Abstract
Proteorhodopsins (PRs) are commonly found in marine prokaryotes and allow microbes to use light as an energy source. In recent studies, it was reported that PR stimulates growth and survival under nutrient-limited conditions. In this study, we tested the effect of nutrient and salinity stress on the extremely psychrophilic sea-ice bacterial species Psychroflexus torquis, which possesses PR. We demonstrated for the first time that light-stimulated growth occurs under conditions of salinity stress rather than nutrient limitation and that elevated salinity is related to increased growth yields, PR levels and associated proton-pumping activity. PR abundance in P. torquis also is post-transcriptionally regulated by both light and salinity and thus could represent an adaptation to its sea-ice habitat. Our findings extend the existing paradigm that light provides an energy source for marine prokaryotes under stress conditions other than nutrient limitation.
Keywords: post-transcriptional regulation, proteomics, proteorhodopsin, salinity stress, sea-ice
Introduction
The discovery of proteorhodopsin (PR) in 2000 challenged the assumption that solar energy is captured mainly through the use of chlorophylls and other analogous pigments (Fuhrman et al., 2008). PR genes are found in various ecosystems (Sabehi et al., 2004; Venter et al., 2004; Giovannoni et al., 2005; Frigaard et al., 2006; Atamna-Ismaeel et al., 2008; Koh et al., 2010) and they are among the most highly expressed and widely distributed proteins in marine bacterial communities (Frias-Lopez et al., 2008). Theoretically, as PR-bearing bacteria can use light energy to supplement energy, it should make them more competitive in situations where other sources of energy are limiting. PR functions as a transmembrane light-driven proton pump that, when exposed to light, creates a proton-motive force that can drive chemical transport processes, motility and generate ATP (Béjà et al., 2000, 2001). To show this, marine-derived PR genes were cloned and expressed in E. coli which then exhibited increased proton pumping (Béjà et al., 2000). Furthermore, when the recombinant strain was provided with retinal, Walter et al. (2007) demonstrated that the recombinant E. coli strain showed increased motility under light and that light energized electron transport, despite cellular respiration being impeded by anoxia and sodium azide. The authors suggested that PR can replace respiration as a cellular energy source under some environmental conditions (Walter et al., 2007).
As the discovery of PR, several works have attempted to demonstrate the physiological role of PR and its effects on bacterial growth. However, laboratory experiments examining light-stimulated growth in PR-containing bacteria have not produced consistent results. For example, no increase in growth rate and/or final cell yield under illuminated conditions was observed in SAR11 and SAR92 strains (Giovannoni et al., 2005; Stingl et al., 2007). In contrast, light-enhanced growth was reported for Dokdonia sp. MED 134 under carbon-limited conditions (Gomez-Consarnau et al., 2007). Later it was demonstrated that PR-containing Vibrio strain AND4 had increased long-term survival when starved if it was exposed to light based on comparisons with a PR-deficient mutant (Gomez-Consarnau et al., 2010). A light/dark growth experiment was conducted on natural mixed communities of oceanic bacteria collected from a low-nutrient ocean environment and most of the marine clades that are thought to possess PR showed different responses to light (Schwalbach et al., 2005).
Currently, nearly all hypotheses argue that PR could provide an adaptive advantage to the bacteria under oligotrophic conditions. To date, all studies have focused on explaining the physiological role of PR that might affect bacterial growth during periods of low-nutrient or carbon-limited conditions (Giovannoni et al., 2005; Gomez-Consarnau et al., 2007; Michelou et al., 2007; Stingl et al., 2007; Lami et al., 2009; DeLong and Béjà, 2010; Gomez-Consarnau et al., 2010; Steindler et al., 2011). Nevertheless, the data to date do not consistently support this hypothesis. Moreover, the non-uniform response between different organisms that all express the PR gene indicates the possibility of multiple functions of PR in these bacteria (Fuhrman et al., 2008). As described above, PR are widespread in natural environments and these environments may have many other conditions in which PR may be advantageous. We hypothesize that PR may be an important provider of light-derived energy under stress conditions that are associated with a specific econiche in a nutrient-independent manner. Here, we report that light-stimulated growth of the PR-containing sea-ice bacterial species Psychroflexus torquis ATCC (Manassas, VA, USA) 70075514 occurs in concordance with increased PR activity. We also show that PR abundance is dependent on osmotically different conditions rather than light/dark and carbon-limited conditions.
Materials and methods
Bacterial strains and media
P. torquis ATCC 700755T was originally isolated from multi-year sea ice collected from Prydz Bay, Vestfold Hills, Antarctica (S 68°36′30′′, E 78°04′19′′) (Bowman et al., 1998). It was routinely grown in modified marine broth (5 g of proteose peptone (Oxoid, Adelaide, SA, Australia), 2 g of yeast extract (Oxoid) 35 g of sea salt (Red Sea) in 1 l of distilled water) aerobically at 5 °C.
Growth experiments
Experiments were carried out to determine the effect of light under conditions of salinity and nutrient stress. The effect of salinity stress was tested with a series of modified marine broths with different salt concentrations: 17.5, 35, 52.5 and 70 g l−1. One hundred milliliter of triplicate cultures were inoculated with 100 μl working culture (6.0 log CFU ml−1) and incubated aerobically at 5 °C. Cultures were constantly exposed to three light intensities: bright light (27.7 μmol photons m−2 s−1); dim light (3.7 μmol photons m−2 s−1) and complete darkness (0 μmol photons m−2 s−1). In all experiments, light was provided from 30 W fluorescent lamps. Nutrient stress experiments were conducted under the same temperature and light conditions described above with modified marine broth diluted 1:10, 1:100, 1:1000 and 1:10 000 with artificial seawater such that the salinity was kept constant but the organic content of the medium was diluted. To determine cell numbers direct counts were performed microscopically using a counting chamber.
RNA extraction
Cells of P. torquis were cultured as described above and harvested by centrifugation at 14 000 r.p.m. for 15 min within the early, middle and late exponential phase. The cell pellets were stored in RNAlater (Ambion Inc, Grand Island, NY, USA) and kept at −20 °C. Total RNA was extracted using the Qiagen RNeasy Mini Kit (Qiagen, Melbourne, VIC, Australia), according to manufacturer's instructions. In addition an on-column DNase treatment using RNase-free DNase (Qiagen) was performed to remove contaminating genomic DNA. The RNA concentration was determined using a Nanodrop 1000 spectrometer (Thermo Fisher Scientific, Melbourne, VIC, Australia).
Analysis of PR expression by quantitative real-time PCR (qRT-PCR)
For qRT-PCR analysis, 16S rRNA was used as a reference gene. The qRT-PCR was performed in duplicates with 10 ng of the total RNA in a final volume of 25 μl using a one-step QuantiTect SYBR Green RT-PCR Kit (Qiagen). PR primers used in this experiment were 5′-TAT GGC CAT GTT GGC TGC AT-3′ (forward) and 5′-CTG AAG CTA GAC CCC ACT GC-3′ (reverse), while 16S rRNA primers were 5′-CAG CMG CCG CCG TAA TAC-3′ (forward) and 5′-CCG TCA ATT CMT TTR AGT TT-3′ (reverse). PCR thermocycling was performed using a Rotor-Gene (Corbet Research, Sydney, NSW, Australia) RG-3000 with reverse transcription performed at 50 °C for 30 min followed by DNA polymerase activation step at 95 °C for 15 min, followed by 35 cycles of amplification at 94 °C for 15 s, annealing temperatures 52 °C for 30 s and extension at 72 °C for 30 s. Only single peak were observed in melting curves for both the 16 S rRNA and PR genes indicating specific amplification with each set of primers. Amplification efficiencies were between 0.85–0.98 and the correlation coefficient for a 10-fold dilution series was between 0.96–1.00. Relative quantification of PR and 16S rRNA gene, mRNA transcripts utilized the Pfaffl equation (Pfaffl, 2001).
Spectral and HPLC analysis of pigment
One hundred microliter of P. torquis working culture was used to inoculate modified marine agar with the range of salinity conditions described above. Agar plates were incubated aerobically at 5 °C for 23 days. Biomass was harvested and freeze dried using a Dyna-Vac freeze dryer (Technolab, Kingston, Tasmania, Australia); 4 mg of freeze-dried biomass were extracted with 1 ml of 3:3:3:1 methanol: acetone: N,N′-dimethylformamide: water; at 4 °C in the dark (Hagerthey et al., 2006). Absorption spectra were measured using a Thermo Fisher Scientific GENESYS 10S UV–vis, utilizing quartz cuvettes. Spectra were recorded in 0.5-nm steps between 250 and 600 nm. The control baseline was established using the same amount of solvent without biomass. The pigment was extracted and analyzed by HPLC using the method described by Sabehi et al. (2005) to identify the presence of carotenoids and retinal.
Light-driven proton pump activity analysis
Fifty milliliter of modified marine broth with different salinities, as outlined above, were inoculated with 50 μl of working culture and incubated aerobically at 5 °C under dark for 23 days. The cells were collected by centrifugation (14 000 r.p.m., 15 min at 4 °C) and then washed twice in artificial seawater. Cell pellets were resuspended in 10 ml of artificial seawater and held for 1 day at 5 °C. Proton pump activity was measured by monitoring pH changes in the suspensions. The suspensions were first placed in darkness and then illuminated by light (∼27.7 μmol photons m−2 s−1) for 30 min. pH changes during incubation were monitored and recorded with a pH meter from BioFlo/CelliGen115 Fermentor/Bioreactor (New Brunswick, NJ, USA). Measurements were repeated under the same conditions after addition of the protonophore carbonylcyanide-3-chlorophenylhydrazone to a final concentration of 10 μM (Yoshizawa et al., 2012).
Liquid Chromatography/tandem MS–MS analysis of PR abundance
Detection and determination of the abundance of PR is described in detail in the Supplementary Material.
Results
The light-stimulated growth response of P. torquis is salinity not nutrient dependent
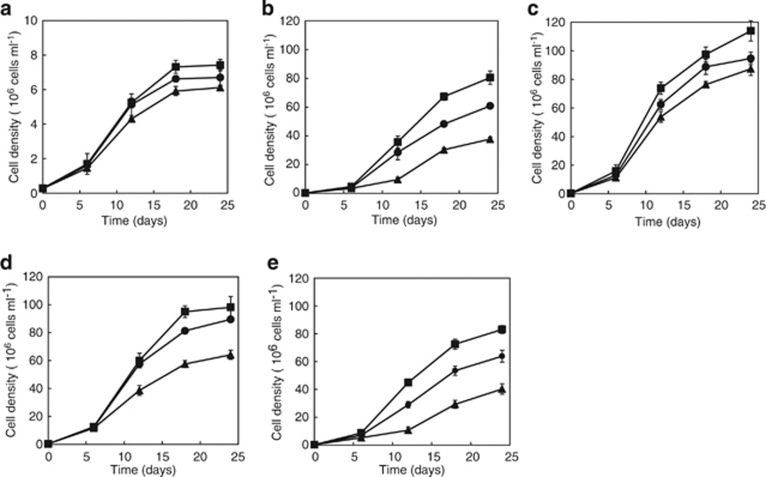
To assess whether light is required for PR-mediated proton translocation, and whether it affected growth rate and increased the final cell population, the psychrophilic species P. torquis ATCC 700755 was grown under nutrient-limited and also under nutrient-rich conditions over a range of growth permissive salinities (Bowman et al., 1998). Preliminary carbon limitation experiments showed that P. torquis did not significantly exhibit differences in cell abundance under the carbon-limited conditions tested (see Supplementary Figure 1). Specifically, under constant carbon limitation (70 mg l−1 organic content) P. torquis grown under different levels of illumination did not significantly differ in cell abundance nor was growth rate altered (Figure 1a). However, when exposed to different levels of salinity under nutrient-rich conditions (5 g l−1 organic content) both growth rate and cell abundance were always higher under illumination compared with when grown in the dark (Figures 1b–e). Maximal differences in growth yield between light and dark incubation occurred at sub- and supra-optimal salinities. Specific growth rates and yields were stimulated about 1.5 per day and twofold under 3.7 μmol photons m−2 s−1 illumination, respectively. Under 27.7 μmol photons m−2 s−1, illumination growth rates were similar but the growth yield differential was markedly reduced suggesting high levels of irradiance become photo-inhibitory to P. torquis.
Figure 1.
Growth of P. torquis in 1:100 diluted modified marine broth at 35 g l−1 (a) and in modified marine broth with salinities of 17.5 (b), 35 (c), 52.5 (d) and 70 g l−1 (e) under 27.7 μmol photons m−2 s−1 (•), 3.7 μmol photons m−2 s−1 (▪) and 0 μmol photons m−2 s−1 (▴). Cell populations were determined microscopically.
P. torquis pigment synthesis is light and salinity dependent
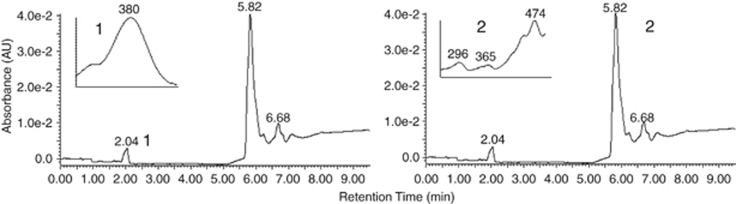
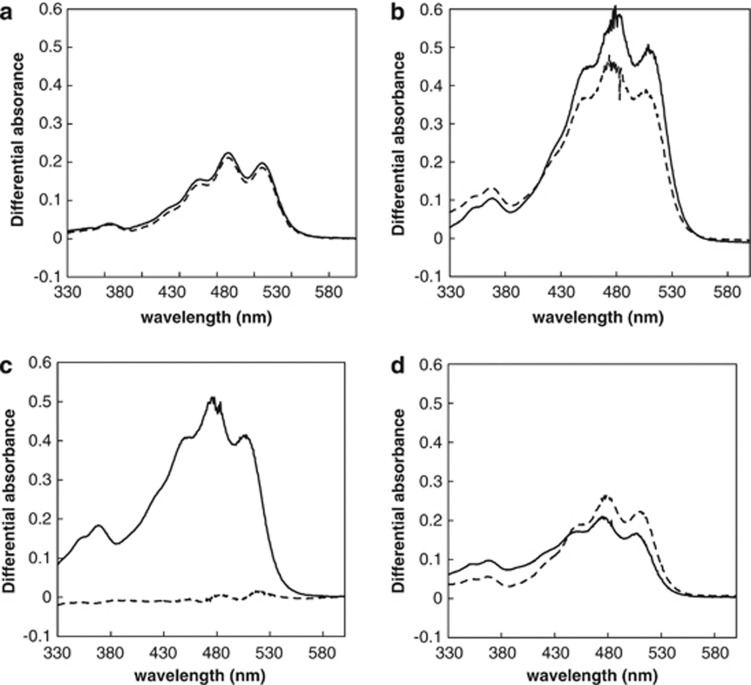
As PR is a retinylidene protein and its function depends on a retinal chromophore derived from carotenoid compounds. The presence of retinal and an unidentified type of carotenoid was confirmed by HPLC (Figures 2a and b). To determine whether illumination stimulates carotenoid biosynthesis, pigment analysis of biomass grown under different levels of illumination and salinities was assessed. Pigment extracts had a major spectral absorbance region between 425–525 nm, which represents carotenoids. When compared with dark-incubated cultures, illumination stimulated pigment production though the effect was primarily influenced by salinity as well as the light intensity (Figures 3a–d). Illumination-driven pigment biosynthesis stimulation occurred maximally at the optimum salinity for growth, which is equivalent to that of seawater; however, at a salinity of 52.5 g l−1 incubation under low irradiance did not correspond to stimulated pigment production (Figure 3c).
Figure 2.
HPLC separation of the carotenoid and retinal from extracted pigment. Insert graphs show the absorption spectra of peaks 1 (retinal) and 2 (unknown carotenoid).
Figure 3.
Pigment levels in P. torquis ATCC 700755 grown under different light levels (bright: 27.7 μmol photons m−2 s−1 dim: 3.7 μmol photons m−2 s−1) and salinity levels. The graph shows absorption spectra of pigment extracted from P. torquis grown in modified marine broth with 17.5 g l−1 (a), 35 g l−1 (b), 52.5 g l−1 (c) and 70 g l−1 (d). The solid line and dot lines indicates the pigment extract absorbance levels in dim and bright light incubated cultures after being substracted from dark-incubated cultures.
Light-driven proton activity of P. torquis is light activated and salinity dependent
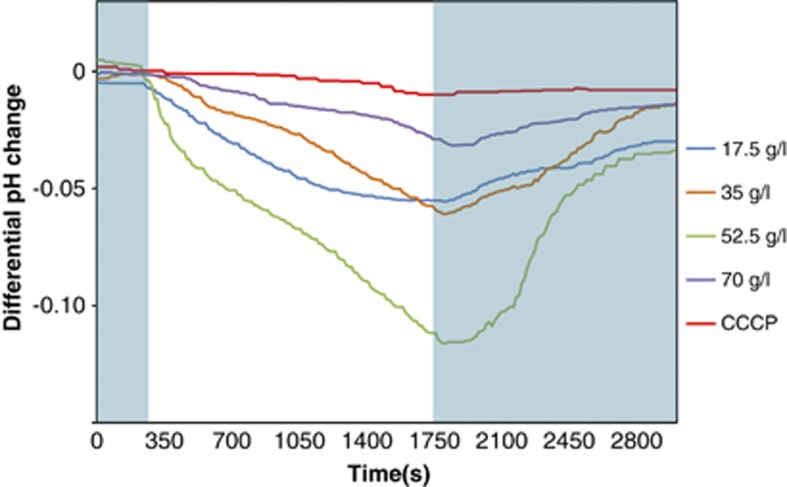
The light-driven proton pump activity of P. torquis was also tested at 1 °C under different levels of salinity. When a suspension of P. torquis (∼108 cells ml−1) was illuminated, the external pH declined rapidly (Figure 4) indicative of proton extrusion. This was confirmed by the use of the carbonylcyanide-3-chlorophenylhydrazone to uncouple H+ translocation (Yoshizawa et al., 2012). The light-driven H+ and translocative-driven pH changes were influenced by the salinity with maximal H+ pumping occurring at 52.5 g l−1 (Figure 4) and under 27.7 μmol photons m−2 s−1 illumination.
Figure 4.
Light-driven proton pump activity in P. torquis cell suspensions. The pH changes in the suspension media were measured (initial pH: 7.7±0.5) under light (27.7 μmol photons m−2 s−1). The shaded area denotes completely dark conditions during the experiment. The incubation temperature was 1 (±0.5)°C. Carbonylcyanide-3-chlorophenylhydrazone (CCCP) was added at a final concentration of 10 μM.
PR gene expression is constitutive in P. torquis regardless of illumination or salinity level
Quantitative RT-PCR was used to determine the expression of the gene encoding PR during growth within early, middle and late exponential growth phases. Dark-incubated cultures were used as controls to allow calculation of the change in PR expression. Different salt concentrations or illumination levels had no significant effect on the expression of the PR gene (see Supplementary Table S1). In addition, the expression of the PR gene was also unaffected during transition from exponential phase into stationary growth. On the basis of these data the PR gene appears to be constitutively expressed in P. torquis ATCC 700755.
PR cellular abundance in P. torquis is influenced by salinity and light
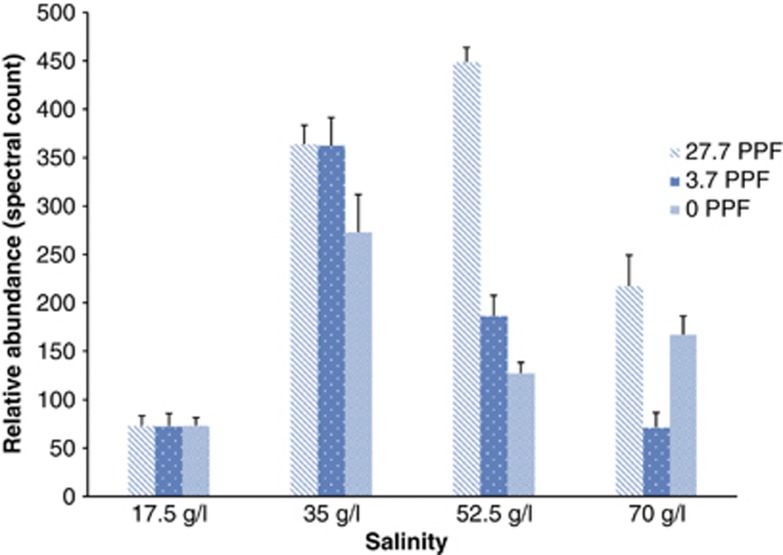
We assessed the abundance of PR by using liquid Chromatography/tandem mass spectrometry (MS)–MS analysis and spectral counting of PR peptides detected in trypsinized protein extracts. Based on the accumulated data of almost 700 000 peptide spectra that passed filtration criteria, PR peptides made up ∼0.4% of the identified peptides. Taking into account the extraction bias that occurs with recovery of transmembrane proteins, PR must be among the most abundant proteins in P. torquis. The cellular abundance of PR was directly influenced by salinity as well as illumination (Figure 5). The relative abundance of PR was stimulated under both illuminated and dark conditions at seawater salinity levels with the differential effect of light levels being small but still noticeable. At 52.5 g l−1, a higher irradiance level lead to a much more pronounced stimulation in PR abundance relative to both dark and more dimly illuminated cultures. At the salinity extremes, PR abundance was not affected by illumination. The results suggest PR abundance is controlled by salinity levels, and at normal seawater salinity levels or slightly above, illumination stimulates PR abundance.
Figure 5.
PR abundance under varying light and salinity levels. Normalized spectral count of PR protein detected by 1D-liquid Chromatography (LC)/MS showed different level of PR protein at different salinity and light conditions (μmol photons m−2 s−1 (PPF)).
Discussion
PR-containing prokaryotes constitute from 13–70% of prokaryotic cells in the marine ecosystem (Venter et al., 2004; Frigaard et al., 2006; Campbell et al., 2008) and thus have an important role in these ecosystems. The current hypothesis to explain the role of PR is that it provides a growth benefit during carbon starvation via H+ translocation-mediated cellular processes. However, in most cases tested so far, the presence of PR does not actually increase cell growth rates or yields. Fuhrman et al. (2008) suggested that PR might be used as a mechanism of survival under harsh conditions and that while there are only a few carbon-limited environments many other elements other than carbon can limit the growth of marine bacteria. This indicates that PR may have a broader range of physiological roles than previously reported, which lead us to hypothesize that PR may stimulate bacterial growth under stressful conditions, such as osmotic pressure. In this study, we observed light-stimulated growth under osmotic pressure rather than carbon limitation in P. torquis ATCC 700755. The results from growth experiments showed a stimulatory influence of PR, similar to that reported for Dokdonia sp. MED 134 (Kimura et al., 2011), on the growth yield of P. torquis under salt stress but not nutrient stress (Figure 1). We also observed that different light levels affected this growth response. According to our study, a light intensity of 3.7 μmol photons m−2 s−1 is most beneficial to the growth of P. torquis.
P. torquis inhabits sea ice and possesses fastidious growth requirements requiring yeast extract in the media to allow growth (Bowman et al., 1998). It is suspected the species is an algal epiphyte. Sea ice is a temporally transient and highly variable environment that experiences extremes of internal temperatures and salinity. It is, however, a relatively carbon rich environment especially for algal-associated bacteria. As the ice crystals form and move in the water column, they trap inorganic and organic materials on and between them, with the incorporation of silt, clay particles, grains, diatom and foraminferal detritus in the ice matrices (Grossmann and Dieckmann, 1994; Arrigo et al., 1997). Algal and bacterial cells that subsequently colonize and flourish in the ice matrices also synthesize copious amounts of exopolysaccharide (Reimnitz et al., 1993; Giannelli et al., 2001; Krembs et al., 2011). The salinity in sea-ice brine channels and pockets formed when salt is extruded during sea ice formation is very variable and can rise as high as 150 g l−1 or drop to <10 g l−1 when the sea ice melts (Thomas and Dieckmann, 2002). The optimum salinity for P. torquis is ∼35 g l−1 (Bowman et al., 1998) and we showed that when the salt concentrations were above or below this level greater growth was recorded in the samples grown under light compared with those incubated in the dark. This is a particularly interesting result as P. torquis is exposed to fluctuating salt levels in its sea-ice habitat. We hypothesize that PR may create a proton gradient allowing the cell to maintain better sodium homeostasis and PR may also contribute to compatible solute uptake or biosynthesis, the latter being a particularly energy-demanding process (Oren, 1999). Kimura et al. (2011) reported that Na+-translocating NADH-quinone oxidoreductase and Na+ transporters were greatly upregulated in Dokdonia sp. MED 134 when exposed to light, which suggests that a sodium gradient may have an essential function in light-enhanced growth in PR-containing marine bacteria and it is even possible that some rhodopsins may act as a sodium-pumps (Kwon et al., 2012). Our study focussed on the effect of salinity, therefore a Na+-transporter may be coupled to the functioning of PR in several ways. It may simply be involved in regulation of osmotic pressure or the Na+ gradient may be used to generate the motive force for the respiratory electron chain instead of just the a H+ gradient (Unemoto and Hayashi, 1993; Gonzalez et al., 2008). In sodium-rich environments, using a sodium-motive force may be more advantageous than using the proton-motive force to power energy transduction or, in some marine bacteria, the sodium-motive force couples with the proton-motive force (Kogure 1998; Albers et al., 2001). It is worth noting that P. torquis cultures incubated under bright light have lower cell abundance and growth rate than dim light treatments indicating that high light levels could be inhibitory and thus counteracting the benefits provided by PR. This could be due to photooxidative stress. However, cell abundance and growth rate still exceeded those found for the dark-incubated samples. This response to light intensity may result from P. torquis being adapted to relatively shaded conditions. In sea ice, photosynthetically available radiation can be very low, down to ∼1 μmol photon m−2 s−1 due to the ice-sheet thickness and obscuration of light due to sea-ice algal assemblages (McMinn et al., 2007).
In some studies, the expression of PR genes is influenced by light/dark conditions (Gomez-Consarnau et al., 2007; Lami et al., 2009). The recent study performed by Akram et al. (2013) suggested that PR expression can be regulated by nutrient limitation in Vibrio sp. AND4. However, other researchers reported in laboratory experiments that in vitro PR gene expression was constant regardless of illumination (Riedel et al., 2010). Our results also indicate that PR expression in P. torquis under different light, salinity and growth stages was expressed at a constant level (see Supplementary Table S1). Our qPCR results suggested that PR gene expression was not regulated either by light intensity or by salinity. High-throughput gel-free label-free proteomic analysis provided a new means to assess PR abundance and our 1D-liquid Chromatography/MS results provided strong evidence that PR abundance is affected by both salinity and illumination levels indicating that in P. torquis PR is likely to be post-transcriptionally regulated. However, responses are clearly complicated by interactions between salinity and illumination. Additionally, high levels of PR do not necessarily translate directly to higher cell abundance or growth rate.
Consistent with the protein abundance data, proton-pumping activity in P. torquis was also strongly affected by salinity (Figures 4 and 5). The data suggest P. torquis responds to light more strongly under osmotic pressure compared with the optimum salinity. However, we suspect there are distinct threshold salinities driving this response. Both PR protein level and proton pump activity increased with increasing salinity peaking at 52.5 g l−1 then dropped sharply at a salinity of 70 g l−1. The variations in activity could be indicative of different energy demands at different salinities.
To be fully functioning, PR requires retinal, a chromophore that binds to the final membrane domain of PR (Béjà et al., 2000, 2001). Retinal is derived from carotenoids, therefore PR is dependent on carotenoid synthesis (McCarren and DeLong, 2007). It can thus be proposed that the level of carotenoids and retinal derivatives can be an indirect indicator of PR abundance. For P. torquis, the results clearly show that at all salt concentrations the production of carotenoids was higher under light compared with dark incubation conditions (Figure 3). The production of carotenoid was associated with both light and salinity levels and the effect of illumination level were smaller than the effect of salinity. However, at 52.5 g l−1 light levels strongly affected the production of carotenoids (Figure 3c). Both light-driven proton pump activity and pigment data indicate that light has greater effect at 52.5 g l−1 than the other salinities tested and this corresponded to the large differences in PR abundances we observed using proteomic analysis (Figure 5).
We theorize that PR may represent an adaptation strategy used by microorganisms to their own specific econiche. This study provides evidence that PR promotes the growth of P. torquis in conditions of different salinity. Our proteomic data show that PR is regulated post-transcriptionally, influenced by both illumination and salinity. Thus, we have extended the existing paradigm that PR provides an alternative source of energy during nutrient stress also include osmotic stress. Furthermore, as many of kinds of stress responses lead to diversion of energy from growth and biosynthesis to maintenance and survival functions, we further suggest that PR may have a broader effect when marine prokaryotes encounter pH, temperature or other kinds of stress conditions.
Acknowledgments
This work was supported by Australian Antarctic Science Grant no. 3127, SEWPac (DEWHA). We thank Professor Tom McMeekin for critical discussion related to the manuscript. We also want to thank Associate/Professor Noel Davies for performing the HPLC analysis.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Akram N, Palovaara J, Forsberg J, Lindh MV, Milton DL, Luo H, et al. Regulation of proteorhodopsin gene expression by nutrient limitation in the marine bacterium Vibrio sp. AND4. Environ Microbiol. 2013;15:1400–1415. doi: 10.1111/1462-2920.12085. [DOI] [PubMed] [Google Scholar]
- Albers SV, Van de Vossenberg J, Driessen AJM, Konings WN. Bioenergetics and solute uptake under extreme conditions. Extremophiles. 2001;5:285–294. doi: 10.1007/s007920100214. [DOI] [PubMed] [Google Scholar]
- Arrigo KR, Worthen DL, Lizotte MP, Dixon P, Dieckmann GS. Primary production in Antarctic sea ice. Science. 1997;276:394–397. doi: 10.1126/science.276.5311.394. [DOI] [PubMed] [Google Scholar]
- Atamna-Ismaeel N, Sabehi G, Sharon I, Witzel KP, Labrenz M, Jurgens K, et al. Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2008;2:656–662. doi: 10.1038/ismej.2008.27. [DOI] [PubMed] [Google Scholar]
- Bowman JP, McCammon SA, Lewis T, Skerratt JH, Brown JL, Nichols DS, et al. Psychroflexus torquis gen. nov., sp. nov., a psychrophilic species from Antarctic sea-ice, and reclassification of Flavobacterium gondwanense (Dobson et al. 1993) as Psychroflexus gondwanense gen. nov., comb. nov. Microbiol. 1998;144:1601–1609. doi: 10.1099/00221287-144-6-1601. [DOI] [PubMed] [Google Scholar]
- Béjà O, Arvind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, et al. Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- Campbell BJ, Waidner LA, Cottrell MT, Kirchman DL. Abundant proteorhodopsin genes in the north atlantic ocean. Environ Microbiol. 2008;10:99–109. doi: 10.1111/j.1462-2920.2007.01436.x. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Béjà O. The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PloS Biol. 2010;8:e1000359. doi: 10.1371/journal.pbio.1000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard NU, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Schwalbach MS, Stingl U. Opinion—Proteorhodopsins: an array of physiological roles. Nat Rev Microbiol. 2008;6:488–494. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- Giannelli V, Thomas DN, Haas C, Kattner G, Kennedy H, Dieckmann GS. Behavior of dissolved organic matter and inorganic nutrients during experimental sea-ice fomation. Ann Glaciol. 2001;33:317–321. [Google Scholar]
- Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL, et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005;438:82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- Gomez-Consarnau L, Akram N, Lindell K, Pedersen A, Neutze R, Milton DL, et al. Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PloS Biol. 2010;8:e1000358. doi: 10.1371/journal.pbio.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Consarnau L, Gonzalez JM, Coll-Llado M, Gourdon P, Pascher T, Neutze R, et al. Light stimulates growth of proteorhodopsin -containing marine Flavobacteria. Nature. 2007;445:210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- Gonzalez JM, Fernandez-Gomez B, Fernandez-Guerra A, Gomez-Consarnau L, Sanchez O, Coll-Llado M, et al. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp MED152 (Flavobacteria) Proc Natl Acad Sci USA. 2008;105:8724–8729. doi: 10.1073/pnas.0712027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann S, Dieckmann GS. Bacterial standing stock, activity and carbon production during formation and growth of sea-ice in the weddell sea, Antarctica. Appl. Environ Microbiol. 1994;60:2746–2753. doi: 10.1128/aem.60.8.2746-2753.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerthey SE, Louda JW, Mongkronsri P. Evaluation of pigment extraction methods and a recommended protocol for periphyton chlorophyll a determination and chemotaxonomic assessment. J Phycol. 2006;42:1125–1136. [Google Scholar]
- Kimura H, Young CR, Martinez A, DeLong EF. Light-induced transcriptional responses associated with proteorhodopsin-enhanced growth in a marine flavobacterium. ISME J. 2011;5:1641–1651. doi: 10.1038/ismej.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure K. Bioenergetics of marine bactera. Curr Opin Biotechnol. 1998;9:278–282. doi: 10.1016/s0958-1669(98)80059-1. [DOI] [PubMed] [Google Scholar]
- Koh EY, Atamna-Ismaeel N, Martin A, Cowie ROM, Béjà O, Davy SK, et al. Proteorhodopsin-bearing bacteria in antarctic sea-ice. Appl Environ Microbiol. 2010;76:5918–5925. doi: 10.1128/AEM.00562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krembs C, Eicken H, Deming JW. Exopolymer alteration of physical properties of sea-ice and implications for ice habitability and biogeochemistry in a warmer Arctic. Proc Natl Acad Sci USA. 2011;108:3653–3658. doi: 10.1073/pnas.1100701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SK, Kim BK, Song JY, Kwak MJ, Lee CH, Yoon JH, et al. Genomic makeup of the marine Flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of rhodopsins. Genome Biol Evol. 2012;5:187–199. doi: 10.1093/gbe/evs134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lami R, Cottrell MT, Campbell BJ, Kirchman DL. Light-dependent growth and proteorhodopsin expression by Flavobacteria and SAR11 in experiments with Delaware coastal waters. Environ Microbiol. 2009;1:3201–3209. doi: 10.1111/j.1462-2920.2009.02028.x. [DOI] [PubMed] [Google Scholar]
- McCarren J, DeLong EF. Proteorhodopsin photosystem gene clusters exhibit co-evolutionary trends and shared ancestry among diverse marine microbial phyla. Environ Microbiol. 2007;9:846–858. doi: 10.1111/j.1462-2920.2006.01203.x. [DOI] [PubMed] [Google Scholar]
- McMinn A, Ryan KG, Ralph PJ, Pankowski A. Spring sea-ice photosynthesis, primary productivity and biomass distribution in eastern Antarctica, 2002–2004. Mar Biol. 2007;151:985–995. [Google Scholar]
- Michelou VK, Cottrell MT, Kirchman DL. Light-stimulated bacterial production and amino acid assimilation by cyanobacteria and other microbes in the North Atlantic Ocean. Appl Environ Microbiol. 2007;73:5539–5546. doi: 10.1128/AEM.00212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev. 1999;63:334. doi: 10.1128/mmbr.63.2.334-348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimnitz E, Clayton JR, Kempema EW, Payne JR, Weber WS. Interaction of rising frazil with suspended particles–tank experiments with applications to nature. Cold Reg Sci Technol. 1993;21:117–135. [Google Scholar]
- Riedel T, Tomasch J, Buchholz I, Jacobs J, Kollenberg M, Gerdts G, et al. Constitutive expression of the proteorhodopsin gene by a Flavobacterium strain representative of the proteorhodopsin-producing microbial community in the North Sea. Appl Environ Microbiol. 2010;76:3187–3197. doi: 10.1128/AEM.02971-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabehi G, Beja O, Suzuki MT, Preston CM, DeLong EF. Different SAR86 subgroups harbour divergent proteorhodopsins. Environ Microbiol. 2004;6:903–910. doi: 10.1111/j.1462-2920.2004.00676.x. [DOI] [PubMed] [Google Scholar]
- Sabehi G, Loy A, Jung KH, Partha R, Spudich JL, Isaacson T, et al. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PloS Biol. 2005;3:1409–1417. doi: 10.1371/journal.pbio.0030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalbach MS, Brown M, Fuhrman JA. Impact of light on marine bacterioplankton community structure. Aquat Microb Ecol. 2005;39:235–245. [Google Scholar]
- Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. Energy starved candidatus pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PloS One. 2011;6:e19725. doi: 10.1371/journal.pone.0019725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl U, Desiderio RA, Cho JC, Vergin KL, Giovannoni SJ. The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl Environ Microbiol. 2007;73:2290–2296. doi: 10.1128/AEM.02559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DN, Dieckmann GS. Antarctic sea-ice—a habitat for extremophiles. Science. 2002;295:641–644. doi: 10.1126/science.1063391. [DOI] [PubMed] [Google Scholar]
- Unemoto T, Hayashi M. Na+-translocating NADH-quinone reductase of marine and halophilic bacteria. J Bioenerg Biomembr. 1993;25:385–391. doi: 10.1007/BF00762464. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelber JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Walter JM, Greenfield D, Bustamante C, Liphardt J. Light-powering Escherichia coli with proteorhodopsin. Proc Natl Acad Sci USA. 2007;104:2408–2412. doi: 10.1073/pnas.0611035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa S, Kawanabe A, Ito H, Kandori H, Kogure K. Diversity and functional analysis of proteorhodopsin in marine Flavobacteria. Environ Microbiol. 2012;14:1240–1248. doi: 10.1111/j.1462-2920.2012.02702.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.