Abstract
Background
Proprotein convertase subtilisin/kexin type 9 (PCSK9) modulates low-density lipoprotein (LDL) receptor (LDLR) degradation, thus influencing serum cholesterol levels. However, dysfunctional LDLR causes hypercholesterolemia without affecting PCSK9 clearance from the circulation.
Methods and Results
To study the reciprocal effects of PCSK9 and LDLR and the resultant effects on serum cholesterol, we produced transgenic mice expressing human (h) PCSK9. Although hPCSK9 was mainly expressed in the kidney, LDLR degradation was more evident in the liver. Adrenal LDLR levels were not affected, likely due to impaired PCSK9 retention in this tissue. In addition, hPCSK9 expression increased hepatic secretion of apoB-containing lipoproteins in an LDLR-independent fashion. Expression of hPCSK9 raised serum murine (m) PCSK9 levels by 4.3-fold in wild-type (WT) mice and not at all in LDLR−/− ice, where mPCSK9 levels were already 10-fold higher than in WT mice. In addition, LDLR+/− mice had 2.7-fold elevation in mPCSK9 levels and no elevation in cholesterol levels. Conversely, acute expression of hLDLR in transgenic mice caused a 70% decrease in serum mPCSK9 levels. Turnover studies using physiological levels of hPCSK9 showed rapid clearance in WT (half-life 5.2 min), faster in hLDLR transgenics (2.9 min), and much slower in LDLR−/− recipients (50.5 min). Supportive results were obtained using an in vitro system. Finally, up to 30% of serum hPCSK9 was associated with LDL regardless of LDLR expression.
Conclusions
Our results support a scenario where LDLR represents the main route of elimination of PCSK9, and a reciprocal regulation between these two proteins controls serum PCSK9 levels, hepatic LDLR expression, and serum LDL levels.
Keywords: cholesterol, lipoproteins, PCSK9, turnover studies, LDL receptor
Introduction
Lowering low-density lipoprotein (LDL) cholesterol (LDL-c) is the most effective single approach for cardiovascular risk reduction, and statins are the drug of choice to achieve target LDL-c levels 1. Statins inhibit 3-hydroxy-3-methylglutaryl-coenzyme A reductase, the rate limiting enzyme for cholesterol biosynthesis, and consequently cause up-regulation of LDL receptor (LDLR) levels via sterol regulatory element binding protein 2 (SREBP-2) activation 2. However, many patients cannot achieve LDL-c goals even on a high dose of statins 3. Human (h) proprotein convertase subtilisin/kexin 9 (PCSK9) is a secreted 692-amino acid protein that promotes the reduction of LDLR protein levels, mainly in the liver 4, 5, without affecting LDLR mRNA levels 6. Gain-of-function mutations in PCSK9 are associated with a rare form of autosomal dominant hypercholesterolemia 7, whereas loss-of-function mutations cause low cholesterol levels and are linked to lower coronary artery disease (CAD) rates 8. The human PCSK9 gene is found on human chromosome 1 (chromosome 4 in mice) 9. The human protein (hPCSK9) is synthesized mainly in the liver, kidney, and small intestine 10, under the regulation of SREBP-2 11 and has 76.6% identity with murine (m) PCSK9 12. By activating the SREPB-2 pathway, statins increase circulating PCSK9 levels, an effect that likely limits the LDL-lowering efficacy of these drugs 13.
The secreted form of PCSK9 binds directly to LDLR, and the complex is internalized and targeted to the lysosomes for destruction 14-16. Although LDLR-dependent uptake is the only known pathway for serum PCSK9 clearance, recent evidence suggests that the converse is not true, and reduced LDLR function does not affect clearance of serum PCSK9 17-19, either due to LDLR-independent clearance routes 20 or because a receptor that is dysfunctional in binding LDL may normally bind PCSK9. The LDLR−/− mouse provides a unique system in which to study metabolism of both LDL and PCSK9 in the complete absence of LDLR.
It is not clear whether PCSK9 has a biological effect other than degrading LDLR or if this effect becomes more obvious or relevant in the absence of LDLR. Some studies reported no effect of PCSK9 overexpression on serum cholesterol levels in the absence of LDLR 21, 22. More recent studies have shown increases in serum cholesterol levels in an LDLR-independent manner 23, 24. Consensus is also lacking on how PCSK9 is transported in the bloodstream, and whether this relates to PCSK9 function. We were the first to show significant association of hPCSK9 with LDL in vivo and in vitro 25, while other laboratories reported that serum PCSK9 was either free or in macromolecular complexes more or less associated with lipoproteins 24-27.
As a secretory protein that regulates LDLR post-transcriptionally, PCSK9 has revolutionized our knowledge of cell-based control of cholesterol homeostasis 28, 29. Unlike the well-defined intracellular pathways that regulate cholesterol metabolism, the function and regulation of serum PCSK9 are not well understood. To characterize PCSK9 levels, distribution, and activity as a function of LDLR expression, we developed a transgenic line of mice that express human PCSK9 (hPCSK9) in addition to their normal expression of murine PCSK9 (mPCSK9). We also performed in vivo and in vitro experiments to determine the reciprocal effects of PCSK9 and LDLR under different conditions, including absence and overexpression of either protein. Our data show that LDLR is the primary route for serum PCSK9 clearance.
Methods
Materials and Reagents
pcDNA3.1 Directional TOPO Expression Kit, ProBond Purification System, and pre gels were purchased from Invitrogen (Grand Island, NY). Aml-12, Y-1 (ATCC; CCL-79) HEK293T and HeLa cell lines were purchased from ATCC (Manassas, VA). The bi-cistronic lentiviral vector pWPI, the lentiviral envelope plasmid pMD2.G, and the packaging plasmid pCMV ΔR8.91 were kindly provided by Dr. Dider Trono (Lausanne, Switzerland). Chicken polyclonal antibody to LDL receptor and rabbit polyclonal antibody to chicken HRP were purchased from Abcam Inc. (Cambridge, MA). Sheep polyclonal antibody toward hPCSK9 was obtained from R&D systems (Minneapolis, MN). Rabbit polyclonal antibody toward hPCSK9 was purchased from MBL-international (Woburn, MA). Mouse anti-Histidine Tag was purchased from eBioscience (San Diego, CA). HRP-conjugated donkey anti-sheep IgG antibody, Goat anti rabbit, Tyloxapol and OptiPrep Density Gradient Medium were purchased from Sigma (St. Louis, MO). Mammalian Transfection System was purchased from Promega (Madosin, WI). Cross linking reagent dithiobis (succinim-idylpropionate) substrate (DSP), was purchased from Thermo-scientific (Logan, UT). ELISA kits for hPCSK9 and mPCSK9 were purchased from MBL- International (Woburn, MA). Liver perfusion and digestion media were purchased from Gibco (Grand Island, NY). OptiSeal™ tubes were purchased from Beckman-Coulter (Kraemer Boulevard, Brea, CA)
Human PCSK9 Expression Construct and Transfection
Methods for the generation of lentiviral PCSK9 construct, lentivirus preparation, and transduction were previously described 30. Briefly, bi-cistronic lentiviral expression plasmids for PCSK9-GFP, along with the packaging plasmid pCMVR8.91 and envelope plasmid pMD2.G, were co-transfected into HEK293T cells. The viral particle suspension was harvested over two days and then filtered and subjected to ultracentrifugation. The titer of resuspended concentrated viral particles was determined by a standard HeLa titer procedure. GFP positive cell were sorted using ARIAIII cell sorter.
Generation of Transgenic Mice
The generation of the hPCSK9 transgenic mice was done in our laboratory as previously described 25. In short, the pcDNA-PCSK9 DNA was digested by BglII and PsiI, and this 3800-bp fragment was used for transgenic injection. The injection was performed by Vanderbilt Transgenic Mouse/Embryonic Stem Cell Shared Resource. The transgenic mice were confirmed by Southern blotting and PCR.
Primary hepatocyte isolation
Primary hepatocytes were isolated as previously described 31. In short, ligatures were made around the portal vein and the inferior vena cava. Livers were perfused through the portal vein using Liver Perfusion Medium™ containing 0.167 mM insulin and 0.5 mM EGTA, for 5 min, and then digested using Liver Digestion Medium™ containing 0.167 mM insulin and 62.7 u/ml collagenase, for 15 min. Livers were dissected, homogenized and filtered through a 70-micron filter. Hepatocytes were cultured in 6-well collagen-coated plates at 1.8×106 cells/well in DMEM with 17% FBS and 0.167 mM insulin. Hepatocytes were cultured for 2 hrs, then the media was changed to 17% FBS without insulin for 4-6 hours prior to the addition of purified human PCSK9.
Protein Purification
His-tagged full-length PCSK9 was purified using the ProBond™ protein purification system as described previously 25. Protein purification was measured using gel electrophoresis with Coomassie blue staining and the concentration was determined by Lowry assays and hPCSK9 ELISA (MBL-international).
Western Blotting
Samples were loaded onto NuPage™ 4–12% Bis-Tris or NativePage™ 4-16% precast gels (Invitrogen) for electrophoresis. The size-separated proteins were then transferred to nitrocellulose membranes. The indicated primary antibodies and HRP-conjugated secondary antibodies were used to detect target proteins. Signal was detected using a mixture of luminol, p-coumaric acid and hydrogen peroxide in 100 mM Tris pH 8.5. Intensity quantification of the bands was obtained using ImageJ software and normalized to β-action or GAPDH.
Mice
WT C57BL/6, LDLR−/−, or hLDLR mice were purchased from the Jackson laboratories (Bar Harbor, ME) and housed at the Vanderbilt University Medical Center. LDLR+/− were bred in house using F1 from WT and LDLR−/−. All animal experiments were carried out in compliance with National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee of Vanderbilt University. Blood and tissues were collected from 6-8 month old mice.
Serum analyses
The levels of hPCSK9 and mPCSK9 were measured in serum of non-fasting mice on chow diet, and determined by ELISA kits (MBL-International), with a measured 5% cross reactivity between mPCSK9 and hPCSK9 within the kit's linear range (not shown). Serum cholesterol concentrations were measured using a colorimetric method. Mouse lipoproteins were isolated by density (high-salt) ultracentrifugation as previously described 31 or using natural gradient media - Optiprep™ using TLN100 rotor and OptiSeal™ tubes at 90,000 rpm for 2.5 hours. Mouse lipoprotein profile was determined by size exclusion chromatography (FPLC).
PCSK9 turnover
His-tagged hPCSK9 was purified from transduced HEK293T cells. Male WT, LDLR−/−, or hLDLR mice were anesthetized, and hPCSK9 (1.5 μg/mouse) was injected into the retro-orbital vein plexus. At the indicated time points, blood was collected from the tail vein. Animals were sacrificed at the end of the experiment and organs were collected for further analysis.
Cross-Linking Studies
For denaturing and non-denaturing gel analyses, FPLC fractions were incubated with dithiobis(succinimidylpropionate) substrate (DSP), (Thermo-scientific), final concentration 0.5mM at ambient temperature for 30 min. Reaction was stopped using 1M Tris, pH 7.5.
Statistical Analyses
GraphPad Prism 4 software was used to carry out statistical analyses. Student's paired t-test was used to compare the means of two groups. Analysis of variance (ANOVA) was used when more than two groups were compared. Results are presented as means±SD or as percent ±CV.
Results
PCSK9 and LDLR expression in WT and hPCSK9 transgenic mice
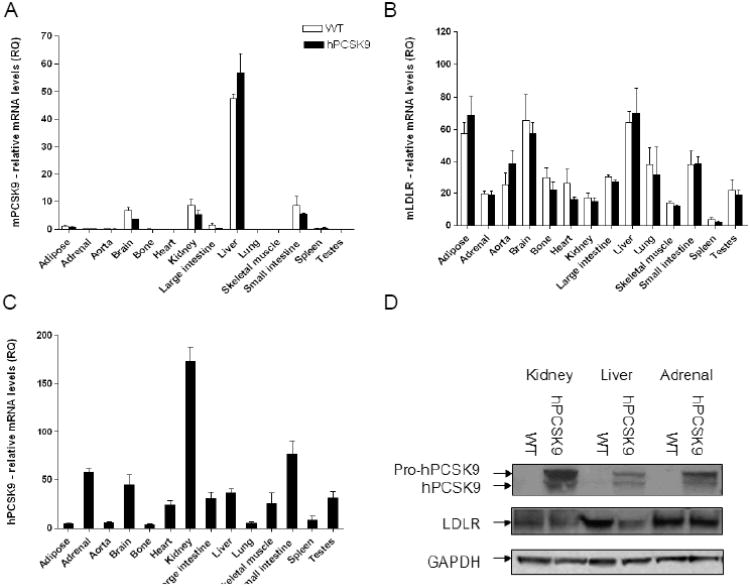
We generated a transgenic line of mice expressing hPCSK9, with a vector designed for expression in multiple tissues 25. First we measured the expression levels of endogenous mPCSK9 and LDLR in hPCSK9 transgenic mice compared to WT controls. As expected, endogenous mPCSK9 was mainly expressed in liver, small intestine, and kidney 10, 32. Figure 1A and 1B show that hPCSK9 expression did not alter the pattern of tissue mPCSK9 and LDLR expression. Expression of hPCSK9 was highest in the kidney (Figure 1C, 1D and Supplementary Figure 1A), whereas LDLR reduction was strongest in liver (−41%), followed by the kidney (−24%). There was no effect of hPCSK9 expression on LDLR levels in the adrenals (Figure 1D, and supplementary Figure 1B). These results support the notion that PCSK9 acts in a tissue-specific manner 26, 33.
Figure 1.
PCSK9 and LDLR levels in WT and hPCSK9 transgenic mice tissues. Relative mRNA expression of (A) mPCSK9, (B) mLDLR, and (C) hPCSK9. Expression levels were calculated using real-time PCR ΔΔCT method. Each tissue was measured in duplicate using 3 mice in each group (n=3). (D) Representative immunoblot of hPCSK9, LDLR and β-actin in WT and hPCSK9 transgenic mice (quantitative analysis is shown in supplementary Figures 1A and 1B).
Cholesterol levels and distribution in hPCSK9 transgenic mice
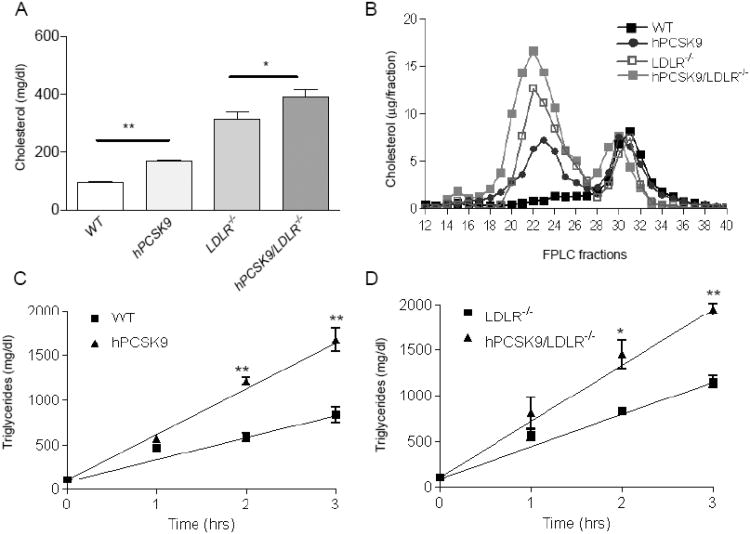
Transgenic mice expressing hPCSK9 were backcrossed into LDLR−/− background to generate hPCSK9/LDLR−/− mice. Figure 2A shows that mice expressing hPCSK9 on WT background have a total cholesterol level of 168±18 mg/dl, as compared to 96±16 mg/dl in WT controls (p<0.01). This was mainly due to increased LDL-c levels (Figure 2B). A more moderate but still significant (p<0.05) effect was observed in LDLR−/− mice, where expression of hPCSK9 increased total cholesterol to 377±98 mg/dl, compared to 315±89 mg/dl in controls. Separation of serum lipoprotein by agarose gel electrophoresis shows that hPCSK9 expression increase the beta band (LDL) both on WT and LDLR−/− background mice, compared to non-transgenic controls (Supplementary Figure 2A). Besides LDLR reduction there was no effect of hPCSK9 on other hepatic lipoprotein receptors, such as LRP1 (data not shown), and thus next we investigated the influence of PCSK9 on lipoprotein production. Our results show that tyloxapol-induced blockade of lipolysis in fasted mice causes a significantly faster and larger serum triglyceride accumulation in hPCSK9 transgenic compared to non-transgenic controls, both on WT and LDLR−/− backgrounds (Figure 3C and D, respectively).
Figure 2.
hPCSK9 effect on serum lipids in WT and LDLR−/− background mice. (A) Total serum cholesterol levels (n=9-16) in mg/dl, (B) serum FPLC lipoprotein profiles, in μg/fraction. Each profile represents pooled serum from 3 mice, Fractions: VLDL 12-17, LDL 18-27, HDL 28-34, lipoprotein-deficient serum (LPDS) 35-40. Changes in triglyceride levels after intravenous injection of 0.5 mg/gr tyloxapol: (C) hPCSK9tg mice vs. WT. (D) hPCSK9tg/LDLR−/− vs. LDLR−/−. The slope indicates the secretion rate of triglyceride-rich lipoproteins.
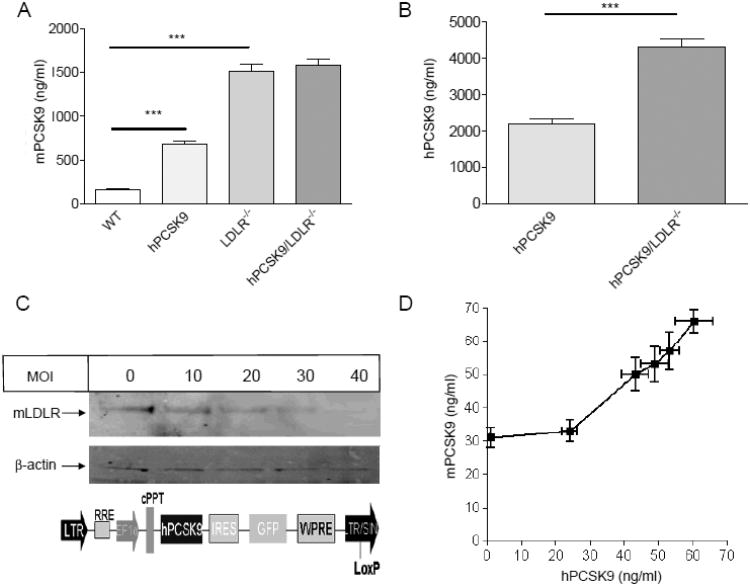
Figure 3.
hPCSK9 effect on mPCSK9 level in WT and LDLR−/− background mice. (A) Serum mPCSK9 levels. Results for mPCSK9 levels in hPCSK9 transgenic mice take into account 5% cross-reactivity with hPCSK9. (B) Serum hPCSK9 levels. Notice the different scale in A vs. B. Mice (n=9-16) were 6-8 month old, non-fasted, and on a chow diet. (C) Immunoblot of LDLR and β-actin in transduced mouse Aml-12 hepatocyte; the bi-cistronic lentiviral DNA construct pWPI-hPCSK9. (D) Secretion of mPCSK9 as a function of hPCSK9 levels in transduced Aml-12 cells. Transduction was performed in triplicate in each virus concentration (MOI); the vertical standard deviation is for measured mPCSK9 in the media and the horizontal standard deviation is for hPCSK9 measured in the same media. (*p<0.05, ** p<0.01, ***p<0.001).
Serum mPCSK9 and hPCSK9 levels
Although mRNA levels of mPCSK9 were not affected by hPCSK9 expression (Figure 1A), serum levels of mPCSK9 were increased by an average of 4.3-fold (682±105 ng/ml vs. 160±30 ng/ml) in transgenic mice compared to WT controls (Figure 3A). Conversely, serum levels of mPCSK9 were extremely elevated in LDLR−/− control compared to WT mice, but did not increase further in hPCSK9/LDLR−/− transgenic mice (Figure 3A). The increase in mPCSK9 levels in transgenic mice is probably due to hPCSK9-induced removal of hepatic LDLR, which in turn reduces mPCSK9 clearance. This mechanism likely explains also the difference in serum mPCSK9 levels between WT and LDLR−/− mice. Figure 3A shows that the complete absence of LDLR resulted in a 9.5-fold increase in mPCSK9 levels (1514±245 ng/ml) compared to WT controls (160±30 ng/ml). Interestingly, heterozygote LDLR+/− mice showed a 2.7-fold increase in serum mPCSK9 levels (to 429±89 ng/ml), whereas acute over-expression of hLDLR reduced serum levels of mPCSK9 by 67% (to 53±19 ng/ml), (Supplementary Figure 2B). Levels of hPCSK9 showed a similar trend, with a 2-fold increase in hPCSK9 levels in transgenic mice on LDLR−/− background (4303±532 ng/ml) compared to transgenic mice on WT background (2181±423 ng/ml), (Figure 3B). Additionally, in hPCSK9 transgenic mice on LDLR−/− background, hepatic intracellular levels of hPCSK9 were lower than those in transgenic mice on WT background, suggesting that absence of LDLR reduces the amount of circulating PCSK9 captured by the cell, and that both newly synthesized and internalized hPCSK9 contribute to its cellular levels (Supplementary Figure 2C).
To further study the effect of hPCSK9 on mPCSK9 accumulation, we transduced murine hepatocytes (Aml-12 cells), which naturally produce mPCSK9, with increasing multiplicities of infection (MOI) of the bi-cistronic lentiviral DNA construct (pWPI-hPCSK9), shown in Figure 3C. Transduced hepatocytes showed reduced LDLR levels with increasing MOI of the pWPI-hPCSK9 lentivirus (Figure 3C). Interestingly, when levels of secreted hPCSK9 were low (below or equal to the levels of secreted mPCSK9) no effects were seen on media mPCSK9 accumulation. However, as the levels of hPCSK9 increased above those of mPCSK9, a linear increase in media mPCSK9 accumulation was observed (Figure 3D). This was the result of a non-genetic effect, as pWPI-hPCSK9 transduction did not alter mRNA levels of mPCSK9 compared to control hepatocytes transduced with GFP (Supplementary Figure 3A).
Turnover of recombinant hPCSK9 in mice
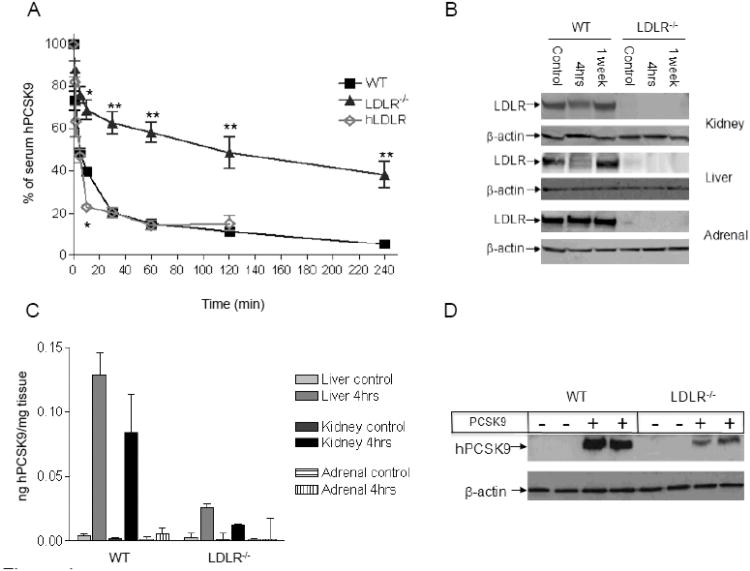
To characterize the effects of acute hPCSK9 exposure on LDLR levels in vivo, we performed a series of turnover studies using unmodified (non-radiolabeled) protein. The hPCSK9 was produced in HEK293T and purified (Supplementary Figure 3B) as previously described 25. In contrast to previous studies that have used PCSK9 at supra-physiologic levels (4-32 μg/mouse) 33, 34, we injected hPCSK9 at a dose of 1.5 μg/mouse into WT, LDLR−/−, and hLDLR transgenic mice. These injections resulted in average serum hPCSK9 peak levels of 714±218 ng/ml, squarely in the physiologic range (50-1000 ng/ml) 35, 36. Serum levels of injected hPCSK9 were then tracked over time (Figure 4A). Rapid removal of hPCSK9 was observed in WT and hLDLR transgenic mice, with a calculated serum half-life of 5.2 and 2.9 min, respectively, and a nearly complete disappearance of the protein from serum after 1 hour. Conversely, hPCSK9 lingered in the serum of LDLR−/− mice, with a calculated serum half-life of 50.5 min. The clearance pattern of hPCSK9 in WT and LDLR−/− mice was similar to that of intravenously injected LDL (Supplementary Figure 4A), suggesting a dominant role for the LDLR in PCSK9 removal. Clearance of serum albumin served as a negative control for LDLR dependent clearance, and was similar in WT and LDLR−/− mice (Supplementary figure 4B).
Figure 4.
Turnover of recombinant hPCSK9 in mice. (A) Serum levels of purified hPCSK9 injected in the retro-orbital vein plexus of WT, LDLR−/−, or hLDLR mice (n=3-4); results are presented as percent ± CV of hPCSK9 relative to time zero. Half-life of hPCSK9 was calculated using exponential decay equation. (B) Representative immunoblot of LDLR and β-actin levels in different tissues of WT or LDLR−/− mice four hours after hPCSK9 injection (quantitative analysis is shown in Supplementary Figure 5A-B). (C) Accumulation of hPCSK9 in tissues of WT or LDLR−/− mice 4 hours after hPCSK9 injection (n=3). hPCSK9 levels were calculated by ELISA assay. (D) Representative immunoblot for hPCSK9 in hepatocytes after a 4-hour incubation with or without 500 ng of hPCSK9. (*p<0.05, ** p<0.01).
In addition to tracking hPCSK9 in the serum, we also monitored hPCSK9 accumulation and LDLR reduction in liver, kidney, and adrenal glands. Four hours after injection, the strongest effect of hPCSK9 in WT mice was seen in the liver, with 44.8% reduction in LDLR levels, whereas the effect in the kidney was a 35% reduction in LDLR levels (Figure 4B and Supplemental Figure 5A). Accumulation of hPCSK9 mirrored that of LDLR reduction, with the highest accumulation in the liver of WT animals. In contrast, livers from LDLR−/− mice accumulated significantly lower levels of hPCSK9 (19.3±4.3%) compared to WT mice (Figure 4C). Despite the changes in LDLR levels following injection of hPCSK9 in WT mice, no effects were seen on serum cholesterol levels, possibly owing to the transient effect and the low amount of PCSK9 used. To confirm the role of LDLR in hPCSK9 uptake directly, we incubated primary hepatocytes from WT or LDLR−/− mice with purified hPCSK9 and found that the uptake of hPCSK9 (500 ng/ml) by LDLR−/− hepatocytes was 72.2% lower than that seen in WT hepatocytes (Figure 4D). As expected, exposure of WT hepatocytes to hPCSK9 reduced LDLR levels (Supplementary Figure 5B).
In agreement with our findings in the hPCSK9 transgenic mice, LDLR levels in the adrenals were not affected by the injected hPCSK9, likely a consequence of the fact that exogenous hPCSK9 did not accumulate in this tissue (Figure 4C). Co-immunoprecipitation of hPCSK9 showed a weaker interaction between LDLR and PCSK9 in the adrenals compared to the liver of hPCSK9 transgenic mice (Supplementary Figure 6A). Such results suggest that tissue-specific penetration or retention of PCSK9 may be involved in its functional regulation. To test this hypothesis in vitro we incubated mouse adrenal cells (Y-1) and mouse hepatocytes (Aml-12) with purified hPCSK9 (500 ng/ml), and found greater LDLR reduction and more cellular hPCSK9 uptake by mouse hepatocytes compared to mouse adrenal cells (Supplementary Figure 6B). To determine whether PCSK9 can influence adrenal cell LDLR levels at all, we studied the effects of overexpression of hPCSK9, or its GOF mutation D374Y, in Y-1 cells. Our results show that transduction of hPCSK9, and particularly the D374Y mutant, results in reduced LDLR levels in Y-1 cell (47% and 77% reduction, respectively) showing that adrenal LDLR can be a target of hPCSK9 action in some circumstances (Supplementary figure 6C).
Serum distribution of human PCSK9
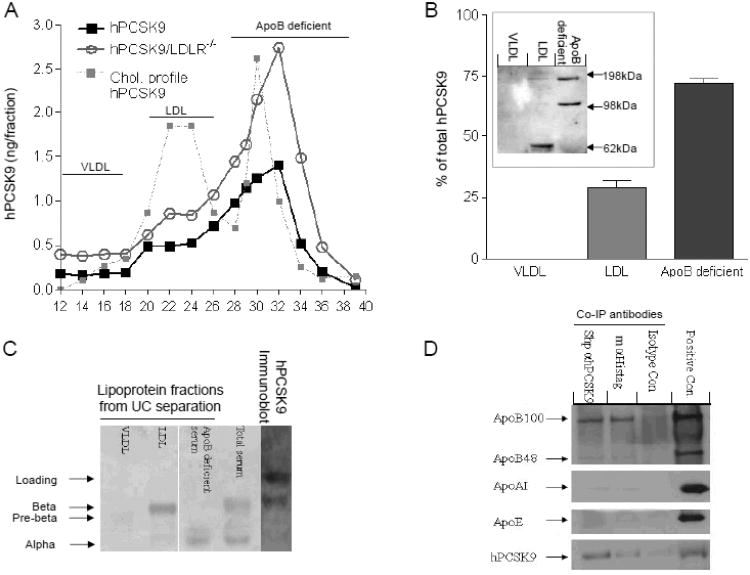
FPLC analysis of transgenic mouse serum shows that 75.2±6.1% of hPCSK9 co-elutes with the apoB-deficient fraction, and the rest (24.8±2.8%) co-elutes with the LDL fraction, regardless of LDLR expression (Figure 5A). Similar results were obtained with ultracentrifugation (UC) of hPCSK9 transgenic mouse serum using neutral density gradient (Figure 5B), with 28.9±4% of total serum hPCSK9 found in the LDL fraction and the rest in the apoB-deficient fraction. Analyses of serum from two normolipidemic subjects confirmed a similar distribution pattern, with 35% and 39% of PCSK9 associated with the LDL fraction (not shown). To analyze the nature of hPCSK9 association with LDL, we treated the FPLC fractions with a crosslinking reagent (DSP, 0.5mM, 30 min) and evaluated PCSK9 distribution after gel electrophoresis (Figure 5B; insert). The results show that VLDL does not contain hPCSK9, LDL contains mainly monomeric hPCSK9, and the apoB-deficient fraction contains hPCSK9 that is mostly of higher molecular weight, likely dimers and trimers 25. In agreement with our FPLC and UC results, direct immunobloting of hPCSK9 transgenic mouse serum after agarose gel electrophoresis showed that ∼30% of hPCSK9 is associated with LDL (Figure 5C). Interestingly, UC lipoprotein separation using high-salt solution (KBr) resulted in >95% of PCSK9 being found in the lipoprotein-deficient serum (Supplementary Table 1), as was previously shown 27. Further analysis of transgenic mouse serum using co-immunoprecipitation of hPCSK9 (with either anti-hPCSK9 or anti-Histidine-tag antibodies) showed that PCSK9 is associated with apoB100, with weak association with apoB48 and no association with apoAI, or apoE (Figure 5D). Native gel immunoblot of the crossed linked FPLC fractions confirmed that PCSK9 is indeed part of the LDL particle (Supplementary Figure 7).
Figure 5.
Lipoprotein association of serum PCSK9. (A) Serum FPLC profile from hPCSK9 transgenic mice on either WT or LDLR−/− backgrounds. Each profile represents pooled serum from 3-4 mice, Fractions: VLDL 12-17, LDL 18-27, apoB-deficient serum (including HDL) 28-39. PCSK9 in each fraction was calculated as the area under the curve (AUC). (B) Relative hPCSK9 levels in transgenic mouse serum after UC using neutral density gradient medium (Optiprep). A pool of 21 mice was used for 3 different separations; Insert: hPCSK9 immunoblot from concentrated FPLC fractions of transgenic mouse serum. (C) Lipid staining of serum from transgenic mice after Optiprep UC (left) or direct immunoblot (right). (D) Apolipoprotein detection via immunoblot of hPCSK9 transgenic mouse serum following immunoprecipitation with anti-PCSK9 antibodies (m=Mouse, Shp=Sheep, Isotype control= Shp IgG). Serum from hPCSK9 transgenic mice served as positive control.
Discussion
We used transgenic expression and turnover studies of human PCSK9 to investigate its serum levels, tissue distribution and activity. Our goal was to investigate the existence and degree of a reciprocal regulation between PCSK9 and LDLR, and its influence on lipid levels in the mouse. Using multiple approaches, we show that clearance of serum PCSK9 is predominantly due to LDLR-mediated uptake. We also found that PCSK9 increases serum cholesterol levels via both LDLR-dependent and LDLR-independent pathways, and that serum PCSK9 associates with LDL in a way that can affect peripheral or hepatic PCSK9 action. Finally, we investigated the basis for the lack of PCSK9 effect on LDLR levels in adrenals, and provide results suggesting that higher levels of PCSK9 are needed to reduce LDLR levels in adrenals compared to liver, a phenomenon that is likely aggravated by the limited retention of circulating PCSK9 in the adrenal tissue. A race to use the inhibition of PCSK9 as a treatment for hypercholesterolemia is underway. Current drug development efforts are directed either at reducing production of PCSK9 37,38, or at blocking circulating PCSK9 via neutralizing antibodies 39-41, as the only known effect of PCSK9 is the binding and degradation of LDLR 42. PCSK9 is expressed in liver, small intestine, and kidneys, but its main target is hepatic LDLR 10, 32. In our transgenic mouse, hPCSK9 was produced mainly in the kidney and affected mostly LDLR levels in liver (Figure 1). The LDLR reducing effect in the kidney was more modest and there was no effect in the adrenals, as previously reported 26, 33. We show that in addition to LDLR reduction, hPCSK9 expression also increases secretion of triglyceride-rich lipoproteins in an LDLR-independent fashion, thus increasing cholesterol levels also in LDLR−/− mice (Figure 2). Recent reports showing that PCSK9 increases apoB secretion in hepatocytes 24 and intestinal cells 43, and decreases VLDLR levels in adipocytes 23, provide a mechanistic basis for an LDLR-independent effect of PCSK9 on cholesterol levels.
Current evidence does not suggest the presence of a correlation between the degree of LDLR function loss and serum PCSK9 levels in humans 20. Similar PCSK9 levels were found among hypercholesterolemia patients with or without LDLR mutations 17. Additionally, PCSK9 levels similar to controls were found in subjects with familial hypercholesterolemia (FH) and cholesterol levels above the 90th percentile, while PCSK9 levels even lower than controls were found in subjects with FH and cholesterol levels below the 75th percentile have 19. Although a recent study reported modestly increased PCSK9 levels in familial combined hyperlipidemia patients compared to controls, LDLR mutations did not have an influence on this small effect 18. Others have reported a minimal (11%) increase in PCSK9 levels of untreated FH patients compared to controls 44. It has to be kept in mind that most FH presentations in humans are due to LDLR mutations that cause dysfunctional interaction with LDL while the receptor protein is intact and appropriately located on the cell surface. Since PCSK9 binds a different domain of the LDLR 16, one would expect normal PCSK9-LDLR interactions in these situations. In contrast, our approach allowed us to study a different scenario, when both LDL and PCSK9 are affected by the absence of LDLR. This scenario is repeated clinically in FH subjects with receptor-negative mutations of the LDLR.
We show that expression of hPCSK9 increases serum mPCSK9 levels in WT but not in LDLR−/− mice, an effect likely due to the reduction in hepatic LDLR caused by hPCSK9, in turn impairing LDLR-mediated clearance of mPCSK9. The massive accumulation of serum mPCSK9 caused by absence of LDLR cannot be obviously modulated further by the expression of hPCSK9 (Figure 3). This phenomenon was also spontaneously evident in LDLR+/− and to a greater extent in LDLR−/− mice (Supplementary Figure 2). A mirror image of this was seen with the acute induction of hepatic hLDLR in transgenic mice, which caused a drastic and prompt two-thirds reduction of circulating mPCSK9 levels. This novel observation also provides the first proof of interaction between hLDLR and mPCSK9, likely explained by the high degree of homology between murine and human PCSK9 12. Additionally, serum hPCSK9 levels in transgenic mice on LDLR−/− background were two-times higher than those of transgenic mice on WT background (Figure 3), again showing that LDLR regulates serum levels of PCSK9 by acting as its primary clearance route. Additionally, hPCSK9/LDLR−/− mice had lower levels of intra-hepatic hPCSK9, suggesting that internalization of PCSK9 by liver cells depends mainly on LDLR.
Similarly, transduced mouse hepatocytes showed a temporal and causal relationship between hPCSK9 expression and levels of both medium mPCSK9 and membrane LDLR. In particular, we showed that mPCSK9 started to accumulate in the medium when the secreted levels of hPCSK9 exceeded basal mPCSK9 levels (>1:1 ratio), (Figure 3). This observation shows that the ability of murine hepatocytes to take up PCSK9 is directly related to the levels of surface LDLR. The above observations support our hypothesis that the relationship between PCSK9 and LDLR is reciprocal rather than unidirectional. It is undisputed that PCSK9 regulates LDLR levels 4, 6, 21, but it is also clear from our studies that LDLR regulates PCSK9 levels. Since both effects are post-transcriptional, it is likely that steady-state serum cholesterol levels in different individuals are affected by the global balance of these reciprocal regulatory influences, where a primary excess of LDLR would deplete the PCSK9 pool and maintain proper LDL clearance, and a primary excess of PCSK9 would deplete the LDLR pool and cause serum LDL elevations. Thus, it is possible that some forms of hypercholesterolemia result from unbalanced homeostasis between levels of serum PCSK9 and those of membrane LDLR. An example of this was recently published, with the demonstration that resistin increases PCSK9 levels and causes hypercholesterolemia in obese subjects 45.
Since transgenic expression of hPCSK9 influences LDLR levels chronically and allows for counter-regulatory mechanisms to help reach a steady state, we also investigated the acute effects of hPCSK9 after intravenous injection in mice. Our results show that LDLR is the dominant clearance route for serum PCSK9. We injected a small amount of hPCSK9 to raise serum hPCSK9 levels to an average of 714±218 ng/ml, well within the physiologic range, and showed a marked difference in the half-life of hPCSK9 between WT (5.2 min) 33, 34 and LDLR−/− mice (50.5 min). In contrast, the acute over-expression of hLDLR decreased hPCSK9 serum half-life by almost half (2.9 min), (Figure 4). The nearly 10-fold increase in hPCSK9 half-life matches the 9.5-fold increase of mPCSK9 in the serum of LDLR−/− mice vs. controls. The significant increase in hPCSK9 half-life we observed in LDLR−/− mice is different from the slightly prolonged increase (15 min) previously reported 33. In addition, our in vivo and in vitro data also show a much larger reduction in hPCSK9 uptake by liver and primary hepatocytes of LDLR−/− mice (19.3% and 27.8% of control, respectively) compared to the milder accumulation defect (∼50%) previously reported 33. These differences may derive from the use of 125I-labeled PCSK9 with its inherent drawbacks due to protein oxidation, or from the supra-physiologic levels of PCSK9 used in previous studies, causing an artificial exaggeration of the LDLR-independent clearance pathway. This latter possibility is also suggested by our data with transgenic mice, showing that mPCSK9 levels are 9.5-fold higher in LDLR−/− mice compared to WT, whereas hPCSK9 levels are only 2-fold higher in hPCSK9/LDLR−/− mice compared to hPCSK9 WT.
Although the clearance of LDL reflected the expected pattern in both in WT mice (fast) and LDLR−/− mice (slow), (Supplementary Figure 4), the absence of LDLR had a stronger effect on the half-life of hPCSK9 (9.7 fold increase over WT) than on that of LDL (5.2 fold increase over WT), suggesting that LDLR dependent clearance is actually more important to PCSK9 than to LDL. This observation was confirmed in heterozygous LDLR+/− mice, where no significant differences were seen in cholesterol levels (not shown) while mPCSK9 levels were up 2.7-fold compared to controls (Supplementary Figure 2)
Transgenic expression of hPCSK9 did not have an effect on adrenal LDLR levels, a finding that confirms a previous report 26, 33. In addition, we show that PCSK9 injected in WT mice accumulates in the liver and kidney but not in the adrenals, and found a weaker association between PCSK9 and LDLR in the adrenals compared to liver. In vitro, adrenal LDLR levels were not affected by the addition of hPCSK9 and these cells internalized much less hPCSK9 compared to hepatocytes. It has been suggested that annexinA2 may act as an extrahepatic inhibitor of PCSK9 46, however the impaired adrenal retention observed in our study appears to be independent of annexinA2 (Supplementary Figure 6B). Moreover, significant LDLR reduction in adrenal cells was seen after over-expression of hPCSK9 or its D374Y GOF mutation. These data combined suggest that the lack of effect of PCSK9 in adrenals is due to its limited retention in this tissue (Supplementary Figure 6).
Finally, we have studied the association of PCSK9 with serum lipoproteins, a controversial issue. We were the first to report PCSK9 association with LDL particles in vivo and in vitro 25, whereas others reported that PCSK9 either simply co-elutes with lipoproteins (using size exclusion chromatography) 24-26, or does not bind at all to lipoproteins (using high-salt ultracentrifugal separation), 27. We used multiple approaches to clarify this important issue and explain the discrepancies in the literature: (1) by FPLC separation of serum lipoproteins, we showed that 24.8% of serum hPCSK9 elutes with the LDL fraction, either because of physical association or coincidental co-elution; (2) by direct immunobloting of serum separated on an agarose gel, we were able to show, for the first time, that about 30% of hPCSK9 is directly associated with the beta-migrating LDL band; (3) using a natural gradient solution, we showed that 28.9% of serum hPCSK9 is found in the LDL fraction; (4) by co-immunoprecipitation of total serum, we showed association between hPCSK9 and apoB-100; (5) by native gel immunobloting of FPLC fractions we showed that PCSK9 is associated with the LDL. Taken together, our data allow us to conclude that more than a quarter of serum hPCSK9 in mouse serum (more than a third in human serum) is associated with the LDL, whereas the remainder is found on the apoB-deficient fraction. In addition, hPCSK9 associated with LDL is a monomer, whereas the rest of serum hPCSK9 is found in larger complexes. We have previously reported that self-associated PCSK9 is more active than its' monomer 25, thus suggesting that the LDL particle can directly influence PCSK9 activity in the serum and regulate LDLR levels in peripheral tissues. Of note, with a high-salt ultracentrifugation approach we did not find PCSK9 association with LDL, in line with results from a previous study 27, and suggesting that this harsh method causes PCSK9 dissociation from the LDL. During the revision of this manuscript, a paper by Kosenko et al 47 used a natural density gradient ultracentrifugation approach similar to the one we are describing here and showed that in human serum, PCSK9 associates with LDL and modulates its function.
In summary, modulating hPCSK9 and LDLR levels in mice enabled us to show that LDLR is the primary route for serum PCSK9 clearance. Our data presents a tight, tissue-specific, reciprocal regulation between these interacting partners, which determines serum PCSK9 levels, hepatic LDLR levels, and serum LDL-c levels. We conclude that the lack of correlation between PCSK9 and LDLR functionality in humans stems from the ability of dysfunctional LDLR (impaired interaction with LDL) to still internalize PCSK9, and not because of alternative LDLR-independent internalization pathways. Our results forecast a clinical scenario where LDLR mutations that affect both LDL and PCSK9 clearance (e.g. receptor-negative mutations) aggravate hypercholesterolemia via the effect of accumulated PCSK9 on the normal LDLR allele product. To this regard, it must be noted that carriers of LDLR-negative mutations have been reported to show significantly higher LDL levels compared with carriers of dysfunctional LDLR mutations 48. Patients carrying such mutations would be particularly good targets for PCSK9 inhibition approaches.
Supplementary Material
Supplementary figure 1: (A) ELISA quantification of hCSK9 in tissues of transgenic mice.(B) Effect of hPCSK9 expression in-vivo: quantification of immunoblots for hepatic LDLR levels normalized to β-actin (n=4-6). (*p<0.05, ** p<0.01).
Supplementary figure 2: (A) Agarose gel separation of serum lipoproteins. (B) mPCSK9 levels. (C) Immunoblot and ELISA quantification of hPCSK9 in liver of controls and transgenic mice.
Supplementary figure 3: (A) qRT-PCR for hPCSK9, mPCSK9 and LDLR in Aml-12 cells transduced with pWPI-GFP, pWPI-hPCSK9, or pWPI-D374Y gain of function mutation. (B) Coomassie blue stain of His-tagged eluted fraction from “ProBond” Nickel column of media from HEK293T cell transduced with pWPI-hPCSK9 lentivirus.
Supplementary figure 4: Turnover of human serum albumin and human LDL in mice. (A) Clearance of human LDL injected to WT or LDLR−/− mice (n=3), (B) Clearance of human serum albumin injected to WT or LDLR−/− mice (n=3), (*p<0.05, ** p<0.01).
Supplementary figure 5: (A) Quantification of immunoblots for hepatic and renal LDLR normalized to β-actin (n=3). (B) Immunoblot for LDLR, and beta actin in primary hepatocyte upon addition of hPCSK9 (500 ng/ml) for 4hrs, (p<0.01).
Supplementary figure 6: Effect of hPCSK9 in the adrenals. (A) Immunoblot of LDLR after co-immunoprecipitation with hPCSK9 of adrenal and liver from hPCSK9tg mice. (B) Immunoblot for LDLR, PCSK9, beta actin and annexinA2 in hepatocytes (AML-12) and adrenal (Y-1) cells (C=cell, M=media). (C) immunoblot of mouse adrenal cells (Y-1) transduced with GFP (control), PCSK9, or PCSK9 GOF mutation (D374Y).
Supplementary figure 7: hPCSK9 immunoblot for FPLC fractions treated with cross linker DSP (0.5mM, 30 min) on a native gel (4-16%).
Supplementary table 1: Serum mPCSK9 and hPCSK9 distribution after high-salt (KBr) ultracentrifugal separation of lipoproteins from hPCSK9 transgenic mouse serum, measured by PCSK9 ELISA and presented as % of total PCSK9.
Clinical Summary.
We show two novel post-transcriptional regulatory mechanisms determining serum levels and function of PCSK9: (1) Clearance via the LDL receptor (LDLR); and (2) Transport on LDL in a quantitative significant but less active form. Although PCSK9 is known to regulate LDLR levels, we report here that the reverse is also true, and the main exit route for serum PCSK9 is via LDLR. Thus, LDLR mutations may have larger effects on LDL metabolism if they affect not only LDL internalization but also PCSK9 clearance. We foresee a clinical scenario where LDLR mutations that affect PCSK9 clearance in addition to LDL clearance (e.g., receptor negative) can aggravate hypercholesterolemia via the effect of accumulated PCSK9, resulting from defective clearance, on the normal LDLR allele product. In this line of thinking, it is also possible to forecast hypercholesterolemia due to an LDLR mutation that only causes defective PCSK9 clearance. It is interesting that carriers of LDLR-negative mutations have indeed been reported to show significantly higher LDL levels compared with carriers of dysfunctional LDLR mutations. Our finding that about one third of serum PCSK9 is part of LDL and is in a monomeric, less active form, suggests that peripheral distribution and function of this regulator of LDLR levels is influenced by the lipoprotein that acts as canonical ligand for LDLR. Maneuvers that increase PCSK9 content of LDL in serum may allow us to broaden the search for inhibitory strategies of PCSK9 action, currently limited to blocking the interaction between PCSK9 and LDLR via antibodies.
Acknowledgments
We acknowledge support from the Atherosclerosis Core of Vanderbilt Mouse Metabolic Phenotyping Center.
Funding Sources: This study was supported by the National Institutes of Health (NHLBI) through grant R01-HL106845 to Sergio Fazio.
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Hunt KJ, Resendez RG, Williams K, Haffner SM, Stern MP. National Cholesterol Education Program versus World Health Organization metabolic syndrome in relation to all-cause and cardiovascular mortality in the San Antonio Heart Study. Circulation. 2004;110:1251–1257. doi: 10.1161/01.CIR.0000140762.04598.F9. [DOI] [PubMed] [Google Scholar]
- 2.Horton JD, Goldstein JL, Brown MS. SREBPs: transcriptional mediators of lipid homeostasis. Cold Spring Harb Symp Quant Biol. 2002;67:491–498. doi: 10.1101/sqb.2002.67.491. [DOI] [PubMed] [Google Scholar]
- 3.Glassberg H, Rader DJ. Management of lipids in the prevention of cardiovascular events. Annu Rev Med. 2008;59:79–94. doi: 10.1146/annurev.med.59.121206.112237. [DOI] [PubMed] [Google Scholar]
- 4.Benjannet S, Rhainds D, Essalmani R, Mayne J, Wickham L, Jin W, Asselin MC, Hamelin J, Varret M, Allard D, Trillard M, Abifadel M, Tebon A, Attie AD, Rader DJ, Boileau C, Brissette L, Chretien M, Prat A, Seidah NG. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J Biol Chem. 2004;279:48865–48875. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- 5.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 6.Rashid S, Curtis DE, Garuti R, Anderson NN, Bashmakov Y, Ho YK, Hammer RE, Moon YA, Horton JD. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc Natl Acad Sci U S A. 2005;102:5374–5379. doi: 10.1073/pnas.0501652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, Derre A, Villeger L, Farnier M, Beucler I, Bruckert E, Chambaz J, Chanu B, Lecerf JM, Luc G, Moulin P, Weissenbach J, Prat A, Krempf M, Junien C, Seidah NG, Boileau C. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 8.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 9.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11:367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 10.Seidah NG, Benjannet S, Wickham L, Marcinkiewicz J, Jasmin SB, Stifani S, Basak A, Prat A, Chretien M. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc Natl Acad Sci U S A. 2003;100:928–933. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubuc G, Chamberland A, Wassef H, Davignon J, Seidah NG, Bernier L, Prat A. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2004;24:1454–1459. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- 12.Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- 13.Ason B, Tep S, Davis HR, Jr, Xu Y, Tetzloff G, Galinski B, Soriano F, Dubinina N, Zhu L, Stefanni A, Wong KK, Tadin-Strapps M, Bartz SR, Hubbard B, Ranalletta M, Sachs AB, Flanagan WM, Strack A, Kuklin NA. Improved efficacy for ezetimibe and rosuvastatin by attenuating the induction of PCSK9. J Lipid Res. 2011;52:679–687. doi: 10.1194/jlr.M013664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mousavi SA, Berge KE, Leren TP. The unique role of proprotein convertase subtilisin/kexin 9 in cholesterol homeostasis. J Intern Med. 2009;266:507–519. doi: 10.1111/j.1365-2796.2009.02167.x. [DOI] [PubMed] [Google Scholar]
- 15.Zaid A, Roubtsova A, Essalmani R, Marcinkiewicz J, Chamberland A, Hamelin J, Tremblay M, Jacques H, Jin W, Davignon J, Seidah NG, Prat A. Proprotein convertase subtilisin/kexin type 9 (PCSK9): hepatocyte-specific low-density lipoprotein receptor degradation and critical role in mouse liver regeneration. Hepatology. 2008;48:646–654. doi: 10.1002/hep.22354. [DOI] [PubMed] [Google Scholar]
- 16.Zhang DW, Lagace TA, Garuti R, Zhao Z, McDonald M, Horton JD, Cohen JC, Hobbs HH. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J Biol Chem. 2007;282:18602–18612. doi: 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- 17.Ahmad Z, Adams-Huet B, Chen C, Garg A. Low prevalence of mutations in known Loci for autosomal dominant hypercholesterolemia in a multiethnic patient cohort. Circ Cardiovasc Genet. 2012;5:666–675. doi: 10.1161/CIRCGENETICS.112.963587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brouwers MC, Konrad RJ, van Himbergen TM, Isaacs A, Otokozawa S, Troutt JS, Schaefer EJ, van Greevenbroek MM, Stalenhoef AF, de Graaf J. Plasma proprotein convertase subtilisin kexin type 9 levels are related to markers of cholesterol synthesis in familial combined hyperlipidemia. Nutr Metab Cardiovasc Dis. 2012 doi: 10.1016/j.numecd.2012.1011.1008. [DOI] [PubMed] [Google Scholar]
- 19.Huijgen R, Fouchier SW, Denoun M, Hutten BA, Vissers MN, Lambert G, Kastelein JJ. Plasma levels of PCSK9 and phenotypic variability in familial hypercholesterolemia. J Lipid Res. 2012;53:979–983. doi: 10.1194/jlr.P023994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cameron J, Bogsrud MP, Tveten K, Strom TB, Holven K, Berge KE, Leren TP. Serum levels of proprotein convertase subtilisin/kexin type 9 in subjects with familial hypercholesterolemia indicate that proprotein convertase subtilisin/kexin type 9 is cleared from plasma by low-density lipoprotein receptor-independent pathways. Transl Res. 2012;160:125–130. doi: 10.1016/j.trsl.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Maxwell KN, Breslow JL. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci U S A. 2004;101:7100–7105. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denis M, Marcinkiewicz J, Zaid A, Gauthier D, Poirier S, Lazure C, Seidah NG, Prat A. Gene inactivation of proprotein convertase subtilisin/kexin type 9 reduces atherosclerosis in mice. Circulation. 2012;125:894–901. doi: 10.1161/CIRCULATIONAHA.111.057406. [DOI] [PubMed] [Google Scholar]
- 23.Roubtsova A, Munkonda MN, Awan Z, Marcinkiewicz J, Chamberland A, Lazure C, Cianflone K, Seidah NG, Prat A. Circulating proprotein convertase subtilisin/kexin 9 (PCSK9) regulates VLDLR protein and triglyceride accumulation in visceral adipose tissue. Arterioscler Thromb Vasc Biol. 2010;31:785–791. doi: 10.1161/ATVBAHA.110.220988. [DOI] [PubMed] [Google Scholar]
- 24.Sun H, Samarghandi A, Zhang N, Yao Z, Xiong M, Teng BB. Proprotein Convertase Subtilisin/Kexin Type 9 Interacts With Apolipoprotein B and Prevents Its Intracellular Degradation, Irrespective of the Low-Density Lipoprotein Receptor. Arterioscler Thromb Vasc Biol. 2012;32:1585–1595. doi: 10.1161/ATVBAHA.112.250043. [DOI] [PubMed] [Google Scholar]
- 25.Fan D, Yancey PG, Qiu S, Ding L, Weeber EJ, Linton MF, Fazio S. Self-association of human PCSK9 correlates with its LDLR-degrading activity. Biochemistry. 2008;47:1631–1639. doi: 10.1021/bi7016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Y, Warren L, Xia D, Jensen H, Sand T, Petras S, Qin W, Miller KS, Hawkins J. Function and distribution of circulating human PCSK9 expressed extrahepatically in transgenic mice. J Lipid Res. 2009;50:1581–1588. doi: 10.1194/jlr.M800542-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alborn WE, Cao G, Careskey HE, Qian YW, Subramaniam DR, Davies J, Conner EM, Konrad RJ. Serum proprotein convertase subtilisin kexin type 9 is correlated directly with serum LDL cholesterol. Clin Chem. 2007;53:1814–1819. doi: 10.1373/clinchem.2007.091280. [DOI] [PubMed] [Google Scholar]
- 28.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 29.Poirier S, Mayer G, Benjannet S, Bergeron E, Marcinkiewicz J, Nassoury N, Mayer H, Nimpf J, Prat A, Seidah NG. The proprotein convertase PCSK9 induces the degradation of low density lipoprotein receptor (LDLR) and its closest family members VLDLR and ApoER2. J Biol Chem. 2008;283:2363–2372. doi: 10.1074/jbc.M708098200. [DOI] [PubMed] [Google Scholar]
- 30.Fan D, Qiu S, Overton CD, Yancey PG, Swift LL, Jerome WG, Linton MF, Fazio S. Impaired secretion of apolipoprotein E2 from macrophages. J Biol Chem. 2007;282:13746–13753. doi: 10.1074/jbc.M611754200. [DOI] [PubMed] [Google Scholar]
- 31.Farkas MH, Swift LL, Hasty AH, Linton MF, Fazio S. The recycling of apolipoprotein E in primary cultures of mouse hepatocytes. Evidence for a physiologic connection to high density lipoprotein metabolism. J Biol Chem. 2003;278:9412–9417. doi: 10.1074/jbc.M208026200. [DOI] [PubMed] [Google Scholar]
- 32.Poirier S, Prat A, Marcinkiewicz E, Paquin J, Chitramuthu BP, Baranowski D, Cadieux B, Bennett HP, Seidah NG. Implication of the proprotein convertase NARC-1/PCSK9 in the development of the nervous system. J Neurochem. 2006;98:838–850. doi: 10.1111/j.1471-4159.2006.03928.x. [DOI] [PubMed] [Google Scholar]
- 33.Grefhorst A, McNutt MC, Lagace TA, Horton JD. Plasma PCSK9 preferentially reduces liver LDL receptors in mice. J Lipid Res. 2008;49:1303–1311. doi: 10.1194/jlr.M800027-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt RJ, Beyer TP, Bensch WR, Qian YW, Lin A, Kowala M, Alborn WE, Konrad RJ, Cao G. Secreted proprotein convertase subtilisin/kexin type 9 reduces both hepatic and extrahepatic low-density lipoprotein receptors in vivo. Biochem Biophys Res Commun. 2008;370:634–640. doi: 10.1016/j.bbrc.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Lagace TA, Curtis DE, Garuti R, McNutt MC, Park SW, Prather HB, Anderson NN, Ho YK, Hammer RE, Horton JD. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J Clin Invest. 2006;116:2995–3005. doi: 10.1172/JCI29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konrad RJ, Troutt JS, Cao G. Effects of currently prescribed LDL-C-lowering drugs on PCSK9 and implications for the next generation of LDL-C-lowering agents. Lipids Health Dis. 2011:10–38. doi: 10.1186/1476-511X-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graham MJ, Lemonidis KM, Whipple CP, Subramaniam A, Monia BP, Crooke ST, Crooke RM. Antisense inhibition of proprotein convertase subtilisin/kexin type 9 reduces serum LDL in hyperlipidemic mice. J Lipid Res. 2007;48:763–767. doi: 10.1194/jlr.C600025-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A, Butler D, Charisse K, Dorkin R, Fan Y, Gamba-Vitalo C, Hadwiger P, Jayaraman M, John M, Jayaprakash KN, Maier M, Nechev L, Rajeev KG, Read T, Rohl I, Soutschek J, Tan P, Wong J, Wang G, Zimmermann T, de Fougerolles A, Vornlocher HP, Langer R, Anderson DG, Manoharan M, Koteliansky V, Horton JD, Fitzgerald K. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc Natl Acad Sci U S A. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stein EA, Gipe D, Bergeron J, Gaudet D, Weiss R, Dufour R, Wu R, Pordy R. Effect of a monoclonal antibody to PCSK9, REGN727/SAR236553, to reduce low-density lipoprotein cholesterol in patients with heterozygous familial hypercholesterolaemia on stable statin dose with or without ezetimibe therapy: a phase 2 randomised controlled trial. Lancet. 2012;380:29–36. doi: 10.1016/S0140-6736(12)60771-5. [DOI] [PubMed] [Google Scholar]
- 40.Giugliano Robert P, MD NRD, Kohli Payal, Rogers William J, Somaratne Ransi, Huang Fannie, Liu Thomas, Mohanavelu Satishkumar, Hoffman Elaine B, McDonald Shannon T, Abrahamsen Timothy E, Wasserman Scott M, Scott Robert, Sabatine Marc S. Efficacy, safety, and tolerability of a monoclonal antibody to proprotein convertase subtilisin/kexin type 9 in combination with a statin in patients with hypercholesterolaemia (LAPLACE-TIMI 57): a randomised, placebo-controlled, dose-ranging, phase 2 study. The Lancet. 2012;380:2007–2017. doi: 10.1016/S0140-6736(12)61770-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raal F, Scott R, Somaratne R, Bridges I, Li G, Wasserman SM, Stein EA. Low-Density Lipoprotein Cholesterol-Lowering Effects of AMG 145, a Monoclonal Antibody to Proprotein Convertase Subtilisin/Kexin Type 9 Serine Protease in Patients With Heterozygous Familial Hypercholesterolemia: The Reduction of LDL-C With PCSK9 Inhibition in Heterozygous Familial Hypercholesterolemia Disorder (RUTHERFORD) Randomized Trial. Circulation. 2012;126:2408–2417. doi: 10.1161/CIRCULATIONAHA.112.144055. [DOI] [PubMed] [Google Scholar]
- 42.Poirier S, Mayer G, Poupon V, McPherson PS, Desjardins R, Ly K, Asselin MC, Day R, Duclos FJ, Witmer M, Parker R, Prat A, Seidah NG. Dissection of the endogenous cellular pathways of PCSK9-induced low density lipoprotein receptor degradation: evidence for an intracellular route. J Biol Chem. 2009;284:28856–28864. doi: 10.1074/jbc.M109.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levy E, Ouadda AB, Spahis S, Sane AT, Garofalo C, Grenier E, Emonnot L, Yara S, Couture P, Beaulieu JF, Menard D, Seidah NG, Elchebly M. PCSK9 plays a significant role in cholesterol homeostasis and lipid transport in intestinal epithelial cells. Atherosclerosis. 2013;227:297–306. doi: 10.1016/j.atherosclerosis.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 44.Dubuc G, Tremblay M, Pare G, Jacques H, Hamelin J, Benjannet S, Boulet L, Genest J, Bernier L, Seidah NG, Davignon J. A new method for measurement of total plasma PCSK9: clinical applications. J Lipid Res. 2010;51:140–149. doi: 10.1194/jlr.M900273-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melone M, Wilsie L, Palyha O, Strack A, Rashid S. Discovery of a new role of human resistin in hepatocyte low-density lipoprotein receptor suppression mediated in part by proprotein convertase subtilisin/kexin type 9. J Am Coll Cardiol. 2012;59:1697–1705. doi: 10.1016/j.jacc.2011.11.064. [DOI] [PubMed] [Google Scholar]
- 46.Seidah NG, Poirier S, Denis M, Parker R, Miao B, Mapelli C, Prat A, Wassef H, Davignon J, Hajjar KA, Mayer G. Annexin A2 is a natural extrahepatic inhibitor of the PCSK9-induced LDL receptor degradation. PLoS One. 2012;7:e41865. doi: 10.1371/journal.pone.0041865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kosenko T, Golder M, Leblond G, Weng W, Lagace TA. Low-density lipoprotein binds to proprotein convertase subtilisin/kexin type-9 (PCSK9) in human plasma and inhibits PCSK9-mediated LDL receptor degradation. J Biol Chem. 2013 doi: 10.1074/jbc.M112.421370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guardamagna O, Restagno G, Rolfo E, Pederiva C, Martini S, Abello F, Baracco V, Pisciotta L, Pino E, Calandra S, Bertolini S. The type of LDLR gene mutation predicts cardiovascular risk in children with familial hypercholesterolemia. J Pediatr. 2009;155:199–204. e192. doi: 10.1016/j.jpeds.2009.02.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1: (A) ELISA quantification of hCSK9 in tissues of transgenic mice.(B) Effect of hPCSK9 expression in-vivo: quantification of immunoblots for hepatic LDLR levels normalized to β-actin (n=4-6). (*p<0.05, ** p<0.01).
Supplementary figure 2: (A) Agarose gel separation of serum lipoproteins. (B) mPCSK9 levels. (C) Immunoblot and ELISA quantification of hPCSK9 in liver of controls and transgenic mice.
Supplementary figure 3: (A) qRT-PCR for hPCSK9, mPCSK9 and LDLR in Aml-12 cells transduced with pWPI-GFP, pWPI-hPCSK9, or pWPI-D374Y gain of function mutation. (B) Coomassie blue stain of His-tagged eluted fraction from “ProBond” Nickel column of media from HEK293T cell transduced with pWPI-hPCSK9 lentivirus.
Supplementary figure 4: Turnover of human serum albumin and human LDL in mice. (A) Clearance of human LDL injected to WT or LDLR−/− mice (n=3), (B) Clearance of human serum albumin injected to WT or LDLR−/− mice (n=3), (*p<0.05, ** p<0.01).
Supplementary figure 5: (A) Quantification of immunoblots for hepatic and renal LDLR normalized to β-actin (n=3). (B) Immunoblot for LDLR, and beta actin in primary hepatocyte upon addition of hPCSK9 (500 ng/ml) for 4hrs, (p<0.01).
Supplementary figure 6: Effect of hPCSK9 in the adrenals. (A) Immunoblot of LDLR after co-immunoprecipitation with hPCSK9 of adrenal and liver from hPCSK9tg mice. (B) Immunoblot for LDLR, PCSK9, beta actin and annexinA2 in hepatocytes (AML-12) and adrenal (Y-1) cells (C=cell, M=media). (C) immunoblot of mouse adrenal cells (Y-1) transduced with GFP (control), PCSK9, or PCSK9 GOF mutation (D374Y).
Supplementary figure 7: hPCSK9 immunoblot for FPLC fractions treated with cross linker DSP (0.5mM, 30 min) on a native gel (4-16%).
Supplementary table 1: Serum mPCSK9 and hPCSK9 distribution after high-salt (KBr) ultracentrifugal separation of lipoproteins from hPCSK9 transgenic mouse serum, measured by PCSK9 ELISA and presented as % of total PCSK9.