relevant linkers were attached to primers, as were eight-base barcodes (Hamady et al., 2008) used to sort individual samples; for the 60 samples, we recovered 714 245 sequences with a median read length of 475 bp. We used the approach of Huse et al. (2007) that is commonly used in the literature for quality control (Brown et al., 2009; Galand et al., 2009). Using the program mothur (http://www.mothur.org; Schloss et al. 2009), we discarded sequences:>±100 bp from the median sequence length, containing any ambiguous bases, containing homopolymers >8 bp, of <25 average quality score, and that did not exactly match the forward primer and barcode sequence. The final criterion is where most (21% of all sequences) sequences were removed. (We did not screen sequences based on the reverse primer, as some of the reads are high quality sequences that did not extend to the reverse primer). In total, 282 030 sequences did not meet quality control criteria and were excluded from subsequent analyses.
Bacterial 16S rRNA sequences were aligned to the Greengenes alignment (DeSantis et al., 2006) in mothur. The alignment was optimized to the start position and we discarded sequences starting after the position that 95% of the sequences start; the alignment was then manually curated to remove misaligned sequences (n=25). Community composition of individual samples was determined by classification of 16S rRNA sequences based on the ARB SILVA data set (Pruesse et al., 2007) in mothur, and data are reported for a consensus confidence threshold of 65%. The 8997 Nitrospina 16S rRNA gene sequences identified here were clustered into operational taxonomic units (OTUs) based on 97% sequence identity, and sequences from the 476 OTUs have been deposited in GenBank under accession numbers KC774803-KC775278. We generated representative sequences for each OTU using the get.oturep command in mothur, and BLASTed these sequences against the Genbank nr/nt nucleotide database using blastn and default parameters (database accessed 8 March 2013).
Data analysis
Oceanographic data were visualized in Ocean Data View (http://www.odv.awi.de/). To compute the abundance of different Nitrospina OTUs, we used QPCR data and the proportion of Nitrospina sequences that were affiliated with the different OTUs; that is, the OTU abundance equals the QPCR-based Nitrospina abundance at a particular depth, multiplied by the fraction of sequences drawn from a particular Nitrospina OTU in proportion to the total number of Nitrospina sequences recovered at that depth. This approach is similar to using the relative abundance of terminal-restriction fragment-length polymorphism OTUs and cell counts to examine patterns in SAR11 ecotypes (Carlson et al., 2009), but in this case we focus on Nitrospina and use pyrosequencing and QPCR data. All statistical analyses were conducted in MATLAB version 7.6.0 (R2008a; MathWorks, Natick, MA, USA).
Results and discussion
Oceanographic variability
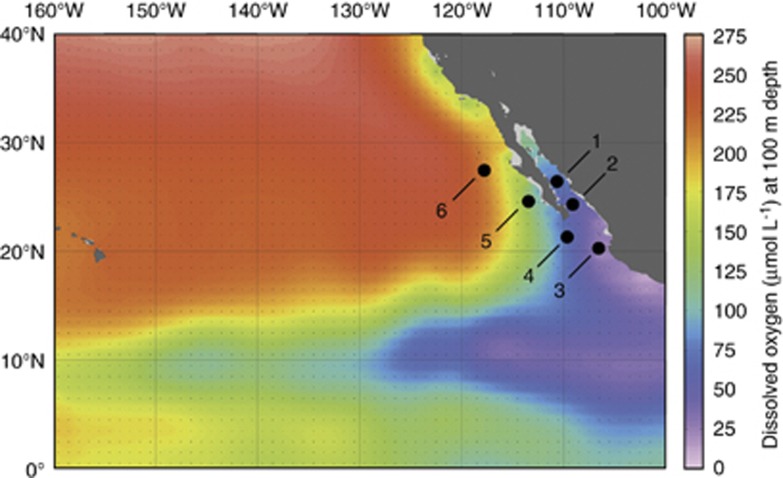
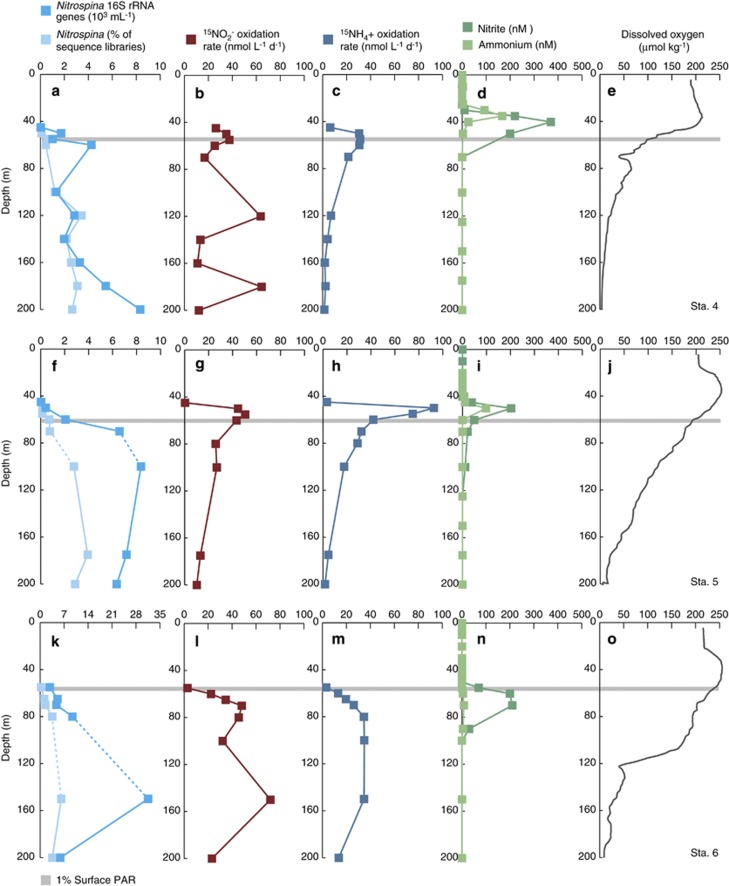
The OMZ of the ETNP extends from the west coast of North America thousands of kilometers across the Pacific Ocean (Figure 1). Low oxygen waters also extend into the GOC and we sampled a series of six stations in the GOC and ETNP; station 1 was located in the Carmen Basin of the GOC, station 2 was located near the mouth of the GOC and station 3 was located in the core of the ETNP OMZ (Figure 1; Supplementary Table S1), where oxygen concentrations declined rapidly with depth and dropped to 20 μmol kg−1 by 111 m (Figure 2o). Three additional stations extended north from the tip of Baja California Sur (station 4) to several hundred kilometers off the coast of northern Baja (Figure 1). (A seventh station sampled for 15NH4+ oxidation rates (Beman et al., 2012) was not sampled for 15NO2− oxidation). General trends in temperature, DO and nutrient concentrations were reported previously (Beman et al., 2012): sea surface temperatures increased from north to south, reaching nearly 30 °C at station 3; the OMZ also broadened and shoaled to the south and DO reached lower absolute values (Figures 2 and 3e). The concentrations of NH4+ and NO2− were highest at discrete depths in the water column (Figures 2 and 3d). These depths were greater at stations 3 and 6 and relatively shallow at stations 1 and 4, with stations 2 and 5 being intermediate. The PNM was offset from the NH4+ maximum by 5–10 m at stations 3 and 4, and was colocated with the NH4+ maximum at stations 1, 2 and 5 (Figures 2 and 3). Prominent secondary NO2− maxima were observed at stations 2 and 3 in OMZ waters, and NO2− concentrations at these depths were an order of magnitude greater than that in the PNM (up to 4.2 μmol l−1 at stations 2 and 3; Figures 2i,n and 4a).
Figure 1.
Cruise track in the GOC and ETNP. Station locations are plotted on DO concentrations (μM) at 100 m depth from the World Ocean Atlas in Ocean Data View.
Figure 2.
Depth profiles from (a–e) station 1, (f–j) station 2 and (k–o) station 3 in the ETNP and GOC. (a, f, k) QPCR data (103 16S rRNA genes per ml) and pyrosequencing data (expressed as % of total 16S rRNA sequence for a given sample) for Nitrospina; (b, g, l) 15NO2− oxidation rates (in nmol l−1 d−1); (c, h, m) 15NH4+ oxidation rates; (d, i, n) NH4+ and NO2− concentrations (nmol l−1); and (e, j, o) DO concentrations (μmol kg−1). s.e. for QPCR fall within the width of the data points, and the gray line denotes the depth of the euphotic zone (that is, 1% of surface photosynthetically active radiation). All data are plotted to 200 m depth, with the exception of station 1 data, which are plotted to 400 m depth. Note that horizontal axes vary between a, f and k, and between b, g and l, but c, h and m are identical to corresponding b, g and l to allow comparison between 15NO2− and 15NH4+ oxidation rates.
Figure 3.
Depth profiles from (a–e) station 4, (f–j) station 5 and (k–o) station 6 in the ETNP and GOC. (a, f, k) QPCR data (103 16 S rRNA genes per ml) and pyrosequencing data (expressed as % of total 16S rRNA sequence for a given sample) for Nitrospina; (b, g, l) 15NO2− oxidation rates (in nmol l−1 d−1); (c, h, m) 15NH4+ oxidation rates; (d, i, n) NH4+ and NO2− (nmol l−1); and (e, j, o) DO concentrations (μmol kg−1). Standard errors for QPCR fall within the width of the data points, and dashed lines connecting data points denote presence of DNA samples with QPCR inhibition (which are subsequently excluded from statistical analyses). The gray line denotes the depth of the euphotic zone, and all data are plotted to 200 m depth. Note that horizontal axes vary between a, f and k, but b, g, l and c, h, m are plotted on identical axes to allow comparison between 15NO2− and 15NH4+ oxidation rates.
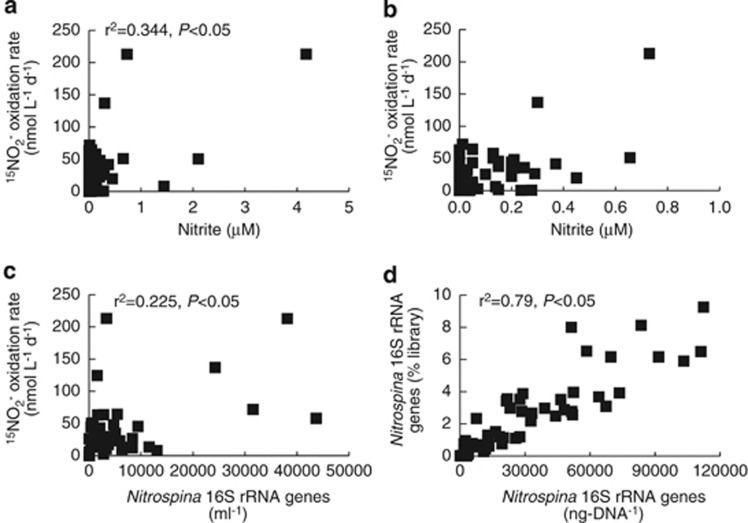
Figure 4.
Correlations between 15NO2− oxidation rates and (a and b) NO2− concentrations and (c) Nitrospina 16S rRNA gene abundance (ml−1) based on QPCR. In b, data points with>1 μM NO2− are not shown in order to show points near the axes. (d) Correlation between Nitrospina as a percentage of pyrosequencing libraries (vertical axis) and Nitrospina 16S rRNA gene abundance (ng-DNA−1) based on QPCR (horizontal axis). QPCR data are normalized per ng-DNA rather than per ml to reflect Nitrospina 16S rRNA genes as a proportion of community DNA.
NO2− oxidation rates and NH3 oxidation rates
Within this oceanographic context, we found that 15NO2− oxidation rates varied strongly with depth and from station to station (Figures 2 and 3b). They typically exhibited peaks at the base of the euphotic zone, decreased slightly with depth, and in some cases, increased again on the edge of the OMZ. Maximum rates at each station ranged from 44.5–213 nmol l−1 d−1, and minimum rates ranged from undetectable to 50.4 nmol l−1 d−1 across stations. These values are in line with previous studies that have measured rates of 0–600 nmol l−1 d−1 across different regions of the ocean (Ward, 1987, Lipschultz et al., 1990; Dore and Karl, 1996; Bianchi et al., 1999; Clark et al., 2008; Füssel et al., 2012). Higher rates have typically been observed in OMZs and other productive regions of the ocean—such as the ETSP, in the Benguela upwelling region and in the Southern California Bight (Ward, 1987; Lipschultz et al., 1990; Füssel et al., 2012)—than within oligotrophic regions (Dore and Karl, 1996; Bianchi et al., 1999; Clark et al., 2008). Overall, 15NO2− oxidation rates were correlated with NO2− concentrations (r2=0.344, P<0.05; Figure 4a) in the GOC and ETNP, and relationships between rates and NO2− were strongest at stations 2 and 5 (Table 1). Most other studies have either not analyzed this relationship or have too few data points to adequately do so, but Ward (1987) proposed that substrate supply may limit NO2− oxidation, and our results are consistent with this suggestion. Lipschultz et al. (1990) also reported strong correspondence between NO2− concentrations and NO2− oxidation rates in the OMZ of the ETSP.
Table 1. r 2 values for the relationships between nitrite oxidation rates, ammonia oxidation rates, NOB abundance and dissolved nitrite.
| Station | Nitrite oxidation vs nitrite | Nitrite oxidation vs ammonia oxidation | Nitrite oxidation vs Nitrospina |
|---|---|---|---|
| 1 | 0.021 | 0.401* | 0.126 |
| 2 | 0.443* | 0.038 (negative) | 0.135 |
| 3 | 0.013 | 0.003 | 0.881* |
| 4 | 0.003 | 0.001 | 0.004 (negative) |
| 5 | 0.443* | 0.790* | 0.085 (negative) |
| 6 | 0.027 | 0.731* | 0.647* |
| Overall | 0.344* | 0.000 | 0.225* |
*P<0.05.
15NO2− oxidation and 15NH4+ oxidation were correlated (r2=0.40–0.79, P<0.05) at stations 1, 5 and 6 consistent with biogeochemical coupling between the two steps of nitrification (Figures 2 and 3). However, 15NH4+ oxidation occurred up to 11 times more rapidly than 15NO2− oxidation in the upper water column at stations 1 and 5, and 56 times more rapidly at station 2. At all of these stations, these rate differences fell within 5 m depth of the PNM where NO2− accumulates to concentrations of hundreds nM. In the most extreme case, 15NH4+ oxidation rates were 342 nmol l−1 d−1 greater than 15NO2− oxidation rates at 45 m at station 2 (Figures 2g–i). At station 1, 15NH4+ oxidation was 119 nmol l−1 d−1 greater than 15NO2− oxidation, and the PNM was 249 nM at 35 m (Figures 2b–d); at station 5, the rate mismatch was 48.1 nmol l−1 d−1 at the 203 nM PNM (Figures 3g–i). At these depths and stations, decoupling between NH3 and NO2− oxidation could potentially produce the PNM in 2–5 days and possibly in <1 day at station 2. This is consistent with previous estimates from the Atlantic by Clark et al. (2008); they calculated that NH3 oxidation could replace >50% of the NO2− pool within 24 h at most of the stations investigated. The data presented here also support our previous contention that NH3 oxidation contributes to generation of the PNM at stations 1 and 5 in the GOC and ETNP (Beman et al., 2012). With our coupled 15NH4+ and 15NO2− oxidation data, we note that differential sensitivity to light does not appear to be a driving factor, because 15NO2− oxidation occurred in the euphotic zone and was correlated with 15NH4+ oxidation in some cases. This is an important distinguishing factor from previous work, which has not measured 15NH4+ and 15NO2− oxidation at high resolution with depth (Ward, 1987, Lipschultz et al., 1990; Dore and Karl, 1996; Bianchi et al., 1999; Clark et al., 2008; Füssel et al., 2012), as we have here (5–10 m intervals in the upper water column). On the basis of our data, the fact that NH3 oxidation rates exceed NO2− oxidation rates appears to be the key in generating the PNM within the upper water column.
OMZ NO2− oxidation
At stations 2, 3 and 4, 15NO2− was oxidized most rapidly (63.7–213 nmol l−1 d−1) at depths >120 m, where oxygen concentrations were low (1.8–8.4 μmol kg−1). Here, 15NO2− oxidation rates were much greater than 15NH4+ oxidation rates; 15NH4+ oxidation was detected in the OMZ, but was typically <5 nmol l−1 d−1 and declined with decreasing DO, such that 15NH4+ oxidation rates were positively correlated with DO concentrations at stations 1–5 (r2=0.51–0.98; Beman et al., 2012). In contrast, 15NO2− oxidation was uncorrelated with DO, and 15NO2− oxidation was up to 50 times more rapid than 15NH4+ oxidation. This represents decoupling between NH3 and NO2− oxidation in the OMZ, and is a probable result of the high concentrations of available NO2−. At stations 2 and 3, NO2− accumulates to concentrations of up to 4.2 μM at 150–200 m; our highest measured rates (213 nmol l−1 d−1) were measured at 200 and 250 m in the OMZ at station 2. NO2− concentrations were not as high in the OMZ at station 4, but they were greater than that at stations 1, 5 and 6 (for example, 114 nM at 400 m at station 4). These accumulations of NO2− evidently allow NOB to oxidize NO2− within the OMZ in spite of low oxygen concentrations, and are consistent with previous work by Lipschulz et al. (1990) and a more recent work by Füssel et al. (2012) in other OMZs. Lipschultz et al. (1990) found rates of up to 600 nmol l−1 d−1 at ∼1 μM DO in ETSP, and Füssel et al. (2012) reported rates of 372 nmol l−1 d−1 at <1 μM DO off Namibia. NO2− oxidation appears to be an important form of microbial metabolism in OMZs (for example, Wright et al., 2012, Ulloa et al., 2012), and the high rates we observed within the oceans' largest OMZ lend further support to this hypothesis.
In previous studies, NO2− oxidation persisted under low DO because these conditions permit NO3− reduction (Lipschulz et al., 1990, Füssel et al., 2012), which produces NO2− that may be oxidized back to NO3−, or used by anammox or denitrification as an oxidant. Alongside measurements of NO2− oxidation, both Lipschultz and colleagues (1990) and Füssel et al. (2012) measured NO3− reduction in the ETSP and Namibian OMZs, finding generally good correspondence between NO3− reduction and NO2− oxidation in magnitude and depth distribution. Lam et al. (2009) also found that NO3− reduction rates were high in the Peruvian OMZ, and detected NO3− reductase gene expression (narG and napA) where NO3− reduction was also measured. We screened OMZ RNA samples where dissolved NO2− exceeded 500 nM for narG and napA expression; this NO2− threshold defines anoxic waters based on recent measurements in the ETSP (Thamdrup et al., 2012, Ulloa et al., 2012), and includes samples from 150–300 m at station 2 and 200–300 m at station 3. We found expression of narG, napA or both, in all of these samples, indicating that NO3− reduction is a possible source of NO2− that may then be oxidized back to NO3−.
Taking a step back, our results indicate that coupling between NH3 and NO2− oxidation is uncommon in the GOC and ETNP. Station 6 was the only station where 15NH4+ and 15NO2− oxidation rates were correlated and similar in magnitude. Instead, quantitative differences in 15NH4+ and 15NO2− oxidation rates in the upper water column, and high 15NO2− oxidation rates measured within the OMZ, indicate that NH3 and NO2− oxidation are not tightly connected, and shift in and out of equilibrium in the ETNP. This does not mean that these processes are decoupled over long time scales: they cannot be, as NO2− would accumulate where NH3 oxidation or NO3− reduction exceeds NO2− oxidation, or be rapidly consumed where NO2− oxidation exceeds NO2− supply.
NOB communities
Of the different NOB that may oxidize NO2−, Nitrospina bacteria are hypothesized to have a key role in the oceanic NO2− oxidation (Mincer et al., 2007) and comprised up to 9.25% 16S rRNA gene sequences recovered in our sequence libraries (250 m at station 2). This indicates that Nitrospina constitute a substantial proportion of the bacterial community in the ETNP, and raises the possibility that Nitrospina may be abundant elsewhere in the ocean. Nitrospina typically increased in relative proportions up to 100–200 m depth (light blue lines in Figures 2 and 3a), and their maximum proportions ranged from 3.09 to 6.52% across the other stations. Throughout our extensive pyrosequencing data set, we found little evidence that other NOB are present in GOC and ETNP. Out of 420 240 sequences, Nitrococcus and Nitrobacter were never detected, and a single Nitrospira 16S rRNA gene sequence was detected at 100 m at station 4. In the Namibian OMZ, Füssel et al. (2012) found that Nitrospina (up to 5.4% of microbial cells) and Nitrococcus (up to 4.9% of microbial cells) were both abundant, and the sum of these groups (0.3–9%) was similar to the relative abundance of Nitrospina that we observed (although their direct counts include Archaea, whereas our data are based on relative proportions within bacterial sequence libraries). We also found strong positive correlation between the proportion of Nitrospina in our pyrosequencing data set and Nitrospina 16S rRNA genes based on QPCR (normalized per ng-DNA, to reflect gene abundance relative to total extracted microbial DNA; r2=0.79, P<0.05; Figure 4d). This correlation with QPCR data indicates that the pyrosequencing data reflect the proportions of different groups within the water column, and that other NOB are therefore substantially less abundant than the Nitrospina in GOC and ETNP.
We examined different Nitrospina ecotypes present in the GOC and ETNP by clustering all classified Nitrospina sequences into OTUs based on 97% sequence identity. We found a few dominant OTUs, with 66.2% of Nitrospina sequences affiliated with just three of them: 24.6% were affiliated with the most abundant OTU, 21.7% with the next most abundant and 19.9% with the third most abundant OTU (Table 2). Nitrospina represented 2.14% of all recovered 16S rRNA sequences, so each of these top three Nitrospina OTUs constitute 0.4–0.5% of all 16S rRNA gene sequences. The remaining 473 Nitrospina OTUs each contained 3.6% or fewer of the Nitrospina sequences. With a few abundant OTUs, and many uncommon OTUs, Nitrospina OTU rank abundance was power law-scaled (r2=0.923; Nitrospina sequences in OTU=1003.1*OTU rank−1.209). This is a general property of many ecological communities (Fuhrman, 2009) that Nitrospina also follow in the GOC and ETNP. Meta-analysis by Wright et al. (2012) showed that other common OMZ bacteria—such as SUP05/ARCTIC96BD-19 and SAR324—are likewise divided into multiple ecotypes, including some that are dominant and nearly ubiquitous, and many others that are less common. Although Nitrospina are frequently detected in OMZs and may have an important role in N cycling within them, Wright et al. (2012) did not analyze Nitrospina in a similar way; our results indicate that Nitrospina communities share similar properties to other typical OMZ bacteria.
Table 2. Dominant Nitrospina operational taxonomic units (OTUs), percentage of sequences, distribution in the water column and similarity to database sequences.
| OTU | % of Nitrospina sequences |
Depth range |
Nitrospina GenBank accession number | Nearest GenBank sequence (accession number) | Depth (m) | Shared identity (%) | Reference | |
|---|---|---|---|---|---|---|---|---|
| Minimum (m) | Maximum (m) | |||||||
| 1 | 24.6 | 60 | 800 | KC775278 | San Pedro Basin, California (DQ009478) | 890 | 99 | Brown et al. (2005) |
| 2 | 21.7 | 55 | 800 | KC775277 | Carmen Basin, Gulf of California (FJ981406) | 2000 | 99 | Dick and Tebo (2010) |
| 3 | 19.9 | 100 | 800 | KC775276 | Eastern Tropical North Pacific (AY726827) | 100 | 99 | Ma et al. (2009) |
| 4 | 3.6 | 50 | 550 | KC775275 | Saanich Inlet, Canada (GQ349069) | 100 | 99 | Walsh et al. (2009) |
| 5 | 2.7 | 70 | 800 | KC775274 | Hawaii Ocean Time-series (HQ675664) | 770 | 99 | Swan et al. (2011) |
| 6 | 2.0 | 375 | 800 | KC775273 | Guaymas Basin, Gulf of California (FJ981193) | 1503 | 99 | Dick and Tebo (2010) |
| 7 | 1.5 | 50 | 550 | KC775272 | Namibian upwelling system, clone N67e_42 (EF646085) | 119–130 | 99 | Woebken et al. (2007) |
| 8 | 1.4 | 55 | 750 | KC775271 | Gulf of Mexico, non-plume sample W65–11 (JN018903) | 800 | 99 | Redmond and Valentine (2012) |
| 9 | 1.1 | 60 | 600 | KC775270 | Saanich Inlet, Canada (GQ349960) | 100 | 99 | Walsh et al. (2009) |
| 10 | 1.1 | 50 | 550 | KC775269 | Arabian Sea, station B high-nucleic acid sample (AY907818) | 25–49 | 98 | Fuchs et al. (2005) |
| 11 | 1.0 | 50 | 500 | KC775268 | Hawaii Ocean Time-series (GU474877) | 200 | 98 | Rich et al. (2011) |
GOC and ETNP Nitrospina OTUs also exhibited 98–99% similarity to database 16S rRNA sequences recovered from other regions of the ocean (Table 2). This includes surface waters (25–49 m depth) of the Arabian Sea (99% identical to OTU 10; Fuchs et al., 2005), suboxic to anoxic waters at 119–130 m depth in the Namibian upwelling system (OTU 7; Woebken et al., 2007), suboxic waters at 890 m depth in the San Pedro Basin (OTU 1; Brown et al., 2005; Beman et al., 2010) and 100 m in the Saanich Inlet, which is typically anoxic below this depth (OTUs 4 and 9; Walsh et al., 2009). OTUs 5, 8 and 11 were similar to sequences recovered from 200 to 800 m depth in the Gulf of Mexico or at Station Aloha in the North Pacific subtropical gyre (Rich et al., 2011; Swan et al., 2011; Redmond and Valentine, 2012), whereas OTUs 2, 3 and 6 were 99% identical to sequences previously recovered in the GOC (Dick and Tebo, 2010) or ETNP (Ma et al., 2009). Given the high sequence similarity to these database sequences and the diversity of oceanic regions represented by them, all of these Nitrospina OTUs appear widely distributed and may be important NO2− oxidizers elsewhere in the ocean. In the ETNP, our data provide an in-depth view of Nitrospina in world's largest OMZ, and indicate that thousands of Nitrospina sequences and hundreds of Nitrospina OTUs are spread across a range of DO concentrations.
Nitrospina and NO2− oxidation
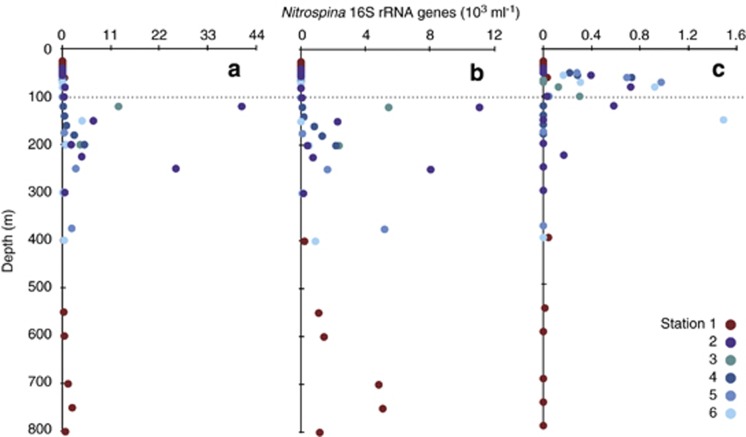
For the entire data set, QPCR-determined Nitrospina 16S rRNA gene abundances (ml−1) were significantly correlated with 15NO2− oxidation rates (r2=0.225 P<0.05; Figure 4c). Like other variables we examined, the strength of this relationship varied from station to station, with strongest correspondence at stations 3 and 6 (Table 1). We computed the distribution of different Nitrospina OTUs throughout the water column based on these QPCR data and the proportion of Nitrospina sequences that were affiliated with the different OTUs (Figure 5; see Materials and Methods and Carlson et al., 2009). Although these PCR-based approaches introduce uncertainties that preclude further quantitative analysis, there are few practical alternatives, as OTU-specific QPCR primers likely have their own biases, as would microarrays with OTU-specific probes; we use these data to demonstrate the vertical distribution of the Nitrospina OTUs.
Figure 5.
Distribution of Nitrospina ecotypes with depth across stations: (a) the most abundant OTU, (b) the third most common OTU and (c) the 7th most abundant OTU. Colors indicate different stations and data are expressed as 103 16S rRNA genes per ml.
Across this data set, no single OTU was found at all depths where NO2− was oxidized, indicating that multiple Nitrospina OTUs must govern the process across the GOC and ETNP. The three dominant Nitrospina OTUs showed slightly different depth distributions, with the third most abundant OTU confined to a smaller depth range than the other two (Table 2), but displaying greater constancy with depth (Figure 5b). All three of these OTUs were uncommon in the upper water column, where high rates of 15NO2− oxidation nevertheless occur; a good example of this is the peak in 15NO2− oxidation rates found at 55 m at station 2, where none of the dominant OTUs were present. OTUs 7, 10 and 11 were the only Nitrospina detected at this depth, as well as in other samples collected at <100 m depth (Figure 5c). Their presence where other Nitrospina are absent suggests that they contribute to NO2− oxidation in the upper water column.
Together with the fact that only a single 16S rRNA gene from other NOB was detected in our pyrosequencing data set, and that Nitrospina abundance based on QPCR was correlated with 15NO2− oxidation rates, these findings indicate that (1) Nitrospina oxidize NO2− in the ETNP and GOC and (2) a mixture of Nitrospina OTUs contribute at different depths. Untangling the relative contributions of these OTUs may be possible through quantifying OTU-specific NO2− oxidoreductase (nxr) gene expression, but requires (meta)genomic/transcriptomic data for the different Nitrospina OTUs. In the absence of such published data, our approach identifies Nitrospina that are candidates for additional study in other OMZs and other regions of the sea. More specifically, the more abundant OTUs identified here are widespread in the ETNP OMZ, whereas OTUs 7, 10 and 11 are less abundant overall, but appear important in the well-oxygenated upper water column.
Conclusions
We report the first 15NO2− oxidation measurements from the ETNP, and show that the two steps of nitrification were rarely connected. NO2− oxidation may be coupled to anaerobic processes that produce NO2− in the OMZ. NO2− concentrations correlated with 15NO2− oxidation rates and the highest rates were observed in the OMZ, where NO2− concentrations were elevated. High Nitrospina abundances (based on pyrosequencing), the lack of other detectable NOB and significant correlations with 15NO2− oxidation rates (based on QPCR) all indicate that Nitrospina are dominant NOB in the GOC and ETNP. For the first time, we show that Nitrospina constitute a large percentage of the bacterial community (up to 9.25%) and are grouped into multiple OTUs that are distributed heterogenously with depth. Many of these OTUs are highly similar to sequences recovered from other areas of the ocean and may represent biogeochemically important populations of Nitrospina. Given this sequence diversity and ecological diversity within the Nitrospina, we suggest that the genomic and metabolic diversity of these putative ecotypes should be explored, such that their relative contributions to NO2− oxidation at different depths and in different oceanic regions may be determined. In the ETNP and other OMZs, NO2− cycling is central and Nitrospina are widespread, abundant and diverse. Our findings support the idea that Nitrospina are key contributors to oceanic NO2− oxidization, and provide new insight into their ecology and rates of N cycling, within the oceans' largest, and expanding OMZ.
Acknowledgments
We thank Fred Prahl (chief scientist), Jackie Mueller, Natalie Wallsgrove, Jason Smith, Julie Fliegler and the officers and crew of the R/V New Horizon for assistance in the field. We also thank Rachel Foster for supplying critical DNA samples, Susan Alford, Molly Carolan and Natalie Wallsgrove for assistance with laboratory analyses. This work was supported by National Science Foundation Oceanography Award 10-34943 to JMB and BNP, and this is School of Ocean and Earth Science and Technology Contribution number 8809.
The authors declare no conflicts of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Beman JM, Popp BN, Alford SE. Quantification of ammonia oxidation rates and ammonia-oxidizing archaea and bacteria at high resolution in the Gulf of California and eastern tropical North Pacific Ocean. Limnol Oceanog. 2012;57:711–726. [Google Scholar]
- Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:429–441. doi: 10.1038/ismej.2007.118. [DOI] [PubMed] [Google Scholar]
- Beman JM, Sachdeva R, Fuhrman JA. Population ecology of nitrifying Archaea and bacteria in the Southern California Bight. Environ Microbiol. 2010;12:1282–1292. doi: 10.1111/j.1462-2920.2010.02172.x. [DOI] [PubMed] [Google Scholar]
- Bianchi M, Fosset C, Conan P. Nitrification rates in the NW Mediterranean Sea. Aquat Microb Ecol. 1999;17:267–278. [Google Scholar]
- Brown MV, Philip GK, Bunge JA, Smith MC, Bissett A, Lauro FM, et al. Microbial community structure in the North Pacific ocean. ISME J. 2009;3:1374–1386. doi: 10.1038/ismej.2009.86. [DOI] [PubMed] [Google Scholar]
- Brown MV, Schwalbach M, Hewson I, Fuhrman JA. Coupling 16S-ITS rDNA clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ Microbiol. 2005;7:1466–1479. doi: 10.1111/j.1462-2920.2005.00835.x. [DOI] [PubMed] [Google Scholar]
- Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 2009;3:283–295. doi: 10.1038/ismej.2008.117. [DOI] [PubMed] [Google Scholar]
- Casciotti KL, McIlvin MR. Isotopic analyses of nitrate and nitrite from reference mixtures and application to Eastern Tropical North Pacific waters. Mar Chem. 2007;107:184–201. [Google Scholar]
- Church MJ, Short CM, Jenkins BD, Karl DM, Zehr JP. Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl Environ Microbiol. 2005;71:5362–5370. doi: 10.1128/AEM.71.9.5362-5370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DR, Rees AP, Joint I. Ammonium regeneration and nitrification rates in the oligotrophic Atlantic Ocean: implications for new production estimates. Limnol Oceanog. 2008;53:52–62. [Google Scholar]
- Costa E, Pérez J, Kreft J. Why is metabolic labour divided in nitrification. Trends Microbiol. 2006;14:213–219. doi: 10.1016/j.tim.2006.03.006. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch C, Brix H, Ito T, Frenzel H, Thompson L. Climate-forced variability of ocean hypoxia. Science. 2011;333:336–339. doi: 10.1126/science.1202422. [DOI] [PubMed] [Google Scholar]
- DeVries T, Deutsch C, Primeau F, Chang B, Devol A. Global rates of water-column denitrification derived from nitrogen gas measurements. Nature Geosci. 2012;5:547–550. [Google Scholar]
- Dick GJ, Tebo BM. Microbial diversity and biogeochemistry of the Guaymas Basin deep-sea hydrothermal plume. Environ Microbiol. 2010;12:1334–1347. doi: 10.1111/j.1462-2920.2010.02177.x. [DOI] [PubMed] [Google Scholar]
- Dore JE, Karl DM. Nitrification in the euphotic zone as a source of nitrite, nitrate and nitrous oxide at station ALOHA. Limnol Oceanogr. 1996;41:1619–1628. [Google Scholar]
- Dore JE, Popp BN, Karl DM, Sansone FJ. A large source of atmospheric nitrous oxide from subtropical North Pacific surface waters. Nature. 1998;396:63–66. [Google Scholar]
- Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, et al. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010;4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- Francis CA, Beman JM, Kuypers MMM. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 2007;1:19–27. doi: 10.1038/ismej.2007.8. [DOI] [PubMed] [Google Scholar]
- Fuchs BM, Woebken D, Zubkov MV, Burkill P, Amann R. Molecular identification of picoplankton populationsin contrasting waters of the Arabian Sea. Aquat Microb Ecol. 2005;39:145–157. [Google Scholar]
- Fuhrman JA. Microbial community structure and its functional implications. Nature. 2009;459:193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- Füssel J, Lam P, Lavik G, Jensen MM, Holtappels M, Gunter M, et al. Nitrite oxidation in the Namibian oxygen minimum zone. ISME J. 2012;6:1200–1209. doi: 10.1038/ismej.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci USA. 2009;106:22427–22432. doi: 10.1073/pnas.0908284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilly WF, Beman JM, Litvin SY, Robison BH. Oceanographic and biological effects of shoaling of the oxygen minimum zone. Annu Rev Marine Sci. 2013;5:393–420. doi: 10.1146/annurev-marine-120710-100849. [DOI] [PubMed] [Google Scholar]
- Graham DW, Knapp CW, Van Vleck ES, Bloor K, Lane TB, Graham CE. Experimental demonstration of chaotic instability in biological nitrification. ISME J. 2007;1:385–393. doi: 10.1038/ismej.2007.45. [DOI] [PubMed] [Google Scholar]
- Granger J, Sigman DM. Removal of nitrite with sulfamic acid for nitrate N and O isotope analysis with the denitrifier method. Rapid Commun Mass Spectrom. 2009;23:3753–3762. doi: 10.1002/rcm.4307. [DOI] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Meth. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RM, Aminot A, Kérouel R, Hooker BA, Peterson BJ. A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can J FIsh Aquat Sci. 1999;56:1801–1808. [Google Scholar]
- Huse S, Huber J, Morrison H, Sogin M, Welch D. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- Keeling RF, Körtzinger A, Gruber N. Ocean deoxygenation in a warming world. Ann Rev Mar Sci. 2010;2:199–229. doi: 10.1146/annurev.marine.010908.163855. [DOI] [PubMed] [Google Scholar]
- Lam P, Jensen MM, Lavik G, McGinnis DF, Muller B, Schubert CJ, et al. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci USA. 2007;104:7104–7109. doi: 10.1073/pnas.0611081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Kuypers MMM. Microbial nitrogen cycling processes in oxygen minimum Zones. Annu Rev Marine Sci. 2011;3:317–345. doi: 10.1146/annurev-marine-120709-142814. [DOI] [PubMed] [Google Scholar]
- Lam P, Lavik G, Jensen MM, van dV, Schmid M, Woebken D, et al. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci USA. 2009;106:4752–4757. doi: 10.1073/pnas.0812444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipschultz F, Wofsy SC, Ward BB, Codispoti LA, Friederich GE, Elkins JW. Bacterial transformations of inorganic nitrogen in the oxygen-deficient waters of the Eastern Tropical South Pacific Ocean. Deep Sea Res A. 1990;37:1513–1541. [Google Scholar]
- Lomas MW, Lipschultz F. Forming the primary nitrite maximum: nitrifiers or phytoplankton. Limnol Oceanog. 2006;51:2453–2467. [Google Scholar]
- Ma Y, Zeng Y, Jiao N, Shi Y, Hong N. Vertical distribution and phylogenetic composition of bacteria in the Eastern Tropical North Pacific Ocean. Microbiol Res. 2009;164:624–633. doi: 10.1016/j.micres.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Mills MM, Ridame C, Davey M, La Roche J, Gelder RJ. Iron and phosphorus co-limit nitrogen fixation in the eastern tropical North Atlantic. Nature. 2004;429:292–294. doi: 10.1038/nature02550. [DOI] [PubMed] [Google Scholar]
- Mincer TJ, Church MJ, Taylor LT, Preston CM, Karl DM, DeLong EF. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- Olson RJ. 15N tracer studies of the primary nitrite maximum. J Mar Res. 1981;39:203–226. [Google Scholar]
- Paulmier A, Ruiz-Pino D. Oxygen minimum zones (OMZs) in the modern ocean. Prog Oceanogr. 2009;80:113–128. [Google Scholar]
- Popp BN, Sansone FJ, Rust TM, Merritt DA. Determination of concentration and carbon isotopic composition of dissolved methane in sediments and nearshore waters. Anal Chem. 1995;67:405–411. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond MC, Valentine DL. Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109:20292–20297. doi: 10.1073/pnas.1108756108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich VI, Pham VD, Eppley J, Shi Y, DeLong EF. Time-series analyses of Monterey Bay coastal microbial picoplankton using a ‘genome proxy' microarray. Environ Microbiol. 2011;13:116–134. doi: 10.1111/j.1462-2920.2010.02314.x. [DOI] [PubMed] [Google Scholar]
- Santoro AE, Casciotti KL, Francis CA. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ Microbiol. 2010;12:1989–2006. doi: 10.1111/j.1462-2920.2010.02205.x. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman DM, Casciotti KL, Andreani M, Barford C, Galanter M, Böhlke JK. A bacterial method for the nitrogen isotopic analysis of nitrate in seawater and freshwater. Anal Chem. 2001;73:4145–4153. doi: 10.1021/ac010088e. [DOI] [PubMed] [Google Scholar]
- Stramma L, Johnson GC, Sprintall J, Mohrholz V. Expanding oxygen-minimum zones in the tropical oceans. Science. 2008;320:655–658. doi: 10.1126/science.1153847. [DOI] [PubMed] [Google Scholar]
- Strickland JH, Parsons TR. A Practical Handbook of Seawater Analysis. Fisheries Research Board of Canada: Ottawa: Ontario, Canada; 1972. [Google Scholar]
- Sutka RL, Ostrom NE, Ostrom PH, Phanikumar MS. Stable nitrogen isotope dynamics of dissolved nitrate in a transect from the North Pacific subtropical gyre to the eastern tropical North Pacific. Geochim Cosmochim Ac. 2004;68:517–527. [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science. 2011;333:1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- Thamdrup B, Dalsgaard T, Revsbech NP. Widespread functional anoxia in the oxygen minimum zone of the Eastern South Pacific. Deep Sea Res I: Oceanog Res Papers. 2012;65:36–45. [Google Scholar]
- Ulloa O, Canfield DE, DeLong EF, Letelier RM, Stewart FJ. Microbial oceanography of anoxic oxygen minimum zones. Proc Natl Acad Sci USA. 2012;109:15996–16003. doi: 10.1073/pnas.1205009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG, et al. Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science. 2009;326:578–582. doi: 10.1126/science.1175309. [DOI] [PubMed] [Google Scholar]
- Ward BB. Nitrogen transformations in the Southern California Bight. Deep Sea Res A. 1987;34:785–805. [Google Scholar]
- Ward BB, Devol AH, Rich JJ, Chang BX, Bulow SE, Naik H, et al. Denitrification as the dominant nitrogen loss process in the Arabian Sea. Nature. 2009;461:78–81. doi: 10.1038/nature08276. [DOI] [PubMed] [Google Scholar]
- Ward BB, Glover HE, Lipschultz F. Chemoautotrophic activity and nitrification in the oxygen minimum zone off Peru. Deep Sea Res A. 1989a;36:1031–1051. [Google Scholar]
- Ward BB, Kilpatrick KA, Renger EH, Eppley RW. Biological nitrogen cycling in the nitracline. Limnol Oceanogr. 1989b;34:493–513. [Google Scholar]
- Ward BB, Zafiriou OC. Nitrification and nitric oxide in the oxygen minimum of the eastern tropical North Pacific. Deep Sea Res A. 1988;35:1127–1142. [Google Scholar]
- Woebken D, Fuchs BM, Kuypers MMM, Amann R. Potential interactions of particle-associated anammox bacteria with bacterial and archaeal partners in the Namibian upwelling system. Appl Environ Microbiol. 2007;73:4648–4657. doi: 10.1128/AEM.02774-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JJ, Konwar KM, Hallam SJ. Microbial ecology of expanding oxygen minimum zones. Nat Rev Micro. 2012;10:381–394. doi: 10.1038/nrmicro2778. [DOI] [PubMed] [Google Scholar]
- Yool A, Martin AP, Fernandez C, Clark DR. The significance of nitrification for oceanic new production. Nature. 2007;447:999–1002. doi: 10.1038/nature05885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.