Abstract
The massive influx of crude oil into the Gulf of Mexico during the Deepwater Horizon (DWH) disaster triggered dramatic microbial community shifts in surface oil slick and deep plume waters. Previous work had shown several taxa, notably DWH Oceanospirillales, Cycloclasticus and Colwellia, were found to be enriched in these waters based on their dominance in conventional clone and pyrosequencing libraries and were thought to have had a significant role in the degradation of the oil. However, this type of community analysis data failed to provide direct evidence on the functional properties, such as hydrocarbon degradation of organisms. Using DNA-based stable-isotope probing with uniformly 13C-labelled hydrocarbons, we identified several aliphatic (Alcanivorax, Marinobacter)- and polycyclic aromatic hydrocarbon (Alteromonas, Cycloclasticus, Colwellia)-degrading bacteria. We also isolated several strains (Alcanivorax, Alteromonas, Cycloclasticus, Halomonas, Marinobacter and Pseudoalteromonas) with demonstrable hydrocarbon-degrading qualities from surface slick and plume water samples collected during the active phase of the spill. Some of these organisms accounted for the majority of sequence reads representing their respective taxa in a pyrosequencing data set constructed from the same and additional water column samples. Hitherto, Alcanivorax was not identified in any of the previous water column studies analysing the microbial response to the spill and we discuss its failure to respond to the oil. Collectively, our data provide unequivocal evidence on the hydrocarbon-degrading qualities for some of the dominant taxa enriched in surface and plume waters during the DWH oil spill, and a more complete understanding of their role in the fate of the oil.
Keywords: biodegradation, Deepwater Horizon, Gulf of Mexico, marine bacteria, polycyclic aromatic hydrocarbons, stable-isotope probing
Introduction
The Deepwater Horizon (DWH) blowout is one of the worst maritime oil spills on record. An estimated 4.1 million barrels of crude oil entered the Gulf of Mexico over a period of 84 days (20 April to 15 July 2010) (McNutt et al., 2011). One distinctive feature that set this spill apart from other historic oil spill incidents at sea was the duration of the leak and depth at which it occurred (1500 m below the sea surface). Mass spectrometric and fluorescence vertical profiling of the water column revealed a spatial and temporal distribution of dissolved and dispersed saturated and aromatic hydrocarbons at discrete depths within the water column. Initial coloured dissolved organic matter maxima measurements at depths between 1000 to 1200 m indicated the formation of a deep water oil plume in early May 2010 (Diercks et al., 2010), which was characterised by local oxygen depletion, elevated concentrations of low-molecular weight polycyclic aromatic hydrocarbons (PAHs), and methane (Camilli et al., 2010; Dierks et al., 2010; Wade et al., 2011). Higher molecular weight PAHs were found to predominate at the sea surface (Camilli et al., 2010; Diercks et al., 2010; Hazen et al., 2010), while high concentrations of n-alkanes were measured in both surface and plume waters (Hazen et al., 2010). In late June 2010, the plume was found to stretch up to 35 km on a southwest-trending horizontal transect (Camilli et al., 2010). The unique nature of the plume had diverted much of the focus on the spill to tracking its movement and succession of its microbial community.
Most of the published literature that interrogated the water column microbial community response to the DWH spill utilised gene sequencing approaches, such as PCR, clone library construction and sequencing of 16S rRNA genes (Hazen et al., 2010; Valentine et al., 2010; Redmond and Valentine, 2012). This revealed an initial enrichment of a specific cluster within the Oceanospirillales—termed DWH Oceanospirillales—that was found to dominate plume water samples and constituted up to 90% of bacterial 16S rRNA gene clone libraries (Redmond and Valentine, 2012) and pyrosequence libraries (T Yang, L Nigro, T Gutierrez, S Joye, R Highsmith, A Teske; submitted for publication) in late May 2010 (ca. 40 days after the spill) and, to a lesser extent, other groups of Gammaproteobacteria. Dominance of DWH Oceanospirillales in the plume was succeeded by members affiliated to Colwellia and Cycloclasticus by early June 2010 (52–62 days after the onset of the spill) (Valentine et al., 2010). This succession from aliphatic hydrocarbon-degrading bacteria (DWH Oceanospirillales) to those with an apparent nutritional preference for utilising aromatic hydrocarbons (Cycloclasticus and to a lesser extent Colwellia) is reminiscent of the microbial community dynamics observed in seawater following enrichment with crude oil (Syutsubo et al., 2001; Röling et al., 2002; Kasai et al., 2002a, 2002b; Röling et al., 2004; Teira et al., 2007). These studies also revealed the enrichment of several other bacterial groups in the water column that included Halomonas, Alteromonas and Pseudoalteromonas (Hazen et al., 2010; Valentine et al., 2010). Conjecture surrounding a role for these plume-enriched taxa in the degradation of the oil during the spill has largely been based on their phylogenetic affiliation to published hydrocarbon-degrading representative strains. Isolation and genome sequencing of DWH Oceanospirillales single cells from deep-sea plume water at DWH revealed genes involved in the degradation of n-alkanes and cycloalkanes (Mason et al., 2012), which is indicative of the hydrocarbon-degrading potential for members of the Oceanospirillales. Hitherto, only a Colwellia strain, RC25, that was isolated from a reportedly uncontaminated deep water sample (at plume depth) near the DWH site, was confirmed by empirical means to be capable of degrading hydrocarbons (Bælum et al., 2012). Compelling evidence that adequately demonstrates the capacity of the various taxa enriched during the spill to degrade hydrocarbons is therefore lacking.
One method that circumvents the requirement to isolate microorganisms in order to assess their metabolic and physiological characteristics is stable-isotope probing (SIP). This method has been used successfully on environmental samples to identify a target microbial group(s) based on their ability to perform a specific metabolic process, thereby being able to link the phylogenetic identity of an organism to its function (Dumont and Murrell, 2005). A major advantage of this technique is its ability to identify target members of a microbial community that are not amenable to cultivation in the laboratory. The use of SIP with hydrocarbon substrates has been scarcely applied to ocean systems, and to the best of our knowledge, the only reported study to have applied SIP to examine hydrocarbon-degrading microbial groups in seawater samples collected during the DWH spill was on bacteria contributing to the degradation of volatile low-molecular weight hydrocarbons (Redmond and Valentine, 2012). A recent study by Gutierrez et al. (2011) employed DNA-based SIP (DNA-SIP) to identify uncultivated taxa associated with low-molecular weight PAH degradation in seawater samples and was the first SIP experiment to identify PAH-degrading bacteria in marine water samples. Here, we used DNA-SIP with uniformly 13C-labelled naphthalene, phenanthrene and n-hexadecane in order to link the phylogenetic identity of bacterial taxa that were present in surface slicks and plume water samples, with their ability to mineralise these substrates. We also performed enrichment experiments with both surface and plume water samples collected during the spill to isolate hydrocarbon-degrading bacteria. 16S rRNA gene clone and pyrosequence libraries, representing total microbial community ‘snap-shots' of contaminated surface seawater and plume waters, were interrogated with the SIP-identified sequences and sequences of isolated strains to reveal their abundance or enrichment during the spill compared with reference uncontaminated water samples, as well as infer their possible contribution to the overall biodegradation of the oil.
Materials and methods
Field samples
During a research cruise on the RV Pelican on 5 May 2010, oil-contaminated water samples were collected from the sea surface near the site of the DWH blowout (28°4.175′ N, 88°22.335′ W) in the Gulf of Mexico. On three follow-up cruises on the RV Walton Smith (31 May 2010), RV Pelican (12 September 2010) and RV Cape Hatteras (18 October 2010), additional water samples were collected from the water column. Sample names and data relating to the respective sampling locations are shown in Supplementary Table S1. At the time of collection, fractions of the samples were filtered onto 0.2-μm pore-size Anodisc or Nucleopore filters and stored frozen for subsequent DNA extraction and barcoded amplicon pyrosequencing, as described later. The remaining volumes of each sample were immediately stored at 4 °C for subsequent use within 2 weeks in enrichment, mineralisation, degradation and SIP experiments (described below).
Enrichment experiments and isolation
Oil-contaminated surface and plume water samples—respectively, PE5 and the B3+B6 samples (Supplementary Table S1)—from the Gulf of Mexico oil spill were used to isolate hydrocarbon-degrading bacteria. Samples (5 μl) of the surface water (PE5) were streaked directly onto ONR7a agar (Dyksterhouse et al., 1995) plates that were then sprayed with phenanthrene, anthracene, pyrene or fluorene (St Louis, MO, USA) dissolved in acetone (ca. 5% w/v) as the sole source of carbon and energy (Kiyohara et al., 1982). Plume water samples (B3+B6) were streaked on ONR7a agar plates supplemented with n-hexadecane (1% v/v) (Acros Organics, NJ, USA) as the sole source of carbon and energy. During the preparation of this medium, the n-hexadecane was added directly to the ONR7a agar while it was still in liquid form after autoclaving. Agar plates were stored in closed plastic bags in the dark at room temperature. Colonies forming clearing zones were picked and subcultured onto fresh ONR7a agar medium amended with the respective PAH until pure cultures were obtained prior to storage in glycerol (30% v/v) at −80 °C.
Enrichments with plume water (B3+B6) were prepared in autoclaved glass test tubes containing 3 ml of liquid ONR7a supplemented with n-hexadecane (5% v/v; ONR+HEX). The tubes were incubated in the dark with shaking (100 r.p.m.) at 21°C. After 1 week, the highest dilutions that yielded growth were selected for dilution-to-extinction culturing in ONR+HEX. After repeating this process four more times, samples (5 μl) from the highest dilutions producing growth were streaked onto ONR+HEX agar plates. The plates were stored as above and after 2 weeks, colonies showing distinct colony morphologies were picked, purified and stored frozen in 30% (v/v) glycerol.
The potential for the strains to degrade phenanthrene or naphthalene was determined using a quantitative spectrophotometric method. For this, acid-washed (0.1 N HCl) steam-sterilised glass test tubes (13 × 100 mm), fitted with screw-caps lined with Teflon-lined silicone septa (Chromacol, Herts, UK) were used. Stock solutions of phenanthrene (ca. 3000 mg l−1) and naphthalene (ca. 9000 mg l−1) dissolved in acetone were prepared. For each strain, two sets of three test tubes were prepared, each containing 2.9 ml of ONR7a medium (Dyksterhouse et al. 1995). To one set of tubes, phenanthrene was added, whereas naphthalene was added to the second set. All six test tubes were then inoculated with 100 μl of the test strain. Uninoculated controls, acid-killed controls and tubes that were inoculated, but without any added PAH, were also prepared. All test tubes were incubated in the dark with gentle shaking (100 r.p.m.) at 21 °C. PAH degradation was determined spectrophotometrically. For this, the test tubes from each PAH incubation were sacrificed after 14 days for extraction with ethyl acetate (high-pressure liquid chromatography (HPLC) grade). This was performed by adding 2 ml of ethyl acetate to each tube and then vortexing for 30 s. Aliquots of the non-aqueous top layer were diluted with ethyl acetate in quartz cuvettes for spectrophotometric analysis at 251 nm for phenanthrene and 275 nm for naphthalene. The A251 and A275 values were converted to concentrations of phenanthrene and naphthalene, respectively, using the molar absorptivity coefficients of 6.3 × 104 and 5.7 × 103 l mol−1 cm−1 (Thomas and Burgess, 2007). A significant decrease (P<0.05) in the PAH concentration measured in the inoculated test tubes, relative to the uninoculated controls, was indicative of degradation.
The ability of the strains to grow on n-hexadecane as the sole carbon and energy source was evaluated in 250-ml Erlenmeyer flasks containing 50 ml of ONR7a medium and supplemented with the hydrocarbon to 0.5% (v/v). Inocula used were cells grown in ONR7a amended with Na-pyruvate (0.1% w/v) and washed three times prior to use. Growth was measured spectrophotometrically by taking measurements of the culture medium periodically at an optical density of 600 nm. An increase in optical density of at least five-fold was indicative of growth on n-hexadecane.
Mineralisation assays
Mineralisation assays using 14C-labelled naphthalene, phenanthrene, anthracene, pyrene, fluoranthene, benz[a]anthracene and n-hexadecane were performed on the collected water samples to determine which of these hydrocarbons could be used in SIP incubations with their 13C-labelled counterparts. [U-14C] Naphthalene (17.8 mCi mmol−1), [9-14C] phenanthrene (8.3 mCi mmol−1), [1, 2, 3, 4, 4a ,9a-14C] anthracene (17.3 mCi mmol−1), [4, 5, 9, 10-14C] pyrene (61 mCi mmol−1), [3-14C] fluoranthene (45 mCi mmol−1) and [1-14C] n-hexadecane (7.5 mCi mmol−1) were from Sigma-Aldrich (St Louis, MO, USA). [5,6-14C] Benz[a]anthracene (54.6 mCi mmol−1) was obtained from Chemsyn Science Laboratories (Lenexa, KS, USA). These experiments were conducted at both 4 °C and 21 °C to evaluate the effect of temperature on the mineralisation of the hydrocarbons. For this, sterile 40-ml amber-glass EPA vials were prepared, each containing a 14C-labelled test compound (to 20 000 d.p.m.) and 2.5 μg of the respective unlabelled test compound in 4.5 ml of ONR7a medium. For the inoculum, 0.5 ml of water sample was inoculated into the vials. Killed controls were prepared by adding 85% phosphoric acid to pH of <1 prior to inoculation. All treatments were conducted in triplicate. For the CO2 trap, a sterile glass test tube (12 × 75 mm) containing a piece of filter paper saturated with 60 μl of 2 M KOH was inserted into each vial. The vials were sealed with foil-covered Teflon-lined caps and incubated with shaking (100 r.p.m.) at 4 °C or 21 °C. The filter paper from each vial was removed daily and the captured 14C from any 14CO2 respired was counted on a Packard (Meriden, CT, USA) Tri-Carb liquid scintillation analyser (model 1900TR). The KOH-saturated filter paper from each vial was replaced at each sampling point for the course of the experiment. The percentage of 14C mineralised for each compound was calculated by subtracting the triplicate values for the acidified controls from those of the experimental and then dividing by the total d.p.m. of 14C added.
SIP incubations
SIP incubations were performed using 125-ml sterilised glass screw-top Erlenmeyer flasks with caps that were lined with aluminium foil to prevent sorption of hydrocarbons. ONR7a medium was used in these incubations in order to prevent the introduction of exogenous and potentially bioavailable sources of carbon, such as in the form of dissolved organic carbon or dissolved organic matter that are inherent to seawater and that could potentially redirect microbial activity away from the labelled substrates. For incubations utilising the surface water inoculum (PE5), each flask contained 18 ml of ONR7a medium, 1 mg of labelled (14C or 13C) and/or unlabelled substrate (naphthalene or phenanthrene) and 2 ml of inoculum. [U-13C] naphthalene and [U-13C] phenanthrene were synthesised by methods described elsewhere (Zhang et al., 2011). For incubations with the deep-water plume inoculum (B3+B6), each flask contained 15 ml of ONR7a medium, 1 mg of labelled (14C or 13C) and/or unlabelled n-hexadecane, and 5 ml of inoculum. [U-13C] n-hexadecane was from Sigma-Aldrich. For SIP, duplicate flasks were prepared with 1 mg of uniformly 13C-labelled hydrocarbon ([U-13C] naphthalene, [U-13C] phenanthrene, or [U-13C] n-hexadecane), and a second set of duplicates was prepared with 1 mg of unlabelled hydrocarbon. To determine the endpoint of each SIP experiment, the mineralisation of the respective [U-14C]-labelled hydrocarbon was measured in triplicate flasks by liquid scintillation counting of 14CO2 trapped in KOH-soaked filter paper over time, as described above. An additional set of triplicate flasks was used to monitor the disappearance of unlabelled hydrocarbon by HPLC for naphthalene and phenanthrene, or by gas chromatography–mass spectrometry (GC–MS) for n-hexadecane. Samples were periodically taken from these flasks for DNA extraction and subsequent measurement of the abundance of target organisms identified through SIP. Triplicate flasks of acid-killed controls (pH<2) containing unlabelled hydrocarbon were prepared by adding ca. 700 μl of 85% phosphoric acid. All flasks were incubated on an orbital shaker (250 r.p.m.; 21 °C) in the dark. At the endpoint of each SIP incubation—defined as the time when the extent of mineralisation of the respective 14C-labelled substrate began to approach an asymptote—whole DNA from the total volume in the paired flasks amended with the [U-13C] hydrocarbon and the corresponding paired set with unlabelled hydrocarbon was extracted using the method of Tillett and Neilan (2000).
CsCl gradient ultracentrifugation and identification of 13C-enriched DNA
To separate 13C-enriched and unenriched DNA, total extracted DNA from each sample (20–30 μl) was added to caesium chloride (CsCl) solutions (1.72 g ml−1) for isopycnic ultracentrifugation and gradient fractionation, as previously described (Jones et al., 2011). Five microlitres of purified Escherichia coli DNA (ca. 40 ng μl−1) was added and mixed into each tube as an internal standard of unlabelled DNA prior to ultracentrifugation. Denaturing gradient gel electrophoresis (DGGE) was then performed on each fraction to visualise the separation of DNA. For this, PCR amplification of each fraction was carried out with primers 63f-GC (Marchesi et al., 1998) and 517r (Muyzer et al., 1993) using a PCR programme as described by Yu and Morrison (2004). PCR products were confirmed on a 1.5% (w/v) agarose gel alongside a HindIII DNA ladder (Invitrogen, Carlsbad, CA, USA). DGGE was performed using 6.5% acrylamide gels containing a denaturant range of 30–60% (100% denaturant contains 7.0 M urea and 40% molecular-grade formamide). After electrophoresis for 16 h at 60 °C and 60 V, gels were stained with ethidium bromide at 1:25 000 dilution for 15 min. Gel image colours were inverted, adjusted for contrast, and cropped to only the regions displaying bands with the GNU Image Manipulation Program (GIMP; version 2.6.8).
16S rRNA gene libraries of 13C-enriched DNA
16S rRNA clone libraries, each comprising 96 clones, were prepared from combined fractions containing the 13C-enriched DNA from each of the three SIP incubations using general eubacterial primers 27f and 1492r and partially sequenced using primer 27f (Wilmotte et al., 1993) at the Beckman Coulter Genomics sequencing facility (Danvers, MA, USA). The 13C-enriched heavy DNA fractions were selected based on the DGGE evidence, which is discussed below. After excluding vector sequences, poor-quality reads and chimeras, the clone sequences were grouped into operational taxonomic units (OTUs) based on applying a 97% sequence identity cutoff. Using the complete linkage clustering and dereplicate tools available at the Pyrosequencing Pipeline tool of RDP-II (Cole et al., 2009), representative sequences were selected to represent dominant OTUs identified in each of the libraries. Near-complete 16S rRNA gene sequences for the represented sequences were obtained at the University of North Carolina-Chapel Hill Genome Analysis Facility. Sequences were edited and assembled using the programme Sequencher 4.8 (Gene Codes Corp., Ann Arbor, MI, USA). The BLASTn search programme and RDP-II (Maidak et al., 1999) were used to check for close relatives and phylogenetic affiliation.
Real-time quantitative PCR
To quantify sequences in the dominant OTUs, primers for real-time quantitative PCR (qPCR) were developed using the Probe Design and Probe Match tools of ARB, as previously described (Gutierrez et al., 2011). Primer specificity was confirmed with the Probe Check tool of RDP-II. The optimal annealing temperature of each primer pair was determined using an Eppendorf (Hauppauge, NY, USA) Mastercycler gradient thermal cycler. The template for these reactions, and for the construction of respective standard curves for quantitative PCR, was a plasmid containing a representative sequence that had been linearised using PstI (New England BioLabs, Ipswich, MA, USA) and purified using the QIAquick nucleotide removal kit (Qiagen, Valencia, CA, USA). The primer pairs, their amplification efficiency (Pfaffl, 2001), optimal annealing temperature, detection limit and RDP hits are shown in Table 1. To confirm the fractions from the DGGE profiles that corresponded to unlabelled DNA, E. coli primers ECP79f (5′-GAAGCTTGCTTCTTTGCT-3′) and ECR620r (5′-GAGCCCGGGGATTTCACA-3′) were used to quantify the abundance of the E. coli 16S rRNA genes in each fraction. The qPCR programme for using these primers used an annealing temperature of 55 °C and an extended extension step of 45 s (Sabat et al., 2000).
Table 1. Quantitative PCR primers developed and used in this study.
| Target OTU | Primer name | Primer sequence (5′→3′) | Tm(°C)a | qPCR standardb | Amplicon length | Amp. Eff.c | Detect.d | RDPe |
|---|---|---|---|---|---|---|---|---|
| 12 | Alt-71f | GAACTTGCCTTTGCGAGG | 60 | SWNAP06 | 94 | 1.97 | 3 | 1690 |
| Alt-163r | TTTGCGCCGGAGCCKAAG | 1465 (1505) | ||||||
| 1, 13 | Cyc-412f | ACCTTAGGCCCTGACGTT | 60 | SWNAP12 | 127 | 1.95 | 7 | 46 |
| Cyc-537r | GTTTAACCGCCTACGCGC | 44 (41) | ||||||
| 2 | Col-541f | CAAGATGTGAAAGCCCTG | 60 | SWPHE03 | 155 | 1.71 | 66 | 115 |
| Col-694r | AGTCTTTGTCCAGGTAGCC | 96 (81) | ||||||
| 15 | Alv-66f | CGTGAGAATCTGCCCATT | 59 | DWHEX05 | 94 | 1.91 | 55 | 332 |
| Alv-158r | CTCATCCATCAGCACAAG | 165 (150) | ||||||
| 16 | Mar-25f | AGCTTGCTCCCCGCTGAC | 58 | DWHEX95 | 59 | 1.95 | 6 | 182 |
| Mar-83r | ACTGGGCAGTTTCCTATG | 465 (158) |
Empirically determined PCR annealing temperature.
Representative clone sequences from which plasmid DNA was used to generate standard curves. Each plasmid was linearised with PstI. Names are as in Figures 1 and 3.
Amp. Eff., amplification efficiency (Pfaffl, 2001) with OTU-specific primers.
Detection limit of each qPCR assay expressed as number of 16 S rRNA gene copies per ml of culture.
Number of sequences returned by the Ribosomal Database Project II release 10.18 (Cole et al., 2009) (excluding sequences from this study) with no mismatches to primer pairs. Values in parentheses are the total hits that each pair of primers target.
Purified DNA from time-series incubations with unlabelled hydrocarbon was quantified using a NanoDrop ND-3300 fluorospectrometer (Thermo, Waltham, MA, USA) and the Quant-iT Picogreen double-stranded DNA (dsDNA) kit (Invitrogen). As duplicates of the separated 12C- and 13C-labelled incubations for each of the three SIP incubations displayed similar distributions of DNA in the fractions, as well as similar DGGE profiles, only the replicate incubation whose fractions contained the highest total amount of DNA, was used for further analyses. SIP-identified sequences were quantified in each separated SIP fraction using a single reaction by qPCR, as described previously (Singleton et al., 2006). Single reactions were performed on each triplicate DNA extraction (from triplicate samples) from the time series containing unlabelled hydrocarbon.
Barcoded amplicon pyrosequencing
DNA was extracted from surface (PE5) and water column (B1, B3, B6, B11) frozen samples using the method of Tillett and Neilan (2000), and barcoded 16S rRNA gene pyrosequencing was performed to analyse the bacterial community of these samples. Ten-fold dilutions of extracted DNA in water were used as a template for triplicate PCR reactions for each sample. Primer pairs 27f and 338r were modified to incorporate an identical 8-base-pair (bp) barcode sequence that was unique to this sample and a 2-bp spacer on the 5′-end of the primer sequence (Hamady et al., 2008). Each 20-μl PCR reaction was run for 25 cycles of 94 °C for 45 s, 55 °C for 45 s and 72 °C for 1 min on the Eppendorf Mastercycler Gradient thermal cycler before verification of the proper amplicon size on a 1% agarose gel. The triplicate reactions were pooled and purified with a QIAquick PCR Purification Kit (Qiagen) and eluted in 30 μl of 10 mM Tris-Cl (pH 8.5) buffer. The DNA concentration of pooled amplicons was then measured using a NanoDrop ND-3300 Fluorospectrometer (Thermo) and Quant-iT Picogreen dsDNA Kit (Invitrogen) prior to combining into a single sample (containing barcoded amplicons from other studies) at a concentration suitable for pyrosequencing. The sample was submitted to the High-Throughput Sequencing Facility at the University of North Carolina-Chapel Hill for sequencing using the 454 Life Sciences Titanium platform (Roche Diagnostics, Branford, CT, USA).
Pyrosequencing reads were trimmed and filtered using the LUCY programme with a minimum PHRED score of 27.5 and minimum length of 200 nt to remove low-quality regions and short reads (Kunin et al., 2010). Reads were de-multiplexed based on an 8-nt barcode identifier and the primer and barcode regions were removed using QIIME (Caporaso et al., 2010). To form OTUs, the reads were clustered at 97% sequence identity with UCLUST (Edgar, 2010) and the most abundant unique read within each cluster was used as its representative sequence. Representative sequences with at least 97% sequence identity to the isolated strains and SIP-identified sequences from this study were identified using BLAST (Altschul et al., 1990).
Phylogenetic tree
The 16S rRNA sequences of the isolated strains and SIP-identified sequences were aligned and manually curated in ARB (Ludwig et al., 2004). The sequences and type strains (from the Living Tree Project) (Yarza et al., 2010) with the highest similarity from the SILVA SSU NR 111 NR database (Quast et al., 2013) were also used for tree construction. A neighbour-joining tree was constructed with Jukes–Cantor correction and bootstrapped replication (n=1000) and Akkermansia muciniphila (CP001071) and Thalassospira xiamenensis (AY189753) were used as an outgroup.
Nucleotide sequence accession numbers
The following accession numbers were submitted to GenBank for 13C-enriched DNA in SIP experiments with phenanthrene (KC242316), naphthalene (KC242318, KC242319) and n-hexadecane (KC242317, KC242320). Accession numbers for the isolated strains are as follows: Alcanivorax sp. strain TY4 (JX467655), Alcanivorax sp. strain TY5 (JX467656), Alcanivorax sp. strain TY6 (JX467657), Alcanivorax sp. strain TK-23 (KC161579), Alteromonas sp. strain TK-46(2) (KC161583), Cycloclasticus sp. strain TK-8 (KC161584), Halomonas sp. strain GOS-2 (JQ246430), Halomonas sp. strain GOS-3a (JQ246431), Halomonas sp. strain TGOS-10 (JQ246432), Marinobacter sp. strain TT1 (JX467654), Marinobacter sp. strain TK-36 (KC161582) and Pseudoalteromonas sp. strain TK-105 (KC161577).
Results
Isolation of hydrocarbon-degrading bacteria
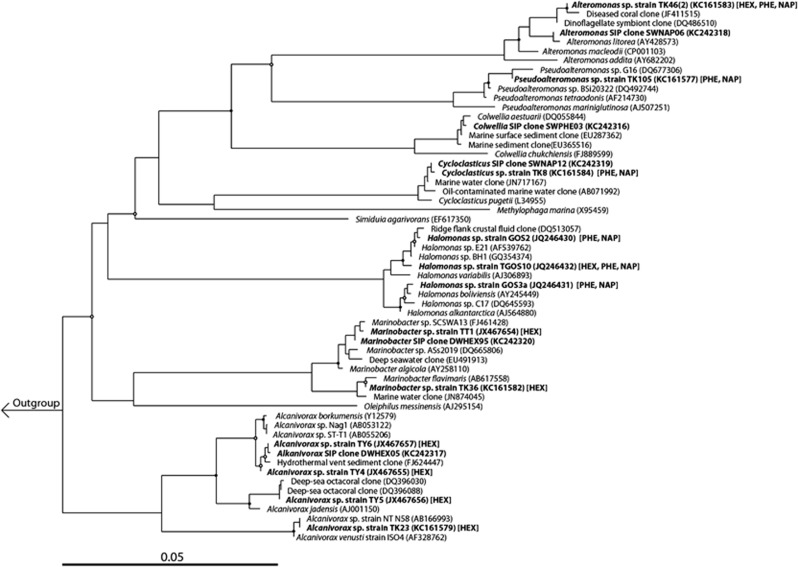
Hydrocarbon-degrading bacteria were isolated from contaminated surface and plume water samples collected during the Gulf oil spill with and without enrichment on phenanthrene, naphthalene or n-hexadecane. The near-complete 16S rRNA gene sequence (>1400 bp) of each of these isolates was used to construct a phylogenetic tree with related sequences (Figure 1). The tree also presents the potential of the isolates to utilise phenanthrene, naphthalene and/or n-hexadecane as a sole source of carbon and energy for growth. Surface water samples that were plated on agar plates and sprayed with anthracene or pyrene did not yield colonies that formed clearing zones. Small inconspicuous colonies with faint clearing zones appeared on plates sprayed with fluorene; however, these organisms proved difficult to culture as colonies did not grow out upon further subculturing. Our attempt to isolate a member of the Oceanospirillales from the plume, in order to interrogate its nutritional profile for various hydrocarbon substrates, was not successful.
Figure 1.
Phylogenetic tree of SIP clones and isolated strains from surface and plume waters. SIP clones and isolates are shown in bold along with the highest similarity sequences and type strains (Yarza et al., 2010) from the SILVA SSU NR 111 NR database (Quast et al., 2013). A neighbor-joining tree was constructed with Jukes–Cantor correction and bootstrap replication (n=1000). Nodes with bootstrap support of at least 65% (○) and 90% (•) are marked. Accession numbers of all sequences and the hydrocarbon(s) utilised (NAP, naphthalene; PHE, phenanthrene; HEX, n-hexadecane) by the isolated strains are given in parentheses. Akkermansia muciniphila (CP001071) and Thalassospira xiamenensis (AY189753) were used as the outgroup. The scale bar indicates the difference of number of substitutions per site.
Exposure of surface and plume water samples to labelled and unlabelled hydrocarbons
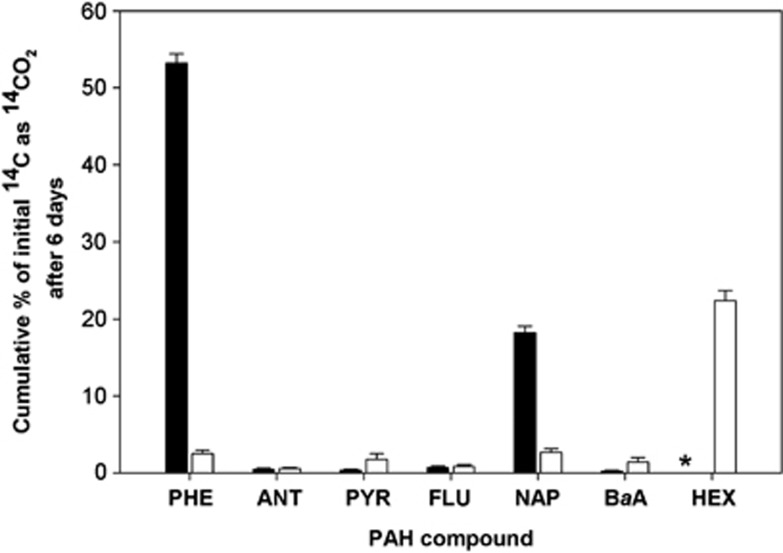
Experiments were conducted to determine the potential of the bacterial community in the surface and plume water samples to mineralise various 14C-labelled hydrocarbons (PHE, ANT, PYR, FLU, NAP, BaA, or HEX) and thereby inform our choice of hydrocarbons that would be most suitable to commence SIP experiments. 14C-hydrocarbon incubations conducted at 4 °C with each of the seven hydrocarbons and water samples yielded very low levels of mineralisation (data not shown) compared with replicate incubations conducted at 21 °C (Figure 2). As shown in Figure 2, 14C incubations using the surface water as inoculum revealed that significant levels of phenanthrene and naphthalene had been mineralised (respectively, 53.2±1.2% and 18.2±0.9% cumulative 14CO2 captured of total initial 14C), whereas low mineralisation levels (<3% cumulative 14CO2 captured of total initial 14C) were measured for ANT, PYR and FLU. Hexadecane was not evaluated with surface water. Conversely, low mineralisation levels were measured for these hydrocarbons by the plume water sample, except for HEX for which 22.4±1.3% of cumulative 14CO2 was captured of total initial [14C] n-hexadecane. For SIP, achieving sufficient incorporation of the labelled carbon under short incubation times is desirable. Hence, based on the significant mineralisation of PHE and NAP measured with the surface water sample, and of HEX with the plume water sample, SIP experiments were subsequently conducted using these combinations of hydrocarbons and water samples.
Figure 2.
Cumulative 14CO2 recovered from incubations with [14C] phenanthrene (PHE), anthracene (ANT), pyrene (PYR), fluoranthene (FLU), naphthalene (NAP), benz[a]anthracene (BaA), or n-hexadecane (HEX) by surface (solid bar) and plume (open bar) water during incubation at 21 °C. Bars are the averages and standard deviation from triplicate incubations. *, no data (surface water was not incubated with n-hexadecane).
The degradation and mineralisation of phenanthrene, naphthalene or n-hexadecane in flasks containing unlabelled or 14C-labelled hydrocarbons incubated in parallel to the respective SIP experiments is shown in Supplementary Figure S1. In the SIP experiments with surface water, complete removal of phenanthrene or naphthalene (respectively, Supplementary Figures S1a and S1b) occurred after day 3, whereas the mineralisation rate of 14C associated with these PAHs significantly decreased after day 5 (the selected endpoint of these two experiments). In the SIP experiment with the plume water sample, complete removal of n-hexadecane occurred by day 5, which coincided with a decrease in the mineralisation of 14C associated with n-hexadecane (Supplementary Figure S1c). Based on these results, the endpoint selected for the extraction of DNA from 13C incubations was 5 days. DNA extractions were performed on each of the duplicate 13C incubations for subsequent isopycnic ultracentrifugation to isolate the 13C-enriched ‘heavy' DNA for analysis.
DNA gradient ultracentrifugation and identification of labelled 16S rRNA genes
DGGE analysis of the fractions derived from the labelled and unlabelled incubations performed for each of the three substrates showed clear evidence of isotopic enrichment of DNA in the 13C incubations, as observed by a clear separation of, and different banding patterns between, the 13C-enriched and unenriched DNA fractions (Supplementary Figure S2). Combined fractions containing 13C-enriched DNA from each of these 13C incubations were used to construct 16S rRNA gene clone libraries. Fractions from the duplicate ultracentrifuge tube from each of the 13C incubations were similarly analysed to confirm our results (data not shown).
Table 2 shows the OTU representation in each of these libraries together with their phylogenetic affiliation based on a BLASTn search in GenBank. Of the 93 sequences representing the PHE clone library (that is, from the [U-13C] phenanthrene incubation), 11 OTUs were identified, with OTU-1 (61 sequences) and OTU-2 (12 sequences) comprising the majority (78%) of these 93 sequences. OTU-1 was found to be affiliated to the genus Cycloclasticus and OTU-2 to the genus Colwellia. OTU-3, represented by five sequences, was also affiliated to Cycloclasticus and differentiated from OTU-1 based on <97% sequence identity. The NAP clone library (that is, from the [U-13C] naphthalene incubation) was represented by 92 sequences that were defined by three OTUs. Two of these, OTU-12 (83 sequences) and OTU-13 (8 sequences), comprised the majority of sequences and were respectively affiliated to Alteromonas and Cycloclasticus. The HEX clone library (that is, from the [U-13C] n-hexadecane incubation) was represented by 87 sequences that were classified into four OTUs, with OTU-15 (73 sequences) and OTU-16 (10 sequences) representing the majority of sequences, and were respectively affiliated to Alcanivorax and Marinobacter. All other OTUs in these clone libraries were represented by ⩽3 sequences and were not further analysed, but are referred to in our discussion below. Singleton sequences are presented in Supplementary Table S2.
Table 2. SIP-identified sequences in each clone library constructed from 13C-enriched DNA.a.
| OTU No. | Rep. seq.b | Closest BLASTn matchc | Accession No. | Clones in library (%)d |
|---|---|---|---|---|
| PHE | ||||
| 1 | SWPHE92 | Cycloclasticus pugetii (99%) | NR25955 | 66 |
| 2 | SWPHE03 | Colwellia aestuarii (99%) | NR43509 | 13 |
| 3 | SWPHE15 | Cycloclasticus pugetii (99%) | NR25955 | 6 |
| 4 | SWPHE27 | Olleya marilimosa (99%) | AY586527 | 3 |
| 5 | SWPHE05 | Neptuniibacter caesariensis (96%) | NR42749 | 3 |
| 6 | SWPHE62 | Pseudoalteromonas tetraodonis (100%) | NR41787 | 3 |
| 7 | SWPHE72 | Halomonas axialensis (98%) | NR27219 | 2 |
| NAP | ||||
| 12 | SWNAP06 | Alteromonas litorea (99%) | NR25780 | 90 |
| 13 | SWNAP12 | Cycloclasticus pugetii (99%) | NR 25955 | 9 |
| HEX | ||||
| 15 | DWHEX05 | Alcanivorax borkumensis (99%) | NR29340 | 84 |
| 16 | DWHEX95 | Marinobacter algicola (99%) | NR 42807 | 12 |
| 17 | DWHEX63 | Oleibacter marinus (97%) | AB435651 | 3 |
PHE, SIP with [U-13C] phenanthrene; NAP, SIP with [U-13C] naphthalene; HEX, SIP with [U-13C] n-hexadecane.
Representative sequence for each OTU. Singleton sequences are listed in Supplementary Table S2.
Results are to the closest-type strain; percentage similarity shown in parentheses.
Total number of sequences in 13C-enriched DNA clone library from phenanthrene, naphthalene and n-hexadecane SIP incubations were, respectively, 93, 92 and 87. A 97% cutoff was used to classify sequences to an OTU.
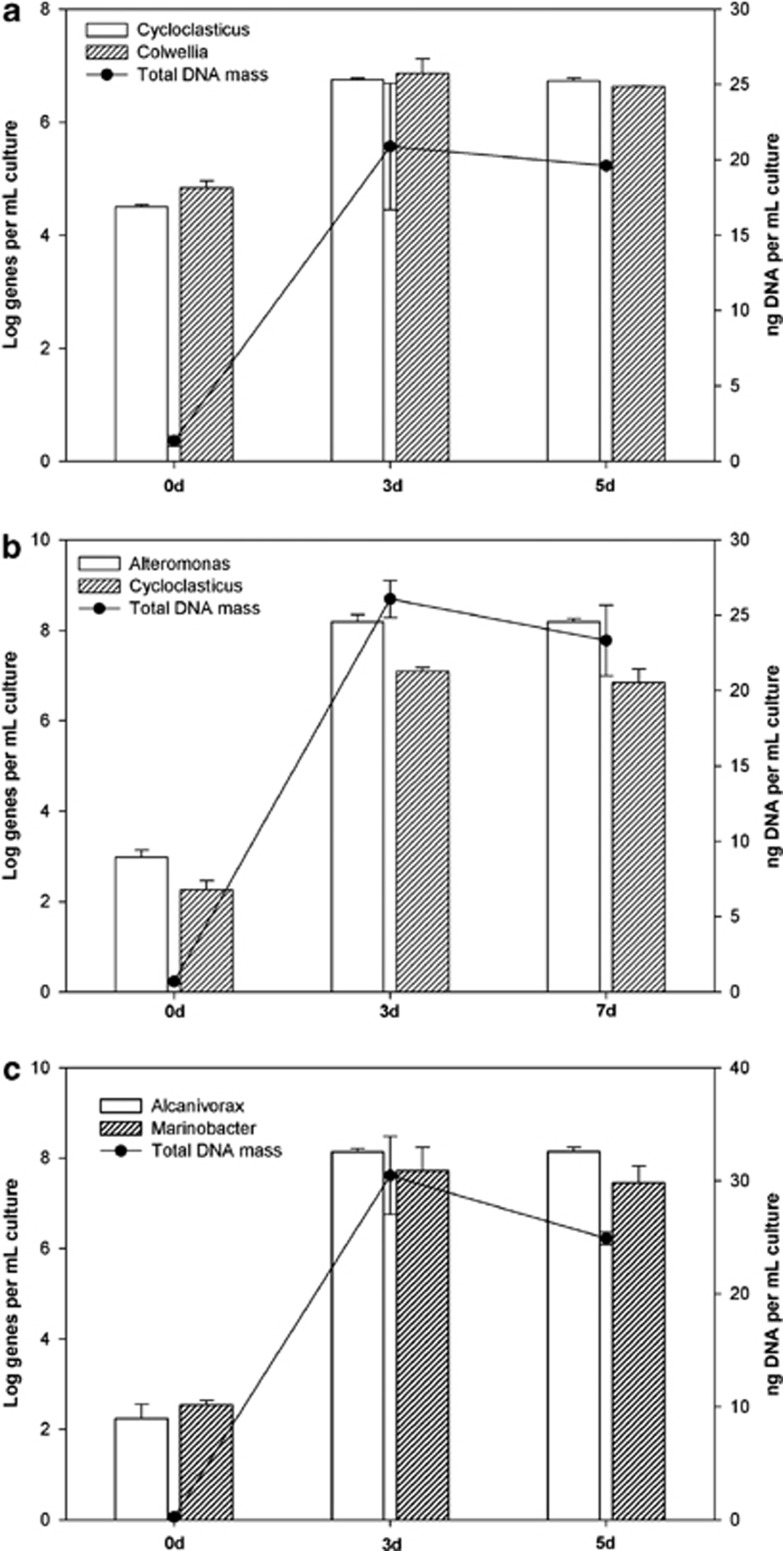
Primers for qPCR targeting the 16S rRNA genes of these major SIP-identified OTUs were developed (Table 1) and used to determine the abundance of each group in response to the corresponding growth substrate. During incubations of the surface water sample with unlabelled phenanthrene, the 16S rRNA gene copy number for the Cycloclasticus OTU-1, Colwellia OTU-2 and Cycloclasticus OTU-3 increased by ca. two orders of magnitude after 3 days (Figure 3a). Similarly, during incubations of the surface water sample with unlabelled naphthalene, the 16S rRNA gene copy number for the Alteromonas OTU-12 and Cycloclasticus OTU-13 increased by four to five orders of magnitude by day 3 (Figure 3b). During incubations of the deep plume water sample with unlabelled n-hexadecane, the 16S rRNA gene copy number for the Alcanivorax OTU-15 and Marinobacter OTU-16 increased by five to six orders of magnitude by day 3 (Figure 3c). An increase in the 16S rRNA gene copy numbers of these OTUs in their respective phenanthrene, naphthalene and n-hexadecane incubations provides further confirmation of their enrichment on these hydrocarbons as growth substrates. These increases also coincided with an increase in the total concentration of DNA (Figure 3), as an indicator of cell growth, and also with the time-frame for disappearance and mineralisation of these hydrocarbons in the respective incubations (Supplementary Figure S1). The growth of these organisms (total DNA as proxy) coupled with hydrocarbon disappearance and the appearance of their 16S rRNA genes only in the most heavily 13C-enriched DNA fractions of incubations containing a 13C-labelled substrate suggest that they performed a primary role in the degradation of the respective hydrocarbons.
Figure 3.
Abundance of 16S rRNA genes of (a) Cycloclasticus (OTU-1) and Colwellia (OTU-2) during incubation with unlabelled phenanthrene, (b) Alteromonas (OTU-12) and Cycloclasticus (OTU-13) during incubation with unlabelled naphthalene, and (c) Alcanivorax (OTU-15) and Marinobacter (OTU-16) during incubation with unlabelled n-hexadecane. Bars are the averages and standard deviations of results from triplicate qPCRs measuring the abundance of group-specific 16S rRNA genes. Circles are the means and standard deviations of triplicate measurements of the total mass of DNA per sample.
Phylogenetic analysis and representation of isolated strains and SIP clone sequences in water column pyrosequencing libraries
The near-complete 16S rRNA gene sequence (>1400 bp) representing each of the major SIP-identified OTUs and isolated hydrocarbon-degrading strains were compared with related GenBank sequences and sequences from related studies (Figure 1; Hazen et al., 2010; Valentine et al., 2010; Bælum et al., 2012; Redmond and Valentine, 2012). The highest sequence identity for any of our SIP-identified clones and isolated strains to published Gulf oil spill sequences that were identified during the active phase of the DWH oil spill was found between the following: Colwellia clone SWPHE03 (OTU-2 of the SIP experiment) and the floc-producing Colwellia strain RC25 (98.7% identity) that was isolated from non-contaminated plume water on 6 June 2010 (Bælum et al., 2012); the cultured Pseudoalteromonas strain TK-105 and a Pseudoalteromonas sequence (GenBank accession no. AY646431) (>98% sequence identity)—the latter was used to represent Pseudoalteromonadaceae-affiliated sequences found enriched in plume water samples collected during late May to early June 2010 (Hazen et al., 2010); Cycloclasticus strain TK-8 and two Cycloclasticus plume clones, H24-8 and H24-11, from plume samples collected during 11–21 June 2010 (Valentine et al., 2010) (>98% and >97% similarity, respectively). The 16S rRNA sequences of Marinobacter clone DWHEX95 (OTU-16 of the SIP experiment), Marinobacter strain TT1 and Halomonas strains GOS-2, GOS-3a and TGOS-10, however, all shared <97% similarity to respective family-level representative sequences reported in other studies (Hazen et al., 2010; Valentine et al., 2010; Redmond and Valentine, 2012).
In order to evaluate the abundance of the isolated strains and SIP-identified organisms in the Gulf of Mexico during and after the spill, we used their respective 16S rRNA gene sequences to interrogate a pyrosequencing data set (Supplementary Table S3) that was produced from a surface oil slick sample (PE5), plume-impacted water column samples (B1, B3, B6, B11), a post-spill water column sample (GIP22), and two additional post-spill water column samples (C4B8 and C4B4). A detailed analysis of this data set is presented elsewhere (T Yang, L Nigro, T Gutierrez, S Joye, R Highsmith, A Teske; submitted for publication). The relative abundance of reads with at least 97% 16S rRNA identity (over at least 300 nt) to each query sequence is given for each sample in Table 3. The sequences representative for Cycloclasticus identified in the phenanthrene and naphthalene SIP experiments—respectively, SWPHE92 (OTU-1) and SWNAP12 (OTU-13)—were found to share >97% 16S rRNA gene sequence identity to 94.1% of all Cycloclasticus pyrosequencing reads in the contaminated surface water sample (PE5) (Table 3). As the microbial community of this sample was dominated by Cycloclasticus (95.4% of total pyrosequencing reads) (Supplementary Table S2), these SIP-identified sequences clearly represented the majority of reads in the surface oil slick pyrosequencing library. Our attempts to isolate a representative strain for this dominant group from the surface oil slick samples were successful as the 16S rRNA sequence of Cycloclasticus strain TK-8 shared >99.9% sequence identity to these SIP-identified clones, and >97% sequence identity to the majority (>94%) of all pyrosequence reads affiliated to this genus in the surface and plume libraries. As a collective group, strain TK-8 and the SWPHE92 (OTU-1) and SWNAP12 (OTU-13) SIP clones accounted for the majority (87–100%) of all Cycloclasticus sequences in water column pyrosequencing libraries constructed from samples collected during the active phase of the oil spill (30 May 2010). In the post-spill pyrosequencing libraries, they accounted for 100%, 93% and 56% in the C4B8, C4B4 and GIP22 samples, respectively.
Table 3. Relative abundance (%) of the major SIP-identified OTUs and of isolated strains in 16 S rRNA gene pyrosequence librariesa.
| SIP OTU/isolateb |
During spill (May 2010) |
After spill (Sep/Oct 2010) |
||||||
|---|---|---|---|---|---|---|---|---|
| PE5 (0 m) | B1 (1320 m) | B3 (1210 m) | B6 (1170 m) | B11 (800 m) | C4B8 (800 m) | C4B4 (1210 m) | GIP22 (1050 m) | |
| From surface water | ||||||||
| Cycloclasticus OTU-1 & 13, TK-8 | 94.1 (95.42)c | 2.8 (3.17) | 6.6 (6.55) | 12.9 (14.38) | 0.79 (0.79) | 0.21 (0.21) | 5.3 (5.7) | 0.5 (0.91) |
| Alteromonas TK-46(2) | 0.98 (1.45) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1.31 (1.48) | 1.85 (2.18) | 0.08 (0.2) |
| Alteromonas OTU-12 | 0.21 | 0 | 0 | 0 | 0 | 0.04 | 0.09 | 0 |
| Halomonas GOS-2, GOS-3a, TGOS-10 | 0.12 (0.14) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0.04 (0.04) | 0.05 (0.08) | 0.02 (0.04) |
| Marinobacter TK-36 | 0 (0.02) | 0 (0) | 0 (0) | 0 (0) | 0 (0.79) | 0.2 (0.7) | 0.01 (0.2) | 0.06 (0.1) |
| From plume water | ||||||||
| Alcanivorax OTU-15, TY4, TY6 | 0.004 (0.004) | 0 (0) | 0 (0.24) | 0 (0) | 0 (0) | 0 (0.12) | 0 (0.17) | 0 (0.28) |
| Alcanivorax TY5 | 0 | 0 | 0 | 0 | 0 | 0.06 | 0.07 | 0.07 |
| Marinobacter OTU-16, TT1 | 0 (0.02) | 0 (0) | 0 (0) | 0 (0) | 0 (0.79) | 0.2 (0.7) | 0.01 (0.2) | 0.06 (0.1) |
| Pseudoalteromonas TK-105 | 1.0 (1.2) | 0 | 0 | 0 | 0 | 0 (0.1) | 0 (0.11) | 0 (0.04) |
Percentage of total reads in each library—refer to Supplementary Table S2. No matches were identified in any of the libraries for Colwellia OTU-2 and Alcanivorax TK-23.
Some OTUs and strains were grouped together based on sharing >97% 16S rRNA sequence identity.
Values in parentheses represent the % of sequences affiliated to the respective genus relative to the total number of sequences in that library.
The representative sequence for Alteromonas identified from incubations with 13C-naphthalene (SWNAP06; OTU-12) shared >97% 16S rRNA sequence identity to 12.3% of the total number of Alteromonas sequence reads in the surface oil slick pyrosequencing library, (OTU-12 sequences and total Alteromonas sequences accounted for 0.2% and 1.62%, respectively, of the total pyrosequence library; Table 3). Alteromonas strain TK-46(2) represented 0.98% of the sequences in the pyrosequenced surface slick sample and shared >97% sequence identity to 60.5% of all Alteromonas reads in that library (Table 3). The collective representation of these Alteromonas sequences among the total Alteromonas pyrosequencing reads in the post-spill water column libraries (C4B8 and C4B4) was >88%, with modest representation in the GIP22 control library (40%).
The three Halomonas strains GOS-2, GOS-3a and TGOS-10 collectively represented 86% of total Halomonas reads in the oil slick pyrosequencing library (Halomonas sequences were 0.14% of the total library; Table 3), and remained undetectable in the subsurface water column libraries (B1, B3, B6, B11). They accounted for 100%, 63% and 50% of the Halomonas pyrosequencing reads in the post-spill samples C4B8, C4B4 and GIP22, respectively. Based on a >97% cutoff for sequence identity, no matches were found across the water column pyrosequencing libraries for the SIP-identified Colwellia sequence SWPHE03 (OTU-2).
Interestingly, none of the previous studies profiling the microbial response to the oil spill had reported a single Alcanivorax sequence in their 16S rRNA pyrosequencing or conventional clone library data. The water column pyrosequencing data presented in Table 3, however, reveal the presence of Alcanivorax sequences in the surface oil slick (PE5) and plume (B3, 1210 m depth) water samples, albeit at very low levels—0.004% and 0.24% of total reads, respectively. Collectively, the 16S rRNA gene sequences of Alcanivorax clone DWHEX05 (OTU-15) and the cultured strains TY4 and TY6 matched, to within 97% identity, the single Alcanivorax read identified in the surface slick pyrosequencing library (Table 3). Pyrosequencing reads affiliated to this genus were also identified in the September 2010 (C4B8, C4B4) and October 2010 (GIP22) post-spill water column samples (Table 3), but exhibited <97% sequence identity to the Alcanivorax clone DWHEX05 (OTU-15) and the cultured strains TY4 and TY6. However, many of the reads in these post-spill libraries showed >97% identity with the sequence of the other cultured Alcanivorax sp. isolated in this study, strain TY5, accounting for 50%, 41% and 25% of total Alcanivorax reads in the C4B8, C4B4 and GIP22 libraries. Marinobacter is a monophyletic group of hydrocarbon-degrading bacteria that are commonly found enriched at marine oil-impacted sites (Head et al., 2006; Yakimov et al., 2007). However, like Alcanivorax, members of this group were poorly represented across all the pyrosequence libraries (0–0.79%, Table 3).
Discussion
The DWH disaster was a significant maritime oil spill and represented a unique opportunity to study the temporal and spatial response of indigenous populations of bacteria to a major influx of oil that originated at depths (1500 m) unprecedented in historic oil spills at sea. Much of the published literature investigating the microbial response in the water column during the spill had focused mainly on a large deep water oil plume that formed at 1000–1200 m (Hazen et al., 2010; Valentine et al., 2010; Kessler et al., 2011; Redmond and Valentine, 2012). These studies provided ‘snap-shots' of the structure and evolution of the microbial communities within the plume from samplings collected at opportune times during the active phase of the spill. Hitherto, there remained a lack of clear evidence to adequately link any of the identified oil-enriched taxa to contributing a role in the degradation of the oil. Coupling our knowledge of the bacterial community structure and response with empirically determined hydrocarbon-degrading microorganisms originating from samples collected during the spill provides a better understanding of the key organisms involved in driving hydrocarbon degradation in the water column during the DWH oil spill.
Using DNA-SIP and cultivation-based methods, we identified several hydrocarbon-degrading organisms, namely phylotypes affiliated to Cyclaclasticus, Alteromonas and Halomonas, from surface oil slick and plume waters. Cross-checking the 16S rRNA sequences of these isolates and SIP-identified clones against water column pyrosequencing data revealed that these were the dominant hydrocarbon-degraders in contaminated surface waters and, to a lesser extent, in the plume during the active phase of the spill. Drawing together our results showing the ability of Cycloclasticus strain TK-8 to degrade hydrocarbons, the observed dominance of highly similar Cycloclasticus sequences in clone libraries constructed from 13C-enriched DNA (from phenanthrene and naphthalene enrichments), and the dominance of this genus in the surface oil slick pyrosequence library, it is clear that Cycloclasticus (strain TK-8 as cultured representative) had a major role in the degradation of aromatic hydrocarbons in surface waters. This concurs with the fact that members belonging to this genus are recognised for their distinct specialisation in degrading aromatic hydrocarbons almost exclusively as a sole source of carbon and energy (Head et al., 2006; Yakimov et al., 2007). Enrichment of these sequences in the plume pyrosequencing libraries indicates that Cycloclasticus may have also contributed significantly to the degradation of aromatic hydrocarbons in deeper waters. Dominance within the plume of the DWH Oceanospirillales (Hazen et al., 2010; Supplementary Table S2) and evidence of their potential to degrade n-alkanes and cycloalkanes from single-cell genome sequencing (Mason et al., 2012) suggests that these organisms likely contributed significantly to the degradation of the aliphatic fraction of the oil. Unfortunately, the recalcitrant response of members of the DWH Oceanospirillales to cultivation in the laboratory (Mason et al., 2012; and this study) has impeded a direct interrogation of their physiological and metabolic potential. This might also explain why they were not identified in our SIP experiments.
Despite a high representation of Colwellia (OTU-2) in the clone library constructed from 13C-enriched DNA (phenanthrene enrichment of oil slick sample), no matching sequence (at >97% identity) was found for this OTU in any of the pyrosequence libraries (Supplementary Table S2), suggesting that these phenanthrene-degraders had not been enriched during the spill. As these SIP incubations were conducted at 21 °C—a temperature reminiscent of that at the sea surface in the Gulf of Mexico during the time that the spill occurred—factors other than temperature would have had a role in suppressing Colwellia and bestowing a competitive advantage to other hydrocarbon-degraders, such as Cycloclasticus and, to a lesser extent, Alteromonas and Halomonas, in the surface oil slicks. Conversely, low temperatures in deeper waters (ca. 5 °C at 1000–1300 m), where the oil plume localized, appeared to have had an important role in enriching for members of psychrophilic hydrocarbon-degrading Colwellia (Bælum et al., 2012; Redmond and Valentine, 2012). While most cultivated strains of Colwellia are psychrophiles (Methé et al., 2005), we have now observed non-psychrophilic strains with hydrocarbon-degrading qualities during the spill. The enrichment of SIP-identified Colwellia in our 13C-enriched DNA from incubations at 21 °C demonstrates that this genus is not obligately psychrophilic. While our rationale for performing the SIP incubations with the plume water sample at 21 °C, rather than at in situ temperatures (for example, 4 °C), was based on the significantly higher levels of n-hexadecane that were mineralised at the higher temperature—which was desirable to achieve sufficient enrichment of the 13C-label into DNA—this did not preclude our identification of hydrocarbon-degrading taxa, notably Cycloclasticus, that were enriched in the plume.
To our knowledge, no report has hitherto emerged identifying Alcanivorax in the water column during the spill. This is interesting from the point of view that members of this group are often strongly selected for in oil-impacted environments where they can increase in numbers from near undetectable levels to constituting up to 70–90% of the total bacterial community (Harayama et al., 2004; Head et al., 2006; Yakimov et al., 2007). Based on the knowledge that Alcanivorax preferentially degrade branched- and/or straight-chain saturated hydrocarbons (Kasai et al., 2002a; Head et al., 2003) that constitute a large fraction of light crude oils, such as the one that entered into the Gulf of Mexico from the leaky Macondo MC 252 well (Reddy et al., 2012), members of this genus could be expected to have bloomed during the DWH spill. Using SIP and cultivation-based methods, we identified a number of Alcanivorax phylotypes in both surface slick and plume water samples, hence, revealing that members of this genus were present in the water column during the spill. Cross-checking their abundance against the water column pyrosequence data showed that members of this genus were present at very low levels in the surface slick (PE5) and plume (B3) water samples, and similarly in all the post-spill samples (Table 3). This reflects background levels for these and other fastidious hydrocarbon-degraders in unpolluted marine waters (Yakimov et al., 2007). Though low cell abundance for any particular microbial group in situ does not always correlate with inactivity, it is unlikely that Alcanivorax had contributed a significant role to the fate of the oil. The absence of Alcanivorax in pyrosequencing libraries constructed by other authors from Gulf spill water column samples may reflect differences in the parameters used for analysing sequence data (for example, distance threshold setting; type of bioinformatics software package) and the type of sequencing platform they used (Shakya et al., 2013).
The enrichment of other hydrocarbon-degrading specialists, notably Cycloclasticus and DWH Oceanospirillales, apparently reflects a competitive advantage that enabled them to respond quickly and bloom soon after the initial influx of oil into the Gulf of Mexico. In deep waters where low in situ temperatures (ca. 5 °C) were distinctive features of the plume (Camilli et al., 2010), these factors may have arrested the potential for Alcanivorax to bloom. Alternatively, their suppressed growth may be attributed to a limit in the availability of nitrogen and phosphorous (Harayama et al., 1999; Kasai et al., 2001, 2002a; Syutsubo et al., 2001). Alcanivorax, however, was the dominant hydrocarbon-degrading bacterial group enriched by oil contamination of Gulf beach sands from the DWH spill (Kostka et al., 2011). The 16S rRNA gene sequence of these organisms did not match any of our Alcanivorax or Marinobacter sequences (<97% identity), suggesting that these beach sand communities were probably not derived through transport on oil droplets or via wind and currents from the water column where the spill originated >77 km offshore.
To the best of our knowledge, this study represents the first application of SIP to identify bacterial taxa with the ability to degrade aliphatic and low-molecular weight PAHs in the oil-contaminated waters at DWH. It also demonstrates the importance of applying this method in linking the phylogenetic identity of targeted microorganisms with their ability to carry out a specific metabolic function (for example, degradation of hydrocarbons), including those not readily amenable to cultivation. Moreover, by cross-checking sequences from isolated strains and from SIP experiments against a temporal and spatial pyrosequencing data set, here we have provided a more complete understanding of the bacterial community that contributed to the degradation of the oil in surface and plume waters. Conjecture on the role of oil-enriched taxa during the DWH spill by previous studies was based almost entirely on metagenomic information. We have now provided unequivocal evidence that some of those taxa—Cycloclasticus, Alteromonas, Colwellia, Pseudoalteromonas, Marinobacter and Halomonas—are in fact capable of degrading hydrocarbons. This work has added to our current knowledge on the diversity of hydrocarbon-degrading communities in the Gulf of Mexico and their capacity to respond to an influx of crude oil from deep waters.
Acknowledgments
We thank chief scientist Arne Diercks and the ship and scientific crew of R/V Pelican (leg PE-1031) for a successful impromptu sampling operation shortly after the DWH blowout, and Luke McKay for collecting the 5 May 2010 samples. We also thank the ship and science crews of R/V Walton Smith, R/V Pelican and R/V Cape Hatteras for their hard work and constant effort to accommodate our research needs. This work was supported by a Marie Curie International Outgoing Fellowship (PIOF-GA-2008-220129) within the 7th European Community Framework Programme. Partial support was also provided through the US National Institute of Environmental Health Sciences, grant 5 P42ES005948, the National Oceanic and Atmospheric Administration via the National Institute for Undersea Science and Technology, which supported the 5 May 2010 cruise, and the BP/Gulf of Mexico Research Initiative to support consortium research entitled ‘Ecosystem Impacts of Oil and Gas Inputs to the Gulf (ECOGIG)' administered by the University of Mississippi. The GRIIDC data set IDs for this manuscript are ‘R1.x132.135:0001 R/V Walton Smith May–July 2010 Microbiological Genetics', ‘R1.x132.135:0002 R/V Walton Smith May–July 2010 Physiological Activity', ‘R1.x132.135:0003 R/V Pelican May 5–9 2010 Microbial Genetics' and ‘R1.x132.135:0004 R/V Pelican May 5–9 2010 Microbial Physiological Activity'. We also thank Stephen Richardson and Scott Hauswirth for assistance with the HPLC and GC–MS analysis, respectively, and Kirk T Semple for providing [1-14C]n-hexadecane. We also thank three anonymous reviewers for their valuable comments during the preparation of the manuscrip. AT, TG and TY were also supported by NSF (RAPID Response: the microbial response to the Deepwater Horizon Oil Spill; NSF-OCE 1045115).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bælum J, Borglin S, Chakraborty R, Fortney JL, Lamendella R, Mason OU, et al. Deep-sea bacteria enriched by oil and dispersant from the Deepwater Horizon Spill. Environ Microbiol. 2012;14:2405–2416. doi: 10.1111/j.1462-2920.2012.02780.x. [DOI] [PubMed] [Google Scholar]
- Camilli R, Reddy CM, Yoerger DR, Van Mooy BAS, Jakuba MV, Kinsey JC, et al. Tracking hydrocarbon plume transport and biodegradation at Deepwater Horizon. Science. 2010;330:201–204. doi: 10.1126/science.1195223. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diercks A-R, Highsmith RC, Asper VL, Joung DJ, Zhou Z, Guo L, et al. Characterization of subsurface polycyclic aromatic hydrocarbons at the Deepwater Horizon wellhead site. Geophys Res Lett. 2010;37:L20602. [Google Scholar]
- Dumont MG, Murrell JC. Stable isotope probing—linking microbial identity to function. Nat Rev Microbiol. 2005;3:499–504. doi: 10.1038/nrmicro1162. [DOI] [PubMed] [Google Scholar]
- Dyksterhouse SE, Gray JP, Herwig RP, Cano Lara J, Staley JT. Cycloclasticus pugetii gen. nov., sp.nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int J Syst Bacteriol. 1995;45:116–123. doi: 10.1099/00207713-45-1-116. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Gutierrez T, Singleton DR, Aitken MD, Semple KT. Stable-isotope probing of an algal bloom identifies uncultivated members of the Rhodobacteraceae associated with low molecular-weight PAH degradation. Appl Environ Microbiol. 2011;77:7856–7860. doi: 10.1128/AEM.06200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S, Harayama S, Kishira H, Kasai Y, Shutsubo K. Petroleum biodegradation in marine environments. J Mol Microbiol Biotechnol. 1999;1:63–70. [PubMed] [Google Scholar]
- Harayama S, Kasai Y, Hara A. Microbial communities in oil-contaminated seawater. Curr Opin Biotechnol. 2004;15:205–214. doi: 10.1016/j.copbio.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- Head IM, Jones DM, Larter SR. Biological activity in the deep subsurface and the origin of heavy oil. Nature. 2003;426:344–352. doi: 10.1038/nature02134. [DOI] [PubMed] [Google Scholar]
- Head IM, Jones DM, Röling WF. Marine microorganisms make a meal of oil. Nat Rev Microbiol. 2006;4:173–182. doi: 10.1038/nrmicro1348. [DOI] [PubMed] [Google Scholar]
- Jones MD, Singleton DR, Sun W, Aitken MD. Multiple DNA extractions coupled with stable-isotope probing of anthracene-degrading bacteria in contaminated soil. Appl Environ Microbiol. 2011;77:2984–2991. doi: 10.1128/AEM.01942-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai Y, Kishira H, Syutsubo K, Harayama S. Molecular detection of marine bacterial populations on beaches contaminated by the Nakhodka tanker oil-spill accident. Environ Microbiol. 2001;3:246–255. doi: 10.1046/j.1462-2920.2001.00185.x. [DOI] [PubMed] [Google Scholar]
- Kasai Y, Kishira H, Sasaki T, Syutsubo K, Watanabe K, Harayama S. Predominant growth of Alcanivorax strains in oil-contaminated and nutrient supplemented sea water. Environ Microbiol. 2002a;4:141–147. doi: 10.1046/j.1462-2920.2002.00275.x. [DOI] [PubMed] [Google Scholar]
- Kasai Y, Kishira H, Harayama S. Bacteria belonging to the genus Cycloclasticus play a primary role in the degradation of aromatic hydrocarbons released in a marine environment. Appl Environ Microbiol. 2002b;68:5625–5633. doi: 10.1128/AEM.68.11.5625-5633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler JD, Valentine DL, Redmond MC, Du M, Chan EW, Mendes SD, et al. A persistent oxygen anomaly reveals the fate of spilled methane in the deep Gulf of Mexico. Sci. 2011;331:312–315. doi: 10.1126/science.1199697. [DOI] [PubMed] [Google Scholar]
- Kiyohara H, Nagao K, Yana K. Rapid screen for bacteria degrading water-insoluble, solid hydrocarbons on agar plates. Appl Environ Microbiol. 1982;43:454–457. doi: 10.1128/aem.43.2.454-457.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, et al. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Appl Environ Microbiol. 2011;77:7962–7974. doi: 10.1128/AEM.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidak BL, Cole JR, Parker CT, Jr, Garrity GM, Larsen N, Li B, et al. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason OU, Hazen TC, Borglin S, Chain PSG, Dubinsky EA, Fortney JL, et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 2012;6:1715–1727. doi: 10.1038/ismej.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNutt M, Camilli R, Guthrie G, Hsieh P, Labson V, Lehr B, et al. 2011Assessment of flow rate estimates for the Deepwater Horizon/Macondo well oil spill Flow rate technical group report to the national incident command, interagency solutions group10 March 2011.
- Methé BA, Nelson KE, Deming JW, Momen B, Melamud E, Zhang X, et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci USA. 2005;102:10913–10918. doi: 10.1073/pnas.0504766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy CM, Arey JS, Seewald JS, Sylva SP, Lemkau KL, Nelson RK, et al. Composition and fate of gas and oil released to the water column during the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109:20229–20234. doi: 10.1073/pnas.1101242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond MC, Valentine DL. Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109:20292–20297. doi: 10.1073/pnas.1108756108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röling WFM, Milner MG, Jones DM, Lee K, Daniel F, Swannell RJP, et al. Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl Environ Microbiol. 2002;68:5537–5548. doi: 10.1128/AEM.68.11.5537-5548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röling WFM, Milner MG, Jones DM, Fratepietro F, Swannell RPJ, Daniel F, et al. Bacterial community dynamics and hydrocarbon degradation during a field-scale evaluation of bioremediation on a mudflat beach contaminated with buried oil. Appl Environ Microbiol. 2004;70:2603–2613. doi: 10.1128/AEM.70.5.2603-2613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabat G, Rose P, Hickey WJ, Harkin JM. Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl Environ Microbiol. 2000;66:844–849. doi: 10.1128/aem.66.2.844-849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya M, Quince C, Campbell JH, Yang ZK, Schadt CW, Podar M. Comparative metagenomic and rRNA microbial diversity characterization using Archaeal and Bacterial synthetic communities. Environ Microbiol. 2013;15:1882–1899. doi: 10.1111/1462-2920.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Sangaiah R, Gold A, Ball LM, Aitken MD. Identification and quantification of uncultivated Proteobacteria associated with pyrene degradation in a bioreactor treating PAH-contaminated soil. Environ Microbiol. 2006;8:1736–1745. doi: 10.1111/j.1462-2920.2006.01112.x. [DOI] [PubMed] [Google Scholar]
- Syutsubo K, Kishira H, Harayama S. Development of specific oligonucleotide probes for the identification and in situ detection of hydrocarbon-degrading Alcanivorax strains. Environ Microbiol. 2001;3:371–379. doi: 10.1046/j.1462-2920.2001.00204.x. [DOI] [PubMed] [Google Scholar]
- Teira E, Lekunberri I, Gasol JM, Nieto-Cid M, Alvarez-Salgado XA, Figueiras FG. Dynamics of the hydrocarbon-degrading Cycloclasticus bacteria during mesocosm-simulated oil spills. Environ Microbiol. 2007;9:2551–2562. doi: 10.1111/j.1462-2920.2007.01373.x. [DOI] [PubMed] [Google Scholar]
- Tillett D, Neilan BA. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J Phycol. 2000;36:251–258. [Google Scholar]
- Thomas O, Burgess C. Techniques and instrumentation in analytical chemistry. Elsevier: Amsterdam, The Netherlands; UV Visible Spectrophotometry of Water and Wastewater. 2007;(vol 27) [Google Scholar]
- Valentine DL, Kessler JD, Redmond MC, Mendes SD, Heintz MB, Farwell C, et al. Propane respiration jump-starts microbial response to a deep oil spill. Sci. 2010;330:208–211. doi: 10.1126/science.1196830. [DOI] [PubMed] [Google Scholar]
- Wade TL, Sweet ST, Sericano JL, Guinasso NL, Diercks A-R, Highsmith RC, et al. 2011Analyses of water samples from the Deepwater Horizon Oil spill: documentation of the subsurface plume Monitoring and Modeling the Deepwater Horizon Oil Spill: A Record-Breaking Enterprise 77–82.Geophysical Monograph Series 195. American Geophysical Union. 10.1029/2011GM001103.
- Wilmotte A, van der Auwera G, De Wachter R. Structure of the 16S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (Mastigocladus laminosus HTF) strain PCC7518, and phylogenetic analysis. FEMS Microbiol Lett. 1993;317:96–100. doi: 10.1016/0014-5793(93)81499-p. [DOI] [PubMed] [Google Scholar]
- Yakimov MM, Timmis KN, Golyshin PN. Obligate oil-degrading marine bacteria. Curr Opin Biotechnol. 2007;18:257–266. doi: 10.1016/j.copbio.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Yarza P, Ludwig W, Euzeby J, Amann R, Schleifer K-H, Glockner FO, et al. Update of the all-species living tree project based on 16S and 23S rRNA sequence analysis. Syst Appl Microbiol. 2010;33:291–299. doi: 10.1016/j.syapm.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Yu Z, Morrison M. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2004;70:4800–4806. doi: 10.1128/AEM.70.8.4800-4806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Sangaiah R, Gold A, Ball LM. Synthesis of uniformly 13C-labeled polycyclic aromatic hydrocarbons. Org Biomol Chem. 2011;9:5431–5435. doi: 10.1039/c0ob01107j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.