Abstract
One carbon metabolism involving the folate and methionine cycle integrates carbon units from amino acids, including serine and glycine, and generates diverse outputs, such as the biosynthesis of lipids, nucleotides and proteins, the maintenance of redox status, and the substrates for methylation reactions. Long considered a ‘housekeeping’ process, this pathway has been recently shown to have additional complexity. Recent genetic and functional evidence also suggests that hyperactivation of this pathway is a possible driver of oncogenesis and establishes links to cellular epigenetic status. Given the wealth of clinically available agents that target one carbon metabolism, these new findings could present opportunities for translation into precision cancer medicine.
Introduction
Cell growth and proliferation requires the construction of building blocks for new cellular components including proteins, lipids and nucleic acids, as well the maintenance of cellular redox, genetic and epigenetic status1–6. Amino acid metabolism involving serine, glycine and threonine and the carbon units they provide satisfies many of these requirements7,8,18,19. One-carbon metabolism encompasses a complex metabolic network that is based on the chemical reactions of folate compounds9–11. These reactions proceed in a cyclical nature where a carbon unit is transferred to other metabolic pathways and eventually replenished by several sources. Modern cancer therapy in part arose from the hypothesis that antagonists of folates could reduce the proliferation of malignant blood cells12,13. The antagonism of folate metabolism and its downstream effectors, such as nucleotide metabolism, has been used in chemotherapy for over sixty years14–17 (see the Timeline in Fig 1).
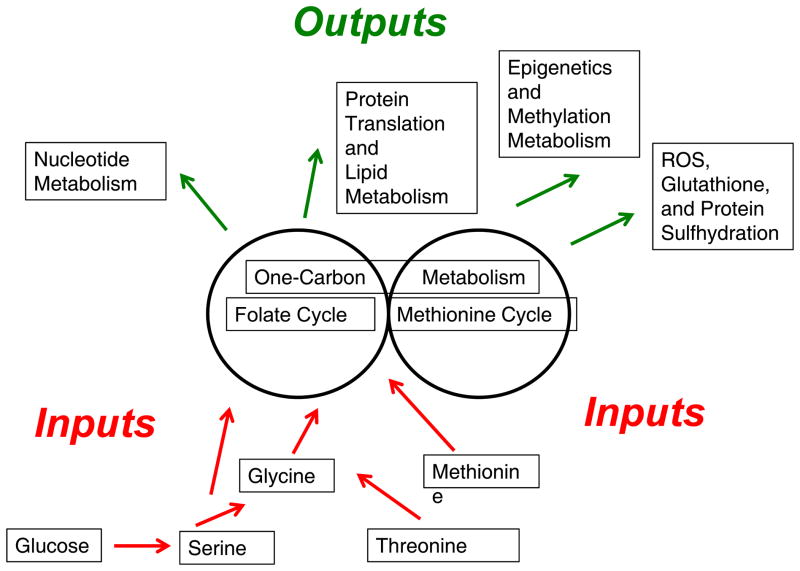
Figure 1. One carbon metabolism is an integrator of nutrient status.
Nutrient sources involving amino acids are either imported or synthesized de novo and enter one carbon metabolism. One carbon metabolism can be viewed as a set of two modular units (i.e. two pathways existing separately from one another) involing the Folate Cycle and Methionine Cycle. Through these metabolic cycles, nutrients are processed. Upon processing, multiple outputs can be generated including nucleotides, proteins, lipids, reducing power, and substrates for methylation reactions.
Recently, there has been a surge of interest in the study of the metabolic processes associated with cancer18. Much of this focus has been on the role of two nutrients, glucose and glutamine, in supporting energy metabolism and anabolic processes19–21. Expanding the compendium of metabolic pathways essential to cancer biology is that of serine and glycine metabolism8,22. Developments in our understanding have led to new biology related to the function of this pathway in cancer. These advances include roles for epigenetics, redox status, genome maintenance, protein translation, and biosynthesis for cell proliferation. Genetic and functional evidence also points to the activity of this pathway as having a role as a driver of oncogenesis.
This Review discusses recent developments that are transforming our understanding of serine, glycine, and one-carbon metabolism in cancer pathogenesis. These advances include genetic and functional evidence for its role as a driver of cancer pathogenesis and new roles for one carbon metabolism in tumor maintenance, including genome integrity and epigenetic maintenance. Ultimately these findings may result in new translational opportunities for drug development, dietary intervention in cancer prevention, biomarkers that indicate in which tumors antimetabolic chemotherapeutic drugs are likely to produce an efficacious response, and new frameworks for approaching the study of the pathogenesis of cancer23.
One-carbon metabolism: an integrator of nutrient status
A way to conceptualize the role of one-carbon metabolism in cellular physiology is that it functions as a metabolic integrator of nutrient status (Fig 2). Inputs in the form of glucose and amino acids enter the pathway, are processed through chemical reactions, and are then output for diverse biological functions. This analogy has been used extensively to describe growth control by the mammalian target of rapamycin (mTOR) signal transduction pathway, but has been used less for metabolic pathways per se24. In the case of mTOR signaling, inputs in the form of amino acids and growth factors are integrated to generate outputs such as protein translation, autophagy inhibition, and anabolic metabolism. For one-carbon metabolism, the integration is carried out through the donation of carbon units from specific amino acids. These carbon units are the substrates for one carbon metabolism and are distributed via a series of chemical reactions for use in diverse cellular processes that include cellular biosynthesis, regulation of redox status, regulation of epigenetics through nucleic acid and protein methylation, and genome maintenance through the regulation of nucleotide pools. The partitioning of carbon units into these different cellular outputs involves three pathways: the folate cycle, the methionine cycle and transsulfuration pathway (Fig 2). Loss of function mutations in enzymes involved in these pathways lead to growth defects both in animals and humans, underscoring the role of one carbon metabolism in modulating cell growth7,25–27.
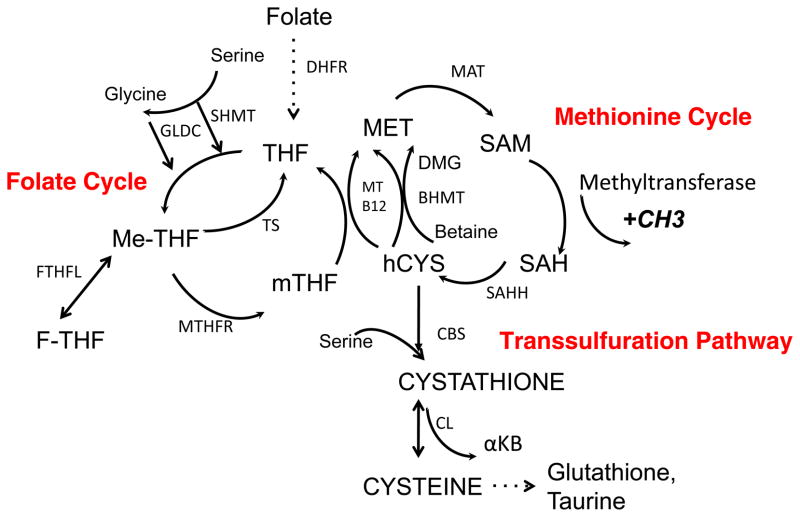
Figure 2. Folate and methionine metabolism comprise one carbon metabolism.
The folate cycle and the methionine comprise of two metabolic pathways that exist independently and thus in modules.
The folate cycle: Folate is imported in cells and reduced to tetrahydrofolate (THF). THF is converted to 5,10-methylene-THF (me-THF) by serine hydroxymethyl transferase (SHMT). Vitamin B6 appears to have an influence on this reaction but the interaction is likely indirect. This product is then either reduced to 5-methyltetrahydrofolate (mTHF) by methylenetetrahydrofolate reductase (MTHFR) or converted to 10-Formyltetrahydrofolate (F-THF) through a sequence of steps.. mTHF is demethylated to complete the folate cycle. With the demethylation of mTHF, the carbon is donated into the methionine cycle through the methylation of homocysteine by methionine synthase and its cofactor Vitamin B12.
The methionine cycle: The methionine cycle begins with homocysteine that accepts the carbon from the folate pool through mTHF to generate methionine. Methionine, through methionine adenyltransferase (MAT) is used to generate S-adenosylmethionine (SAM), which is demethylated to form S-adenosylhomocysteine (SAH). After deadenylation by S-adenosyl homocysteine hydrolase (SAHH), SAH is converted back to homocysteine resulting in a full turn of the methionine cycle.
Another modular unit of one carbon metabolism is the transsulfuration pathway. This pathway is connected to the methionine cycle through the intermediate homocysteine. Serine can condense enzymatically with homocysteine to generate cystathione by cystathionine synthase (CBS). Cystathionine is then cleaved by cystathione lyase (CGL) to generate alpha-ketobutyrate (αKB) and cysteine, which can be shunted into glutathione production and taurine metabolism. The metabolism of cysteine can also lead to its desulfhydration and production of hydrogen sulfide through CBS and CGL.
Abbreviations: SAM – S-adenosylmethionine, SAH – S-adenosylhomomocysteine, hCYS – homocysteine, THF – Tetrahydrofolate, mTHF – 5-methyltetrahydrofolate, Me-THF – 5,10 Methylenetetrahydrofolate, F-THF – 10 Formyltetrahydrofolate, DMG – Dimethylglycine, GLDC – Glycine Decarboxylase, TS – Thymidylate Synthase, MT – Methionine Synthase, B12 – Vitamin B12, MAT – Methionine adenyltransferase, SAHH – S-adenosyl homocysteine hydrolase, GNMT – Glycine N-methyltransferase, BHMT – Betaine hydroxymethyltransferase, SHMT – Serine hydroxymethyltransferase MTHFR – Methyltetrahydrofolate Reductase, DHFR –Dihydrofolate reductase, CDO – Cysteine Dioxygenase. Bi-directional arrows denote reversible steps. Dotted arrows denote multiple biochemical steps.
One carbon metabolism and the transsulfuration pathway
Folic acid is a B vitamin found in and added to foods that are widely available in Western diets. In cells, folic acid is reduced by a series of enzymes leading to the generation of tetrahydrofolate (THF) (Fig 3). THF participates as a scaffold in a set of metabolic reactions that involve the movement of carbon atoms to different positions along the THF-moiety (Fig 3). The folate cycle is coupled to the methionine cycle through the generation of methyl-THF (mTHF). mTHF donates a carbon through methylation of homocysteine and this generates methionine. Thus, the folate cycle coupled to the methionine cycle constitutes a bi-cyclic metabolic pathway that circulates carbon units. These metabolic cycles are collectively referred to as one-carbon metabolism. The transsulfuration pathway is connected to the methionine cycle through the intermediate homocysteine. Serine can be directly metabolized through transsulfuration eventually resulting in the generation of glutathione, one of the major redox-regulating metabolic systems in cells (discussed below).
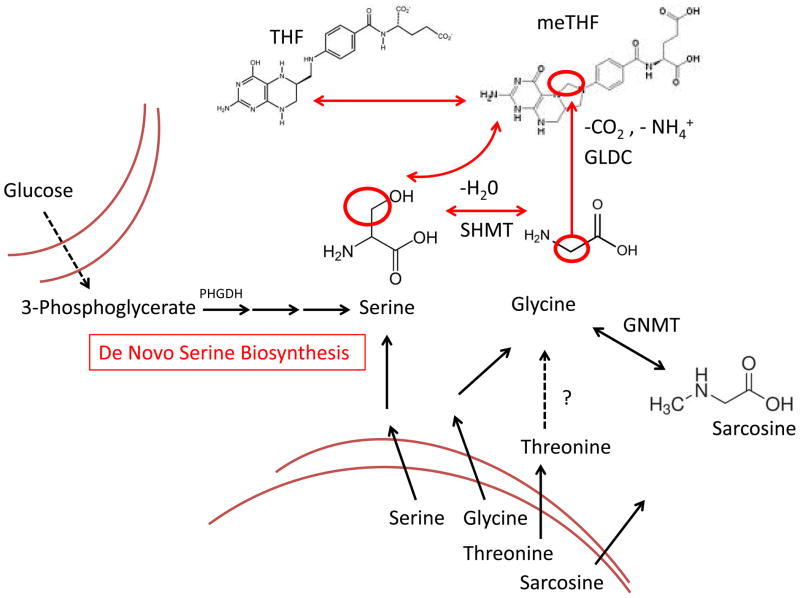
Figure 3. Nutrients that fuel one carbon metabolism.
Serine and glycine metabolism is generated de novo from glycolysis through the oxidation of intermediate 3-phosphoglycerate. Serine and glycine are also transported into cells. Sarcosine and possibly threonine can also enter cells and be converted to glycine. The question mark denotes that threonine catabolism has been found to be utizilized in mammals including mice. Its analog in humans has not been identified. Dashed arrows denote multiple biochemical steps. Bi-directional arrows denote reversible steps. Abbreviations: PHGDH – phosphoglycerate dehydrogenase, GLDC – Glycine Decarboxylase, SHMT – Serine hydroxymethyltransferase, GNMT – Glycine N-methyltransferase, THF – Tetrahydrofolate, Me-THF – 5,10 Methylenetetrahydrofolate.
Inputs to one-carbon metabolism
The carbon units that feed into one-carbon metabolism can be synthesized de novo (Fig 4). For example, an intermediate metabolite involved in glycolysis, 3-phosphoglycerate (3PG), can be converted into serine. Serine donates the carbon atom from its side-chain to folate, converting serine to glycine and THF to mTHF, which starts the folate cycle. Serine can also be directly imported from the extracellular environment by facilitated transport through amino acid transporters. In addition to the side chain of serine, there are other routes of entry into one-carbon metabolism. A glycine cleavage system is active in some cells where the enzymatic cleavage of glycine produces ammonia, carbon dioxide and a carbon unit for the methylation of THF, which also charges the folate cycle. In some cells, threonine can also be converted to glycine through an aldol cleavage28. Glycine can also be generated from many other sources including choline, betaine, dimethylglycine and sarcosine (also known as N-methylglycine) through a series of reactions that involve demethlyation.
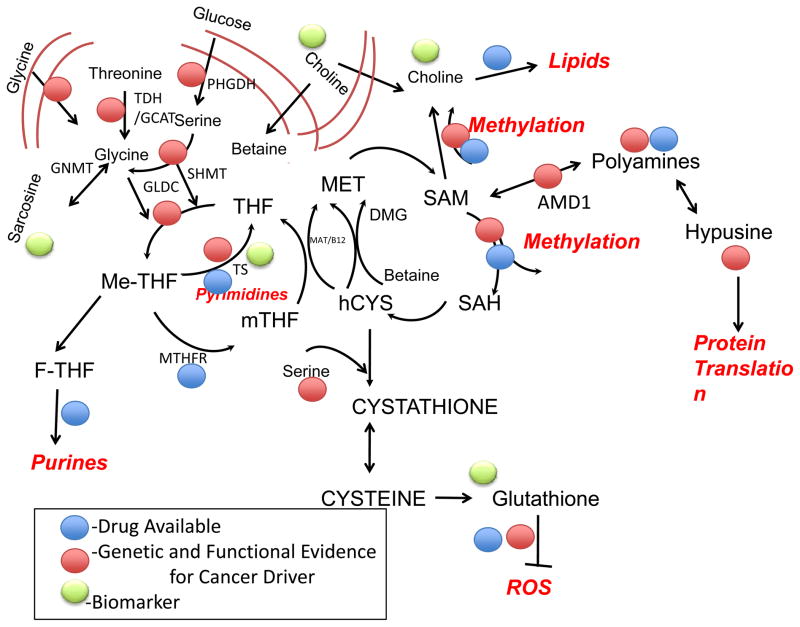
Figure 4. One carbon metabolism and cancer pathology and intervention.
Schematic of one-carbon metabolism and the transsulfuration pathway. Recent findings have identified roles for these pathways in cancer. Genetic mutations and functional evidence for the existence of a cause of cancer (driver) at this point in the pathway (red), currently available drugs (yellow), and biomarker development (green) are highlighted. The specifics are indicated in Tables 1, 2 and 3. Causality is defined as either the presence of a genetic lesion or functional evidence (e.g. overexpression of a pathway component) that enhanced activity at this point in the pathway promotes oncogenesis. The biological outputs of the pathway are in a bold-italic red font. Abbreviations: SAM – S-adenosylmethionine, SAH – S-adenosylhomomocysteine, hCYS – homocysteine, THF – Tetrahydrofolate, mTHF – 5-methyltetrahydrofolate, Me-THF – 5,10 Methylenetetrahydrofolate, F-THF – 10 Formyltetrahydrofolate, DMG – Dimethylglycine, PC – Phosphatidylcholine; PHGDH – phosphoglycerate dehydrogenase, GLDC – Glycine Decarboxylase, TS – Thymidylate Synthase, MAT – Methionine Synthase, B12 – Vitamin B12, GNMT – Glycine N-methyltransferase, MTHFR – Methylenetetrahydrofolate Reductase, TDH/GCAT – Threonine Dehydrogenase/Glycine C-acetyltransferase, ROS – Reactive Oxygen Species. Bidirectional arrows denote reversible steps.
Outputs of one-carbon metabolism
Biosynthesis
All cells require the synthesis of macromolecules, such as proteins, lipids and nucleic acids for cellular renewal and proliferation29,30. Amino acids, such a methionine, can be generated from one-carbon metabolism and used to generate proteins31. Nucleotides required for DNA and RNA are also constructed through reactions that involve the folate cycle 32. Deoxy-thymidine monophosphate is synthesized through the methylation of deoxy-uridine monophosphate by thymidylate synthase. This methylation reaction generates THF from mTHF. Purine nucleotide bases are also generated from the folate pool through the intermediate 10-formyltetrahydrofolate (F-THF), which is derived from 5,10-methylene-THF (me-THF). The ribose moiety of RNA and DNA is derived from the pentose phosphate pathway33. Lipids can also be generated in part through the methionine cycle34. Phosphatidylcholine (PC) is a major component of the cell membrane that can account for up to half of lipid membrane content35. The head group of PC is synthesized from choline36 through the adenylation of methionine to S-adenosylmethionine (SAM). SAM functions as a methyl donor for the three subsequent methylation reactions that generate the lipid head group37,38.
Redox balance
The metabolism of carbon atoms through one carbon metabolism is linked with changes in redox status. These changes largely occur through the reduction of NADPH and oxidation of NADP+. Tetrahydrofolate reductase reduces THF and this reaction consumes one molecule of NADPH for each turn of the folate cycle. Although these reactions are thought to proceed in a reductive manner, it is conceivable that the reverse of these reactions could occur in vivo based on known in vitro activities39. Glutathione, one output of the transsulfuration pathway, is also important for the maintenance of the ratio of NADPH and NADP+. A tripeptide comprised of cysteine, glycine and glutamate, glutathione is one of the most abundant metabolites in cells, often reaching concentrations as high as five millimolar40,41. Thus it serves as a major contributor to the redox balance in cells through its ability to scavenge and reduce reactive oxygen species (ROS) and to maintain the appropriate NADPH/ NADP+ ratio, which is required for anabolic metabolism2. The desulfhydration products in the transsulfuration pathway also lead to the sulfhydration of proteins. These post-translational modifications are considered to be an important and underexplored signal transduction mechanism42.
Methylation reactions
Cells also require substrates derived from metabolism to carry out signal transduction through post-translational modifications. Methyl groups derived from one carbon metabolism provide a major source of substrate for post-translation modifications43–46. As the major methyl donor in cells, SAM is involved in histone, DNA and RNA methylation, and all general protein lysine and arginine methylation47–49. SAM is also involved in other metabolic pathways that require methyl moieties,50 including polyamine synthesis51–53.
Cancer therapy and one carbon metabolism
One of the first modern chemotherapies resulted from the observation that anemic patients could be treated with B vitamins to stimulate red blood cell production12. Sydney Farber also noted that folic acid could stimulate the proliferation of acute lymphoblastic leukaemia (ALL) cells and therefore investigated whether intermediates of the chemical synthesis of B vitamins might act to antagonize cell proliferation. In a landmark study by Farber and colleagues one of these molecules, aminopterin, was shown to induce remissions in children with acute lymphoblastic leukemia (ALL)13,54. To this day, chemical variants, such as methotrexate and pemetrexed, of these initial folate antagonists constitute a major class of cancer chemotherapy agents and are used as frontline chemotherapy for diverse cancers including ALL, breast cancer, bladder cancer and lymphomas14,55–59. These agents inhibit di- and tetrahydrofolate reductase activity in humans resulting in a disruption of one carbon metabolism60,61. It is interesting to note however that disruption of one carbon metabolism by these agents is not efficacious in all cancer types. More recent findings (discussed below) might help in understanding why this variation exists and potentially enable patients who will benefit most from these drugs to be identified.
In addition, multiple pathways downstream of one-carbon metabolism are the targets of numerous cytotoxic chemotherapies. 5-fluorouracil (5-FU) targets nucleotide metabolism that is derived from the folate cycle and is a standard of care agent for many cancers including advanced stage colorectal cancer62,63. 5-FU is an analog of the DNA base pyrimidine. It is a potent inhibitor of thymidine synthase thus blocking the methylation dUMP to dTMP and disrupting the folate cycle64. Gemcitabine, which is used to treat patients with pancreatic cancer65, is another inhibitor of nucleotide metabolism66,67. Gemcitabine is a nucleoside analog that interferes with the biosynthesis of cytidine68 and it inhibits ribonucleotide reductase (RNR), preventing the formation of deoxynucleotides66. Gemcitabine efficacy in pancreatic cancer is variable, but whether this stems from resistance because of alterations in one carbon metabolism, or from differences in drug delivery is yet to be fully resolved.
Other pathways downstream of one-carbon metabolism are also the target of cancer therapy. Targeting the epigenetic status of tumors is a hotly pursued area69–73. Several drugs that target the enzymes that are involved in the post-translational modifications of histones and DNA are being evaluated pre-clinically and in early-stage clinical trials74–76. These include inhibitors of methyltransferases that interfere with SAM-mediated methylation of histones and DNA77,78. Changes in the levels of histone and DNA methylation would be predicted to alter cellular epigenetics through the activation and repression of many genes79,80. Examples of specific enzyme targets include DNA methyltransferases that are targeted by azanucleosides81. Inhibitors of histone methyltransferases have been developed and are being considered pre-clinically76,82. Polyamine metabolism, which involves the decarboxylation of SAM and results in the generation of spermidine, has also been heavily explored as targets for anti-cancer therapy, reaching clinical trials in some cases83. These agents include 2-difluoromethyl ornithine (DMFO), an inhibitor of ornithine decarboxylase and methylglyoxal bis(guanylhydrazone) (MGBG) and 4-amidinoindydrazone-1-(SAM486A), both of which are competitive inhibitors of S-adenosylmethionine decarboxylase. Both enzymes are required for spermidine synthesis.
One-carbon metabolism and cancer pathogenesis
De novo serine and glycine metabolism
An intermediate in glycolysis, 3PG, can be oxidized to form 3-phosphohydroxypyruvate (pPYR). This reaction is the initial and committed step for de novo (that is originating from glucose) serine biosynthesis84,85. Thus, carbon derived from glucose can be shunted from glycolysis into de novo serine metabolism and from here into the folate cycle. It has been known for many years that this pathway correlates with tumorigenesis84,86,87. Initial studies that provided the biochemical characterization of this pathway also showed that it was active in tumors88. Further extensive work by Snell and colleagues showed that flux through this branch point in glycolysis correlated with cancer progression in rat carcinoma models84,86,87. Recent studies using isotope tracing with 13C labeled glucose showed that a subset of cancer cells diverted a substantial amount (approximately 10%) of 3PG away from glycolysis and into one carbon metabolism through phosphoglycerate dehydrogenase (PHGDH)89,90. This resulted in large amounts of de novo serine biosynthesis90,91. PHGDH was also found to be overexpressed in the triple negative subtype of breast cancer90–92.
Despite these observations, it was not known whether the activity of this pathway had any causative role in cancer development or maintenance. One context in which this pathway might be relevant to cancer came from data analyzing the copy number variations in human cancers93. It was observed that the genomic locus encoding PHGDH was the subject of a focal, recurrent gene amplification and this region of the genome harbors no known oncogenes. These data suggested that tumors containing a PHGDH amplification may have gained a selective advantage in expressing many copies of the gene. A small hairpin RNA (shRNA) screen revealed that a breast cancer cell line required PHGDH for in vivo tumorigenesis91. It was also confirmed that breast cancer and melanoma cell lines containing the amplification required PHGDH for proliferation90,91. Furthermore, expression of PHGDH in cells that exhibited no detectable flux into de novo serine metabolism was shown to increase serine biosynthesis and induce phenotypic properties that predispose cells to malignancy. These properties include loss of polarity and proliferation in the absence of extracellular matrix contact 90. Together these findings have provided evidence that de novo serine metabolism could be both necessary and sufficient for tumour maintenance and promotion of oncogenesis22.
Additional studies have identified further roles for this pathway in tumour cells94–99. In a series of studies, it was shown that the activity of an isoform of pyruvate kinase (PKM2), the enzyme that catalyzes the final step in glycolysis, can regulate the flow of glucose into serine metabolism94,97. These studies have claimed that this regulation occurs through the allosteric activation of PKM2. However, serine appears to be a weak activator of PKM2, with activity occurring at concentrations greater than one millimolar94,97. The regulation is likely more complex. Other enzymes appear to also be involved directly in the activity of PHGDH including PKCζ, which has been shown to phosphorylate PHGDH and inhibit its activity. PKCζ’s tumor suppressor activity is thought to occur through this mechanism99. Furthermore, a study revealed that 2-phosphoglycerate, the product of phosphoglycerate mutase that uses 3PG as a substrate, can activate PHGDH, providing additional points of regulation in glycolysis95.
Glycine uptake, cleavage and entry into one-carbon metabolism
Recent work has also implicated the glycine cleavage system in cell transformation and tumorigenesis28,100–103. This pathway has been studied extensively in plants and lower eukaryotes but its role in mammalian physiology and pathology has been less well explored. Studies in mouse embryonic stem cells (ESCs) showed that these cells uniquely required threonine for stem cell maintenance and self-renewal28,101,102,104. Withdrawal of threonine from the culture media, but not any of the other nineteen amino acids, induced rapid cell death. Dramatic reductions in methylation on specific histone residues, including lysine 4 on histone 3 (H3), were also observed103,105. Intriguingly, the absence of methionine induced a similar, albeit milder phenotype. Using isotope tracing of the metabolic flux emanating from threonine, it was shown that threonine entered one-carbon metabolism through glycine cleavage. Threonine is converted to glycine by the enzymes threonine dehydrogenase (TDH) and glycine C-acetyltransferase (GCAT). The activity of glycine dehydrogenase (decarboxylating) (GLDC) mediates glycine cleavage and the charging of the folate cycle. Additional studies also confirmed that this pathway was essential for the maintenance of stem cell pluripotency in mice103.
Recent work on cancer metabolomics has shown that glycine metabolism is associated with cancer cell proliferation. In a survey of the NCI-60 cell line panel, the uptake and release rates of over two hundred metabolites was measured106. Surprisingly, glucose uptake and lactate production (i.e. the Warburg Effect) were not associated with cell proliferation. By correlating individual metabolic fluxes with cell proliferation, it was found that glycine uptake was most strongly associated with cancer cell proliferation106. Isotope tracing revealed that glycine cleavage was involved in the catabolism of the glycine taken up from the media. This pathway was also shown to be required in rapidly dividing cells106. GLDC activity has also been implicated as having a causal role in tumorigenesis. One study found that a subpopulation of tumor-promoting cells expressed high levels of GLDC and that ectopic expression of GLDC was sufficient to induce tumor formation in xenografts of NIH 3T3 cells100. Interestingly, this study also reported that ectopic expression of two other enzymes involved in serine and glycine catabolism phosphoserine aminotransferase (PSAT) and serine hydroxymethyltransferase (SHMT) in NIH 3T3 cells could induce tumor formation in vivo. Importantly, the induction of tumorigenesis depended on the enhanced enzyme activity of GLDC. Another study also reported that increased availability of glycine or sarcosine could increase the invasiveness of prostate cancer cells107. Together, these findings provide evidence that glycine uptake and catabolism promotes tumorigenesis and malignancy.
In addition to the newly appreciated functional roles of one carbon metabolism in cancer progression and maintenance, several biological pathways that are involved in tumorigenesis, including those related to protein translation, genome maintenance, epigenetic status and cellular redox status, are linked to one carbon metabolism.
Protein translation
Recently, an in vivo screen for new tumor suppressor genes in lymphoma identified two genes, S-adenosylmethionine decarboxylase (AMD1) and eukaryotic initiating factor 5α (EIF5A), which are associated with methionine metabolism and hypusine biosynthesis108. AMD1 decarboxylates SAM, diverting SAM into a pathway involving the biosynthesis of spermine and spermidine109. This pathway eventually leads to the production hypusine, which is required for the hypusination of proteins. So far, only two eukaryotic initiating factors, including EIF5α, have been shown to be modified by hypusination 110–113. Subsequent work demonstrated that other genes involved in the production of hypusine were also sufficient to induce lymphomas. The hypusination of EIF5α is thought to be required for its physical association with ribosomes and is essential for translation elongation111,114. Together these findings link tumor suppression with methionine metabolism through its ability to directly modify protein translation.
Nucleotide metabolism and genome maintenance
Genome maintenance requires the successful incorporation of the appropriate nucleotides during both DNA replication and repair115,116. Nucleotide metabolism has been directly implicated in tumorigenesis117,118. Recent studies have demonstrated that a decrease in nucleotide levels is sufficient to induce genome instability and increase mutagenesis rates119,120. The mechanism involves the incorporation of uracil into DNA due to its increased relative abundance to thymidine. Several enzymes involved in nucleotide metabolism are considered to be bona fide tumor suppressor genes. FHIT, a gene that codes for an enzyme with dinucleotide hydrolase activity involved in purine metabolism, is one of the most frequently deleted genes in human cancer — almost half of all human colon cancers contain a focal deletion of this gene121–123. Fhit deletion in mice has been shown to induce genome instability and spontaneous tumor formation, which can be rescued by the introduction of an FHIT transgene124,125. The mechanism through which this phenomenon occurs remains unclear but probably has some connection to genome maintenance and DNA repair through the ability of repair enzymes to appropriately incorporate nucleotide bases into DNA.
Other cancer-associated genes involved in nucleotide metabolism include thymidylate synthase and RNR126–128. Overexpression of RNR induces lung tumors in mice through alterations in DNA repair, indicating that it can function as an oncogene129. Additionally, thymidylate synthase can also function as an oncogene126,130. The ability of its overexpression to induce cell transformation and the growth of cells as tumour xenografts in mice is dependent on its catalytic activity. More studies are needed to define the mechanistic principles of these phenomena but a possible mechanism lies in the maintenance of genome integrity.
Epigenetic alterations
The methylation reactions mediated by SAM involve the transfer of methyl groups onto the lysine and arginine residues of proteins, DNA, RNA and intermediary metabolites131–133. These modifications have long been observed to affect gene regulation, but the extent to which they are reversible has been less clear73. Much of the recent interest in this area of cancer biology has come from multiple genetic and functional studies that have identified methyltransferases and demethylases as being recurrently mutated and having causal roles in cancer development134–139.
Recent pulse-chase experiments using isotopically labeled SAM have revealed that methylation modifications are dynamic140,141 and tightly regulated 73,80,142,143. As SAM concentrations fluctuate in cells and affect the activity of methyltransferases, the levels of SAM influence the levels of histone methylation3,20,45,47,105.
Translational opportunities for cancer therapy
Drug development
The surge of work carried out in the study of cancer metabolism has created the expectation that the mechanistic understanding will lead to the development of new therapeutics that target key nodes within the cancer metabolic network. Metabolic enzymes, having evolved to carry out chemical catalysis, are thought to be druggable56,144–146. In addition to targeting the catalytic site of metabolic enzymes, it is also possible to design small molecules that target allosteric binding sites that naturally fit endogenous metabolites. Pre-clinical studies are underway that are currently evaluating the promise of targeting multiple nodes in one carbon metabolism22,147. These targets include, but are not limited to, PHGDH, PSAT, PSPH, GNMT, GLDC and GCAT. Drugs that target the generation of ROS, an indirect product of metabolism, are also actively being pursued148. One question that is often raised is what improvements would these new targets offer over currently existing drugs, such as methotrexate and pemetrexed, that already target one carbon metabolism? One possible advantage to targeting these nodes lies in the potential for an improved therapeutic window. For example, since some tumors and most tissues appear not to require PHGDH and thus serine synthesis from glucose, targeting this pathway might prove less toxic in some contexts. Furthermore, some tumors display hyperactivation of this pathway and thus, since other pathways in addition to de novo serine biosynthesis enter into folate metabolism, these tumors might be more susceptible to PHGDH inhibition than inhibition of MTHFR by methotrexate and pemetrexed. Nevertheless more data are needed to resolve the contexts in which directly targeting enzymes involved in one carbon metabolism would provide greater efficacy than the administration of anti-folate chemotherapy.
Metformin, cancer and one carbon metabolism
Metformin has recently come into focus as a promising agent for cancer therapy. Metformin is the most commonly used treatment for type II diabetes149 and also exhibits other activities, such as an anti-aging effect in Caenorhabditis elegans through inhibition of one carbon metabolism in the gut microbiota150. Epidemiological evidence has suggested that metformin may have anti-cancer effects151. These data have been augmented with pre-clinical studies showing an anti-tumor activity for metformin152 and so this drug has advanced to clinical trials for both cancer treatment and prevention. The mechanism of action of metformin has proven controversial with numerous mechanisms having been proposed153. Recent work has provided evidence that metformin acts on one carbon metabolism in patients. A metabolomics study of a cohort of patients treated with metformin revealed that the signature of metformin response was remarkably similar to that obtained from patients treated with anti-metabolite chemotherapies154. These findings suggest that metformin confers some and maybe many of its effects through the alteration of folate and one carbon metabolism. Additional support for this hypothesis would be useful to complement current clinical trials of metformin in cancer patients.
Dietary intervention in cancer treatment and serine and glycine metabolism
A complementary strategy to targeting cancer metabolism with pharmacological agents is to affect the same nodes through changes in nutrient uptake. This intervention could occur through changes in diet, especially as high carbohydrate intake is positively correlated with cancer incidence155–158. Pre-clinical 159 and clinical160,161 studies have shown that reducing carbohydrate (glucose) intake can have negative effects on tumour biology.
Pre-clinical work has also explored the possibility of restricting serine and glycine metabolism for cancer intervention. Isogenic colon cancer cells containing wild-type or null alleles of Trp53 were studied in vitro and grafted into mice fed diets containing no serine and no glycine98,162. The removal of serine and glycine dramatically affected cell proliferation and tumor growth. In the absence of p53, serine and glycine withdrawal had an even greater effect, suggesting an epistatic interaction between p53 and the availability of serine and glycine. Mechanistically, the absence of serine and glycine increased de novo serine and glycine metabolism and this was found to decrease glutathione synthesis and increase ROS levels suggesting that this effect could contribute to tumor growth. Strikingly, it was observed that removal of serine and glycine had an even larger effect on the reduction of in vivo tumor growth than the re-introduction of wild-type p53 alleles into the tumour cells. Other mechanisms are likely to also be relevant, such as a reduced if not disrupted rate of biosynthesis, nucleotide metabolism that may affect AMPK activity163, and possibly changes in epigenetic status. Removing serine and glycine from a natural diet seems difficult. However, specific diets might be constructed that could circumvent this problem, as is the case for dietary intervention for several diseases, such as the ketogenic diet which is used to prevent seizures for epileptic patients or diets such as a gluten-free diet that are used to alleviate autoimmune disorders.
In light of these findings, it is tempting to speculate that similar activities could be obtained with the restriction of B vitamins that are readily available in food. This seems particularly relevant given the initial evidence that the effects of metformin might work by targeting folate metabolism. However, numerous longitudinal studies have associated folate and vitamin B12 intake with alterations in DNA methylation and cancer risk. Lack of adequate intake of folate during pregnancy is associated with improper germline transmission of methylation patterns. Folate consumption has also been shown to affect DNA methylation during development26,27,184,185. Decreased folate intake is associated with cancer, most notably colorectal cancer186–194. In breast cancer, reduced folate intake is associated with cancer development and global hypomethylation195,196. Given data on serine and glycine deprivation and its anti-tumor effects, and the metformin data, the relationship between diets related to one carbon metabolism is apparently complex with more work needed to define the mechanisms related to one-carbon metabolism activity and cancer susceptibility.
Biomarkers for precision medicine and diagnosis
Many of the intermediary metabolites in serine, glycine and one-carbon metabolism are water soluble and detectable in biological fluids such as serum and urine. These properties allow for the possibility of innovations in diagnostics. For example, increased homocysteine levels in serum are used as a biomarker for folate deficiency164. The buildup of homocysteine results from the lack of available methyl donors to complete a turn of the folate cycle. The use of anti-metabolite chemotherapies has identified biomarkers, mostly in the form of the enzymes, which these drugs target, that predict response or resistance. For example, expression of thymidylate synthase has been shown to predict response to 5-FU165–169. Biomarkers of response to methotrexate have been found in serum metabolites170,171 and a recent meta-analysis reported that expression of folate-metabolizing enzymes and those involved in serine and glycine metabolism could predict tumor response to methotrexate in diverse tumor types172.
A metabolomics study of urine from patients with benign prostatic disease, localized prostate cancer and metastatic prostate cancer revealed that glycine metabolismis a predictor of metastatic cancer107. Glycine and sarcosine were identified in the metastatic urine samples107. Subsequent studies have found conflicting results and the probability that sarcosine is a biomarker of metastatic disease is likely to depend on additional variables173–175. Nevertheless, given the non-invasiveness of this metabolomics assay, it is possible that these results could eventually be used clinically. In some contexts, choline metabolism has been found to be increased during tumor progression176,177. Positron emission tomography (PET) imaging of C11 choline has been approved by the US Food and Drug administration (FDA) as a biomarker for advanced malignancy178,179. Gene expression levels of glutathione-metabolizing enzymes is also an FDA-approved biomarker for treatment decisions in node-negative, estrogen receptor-positive breast cancers180. Together, these findings currently have and will probably continue to result in clinical impact. With the new found molecular mechanisms connecting one carbon metabolism to cancer pathogenesis that have been recently characterized, additional advances in biomarker discovery for precision medicine surrounding anti-folate agents could be obtained.
Summary and future directions
Once thought to be the subject of mundane biochemistry lectures and the target of non-specific cytotoxic chemotherapies, amino acid and one carbon metabolism has reemerged as a core feature in the biology of cancer (Figure 5). These findings have occurred alongside the discovery of an ‘oncometabolite’ the R enantiomer of 2-hydroxyglutarate (2HG), a product of mutant isocitrate dehydrogenase (IDH) enzymes181. IDH1 and IDH2 are recurrently mutated in leukemia and gliomas, as well as other cancer types182. 2HG has been shown to have numerous functions, including the alteration of epigenetic marks through the inhibition of histone demethylases183. Perhaps the intermediates in one carbon metabolism are also oncometabolites, whose aberrant activity promotes cancer pathogenesis.
Many other questions also remain. For example, how one carbon metabolism is integrated with signals from diverse nutrient inputs to generate the appropriate downstream carbon portioning is not understood. The extent to and context under which this pathway modulates epigenetics, genome maintenance, redox status and anabolic metabolism are only just beginning to be understood. Whether any of these newly appreciated roles in cancer pathogenesis will lead to further clinical benefit is a subject for further exploration. Nevertheless, with technological advances, it is expected that we will uncover many other unknown angles that connect epigenetics, nucleic acid metabolism and redox biology with one carbon metabolism and the pathology of cancer. Metabolomics, computational models and integrative bioinformatics approaches will hopefully allow for rapid progress in this area.
Another appealing aspect of studying serine, glycine and one-carbon metabolism in cancer pathogenesis is the wealth of drugs already clinically available and dietary options that may be further available. For example, chemotherapies such as methotrexate, 5-FU and gemcitabine have a dramatic response in subsets of patients with cancer, but our ability to predict these responses remains poor. If any of this new found knowledge could be harnessed for advances in precision medicine, dramatic inroads would be made.
Table 1.
Candidate Drivers of Cell Transformation
Candidate oncogenes and tumor suppressor genes in one carbon metabolism
| Pathway | Genes or Enzymes | Evidence |
|---|---|---|
| De Novo Serine Biosynthesis | PHGDH, PSAT, PSPH, SHMT1 | Genetic Aberrations, Functional Data |
| Glycine Cleavage | GLDC, GCAT | Overexpression, Functional Data |
| Polyamine Synthesis | AMD1, EIF5A, SRM, DHPS | Underexpression, Functional Data |
| Methylation Metabolism | IDH1, EZH2, SET9, DNMT1, ARID1A, JARID1C, UTX, SETD2 | Genetic Aberrations, Functional Data |
Table 2.
Oncology biomarkers in one carbon metabolism
| Biomarker | Source | Use |
|---|---|---|
| Choline | C11 Pet Imaging | Detection of advanced malignancies |
| Sarcosine | Urine Metabolite | Possible predictor of metastatic prostate cancer |
| Thymidylate Synthase | Tumor mRNA expression | Expression Correlates with response to 5-Fluorouracil |
| Glutathione sulfer transferase | Tumor mRNA expression | FDA-approved as part of assay for decision to use chemotherapy in Estrogen Receptor Positive Breast Cancer |
Table 3.
Drug targets in one carbon metabolism
| Enzymes | Compounds | Status |
|---|---|---|
| Methotetrahydrofolate Reductase | Methotrexate, Pemetrexed | Approved for multiple cancers |
| Thymidylate Synthase | 5-Fluorouracil (5-FU) | Approved for multiple cancers most notably Colorectal Cancer |
| Ribonucleotide Reductase | Gemcitabine | Approved for multiple cancers most notably Pancreatic Cancer |
| Polyamine Inhibitors | Various | Clinical trials ongoing |
| DNA Methyltransferase Inihibitors | Azanucleosides | Approved for Myeloid Leukemias |
| Histone Methyltransferase Inhibitors | Various (SAM-analogs) | Clinical trials ongoing |
| Histone Demethylase Inhibitors | Various | Preclinical studies |
| Ornithine Decarboxylase | 2-difluoromethyl ornithine (DMFO) | Preclinical studies |
| S-adenosyl decarboxylase inhibitors | methylglyoxal bis(guanylhydrazone) (MGBG), 4-amidinoindydrazone-1-(SAM486A) | Preclinical studies |
Online ‘at-a-glance’ summary.
One carbon metabolism integrates cellular nutrient status by cycling carbon units from amino acid inputs to generate diverse outputs including redox maintenance and cellular biosynthesis.
The epigenetic status of cells also seems directly linked with one carbon metabolism through protein and nucleic acid methylation.
One carbon metabolism has long been the focus of anti-metabolite based chemotherapy and includes the agents methotrexate and 5-Fluorouracil – two of the most widely used chemotherapies. Additional therapies are currently being explored.
Recent findings have provided genetic and functional evidence that multiple nodes within the pathway contain candidate driver genes for oncogenesis.
Additional research in one carbon metabolism may provide biomarkers that would enable advances in patient selection for antimetabolite chemotherapy.
Acknowledgments
I am grateful to members of my laboratory for helpful discussions. I also thank Lew Cantley for stimulating conversations in this area and the anonymous reviewers for their helpful comments. The work was supported through funding from the American Cancer Society and the National Cancer Institute.
Glossary
- Aldol Cleavage
A chemical reaction that can be catalyzed enzymatically resulting in the splitting of a betahydroxy ketone
- NCI-60
A panel of tumor-derived cell lines originating from diverse tissue types. Extensive genomic, biochemical, and pharmacological data have been obtained on these cell lines
- Therapeutic Window
A term used in drug development and medical practice that refers to range of drug concentrations that satisfy the tradeoff between an efficacious clinical response and unwanted toxicities
Biography
Jason W. Locasale, Ph.D. is an Assistant Professor at Cornell University. He graduated from Rutgers University with degrees in Chemistry and Physics. He received his Ph.D. at MIT in Biological Engineering. He then studied cancer metabolism at Harvard Medical School as a postdoctoral fellow with Lew Cantley. Dr. Locasale’s research focuses on the roles of metabolism in cell growth and cancer. At the core of this effort lies the use of computational modeling and mass spectrometry-based metabolomics. This systems biology approach combines these tools with an integration of genetics, biochemistry, and cell biology.
Footnotes
Links
Database of the human metabolic network:humanmetabolism.org
Status of Cancer clinical trials in the United States: cancer.gov/clinicaltrials
Encyclopedia of metabolic pathways: metacyc.org
Human Metabolite Database: hmdb.ca
Jason Locasale’s website: Jlocasale.human.cornell.edu
Metabolomics resources on behalf of Dr. Gary Siuzdak’s lab: metlin.scripps.edu
National Institutes of Health initiative on metabolomics: commonfund.nih.gov/Metabolomics/
Timeline
1944 – Soldiers in the Pacific Islands with tropical anemia are treated with B vitamins.
1949 – Sydney Farber successfully uses anti-folate agents to induce remission in children with Acute Lymphoblastic Leukemia.
1955 – De novo serine biosynthesis is observed in tumors.
1956 - The folate antagonoist, Methotrexate becomes widely used in oncology
1957–1970 - 5-FU is discovered and later approved for the treatment of advanced colorectal cancer.
1984 - Gemcitabine is approved for metastatic cancer and shows activity in pancreatic cancer.
1987 – Snell and colleagues demonstrate that serine biosynthesis is increased during tumor progression in rat models.
1996 – FHIT is cloned as a tumor suppressor gene.
2000 - Methyltransferase inhbitors are tested pre-clinically.
2004 – Thymidylate synthase is identified as an oncogene.
2009 - Sarcosine identified as candidate biomarker for metastatic prostate cancer.
2009–2013 – One carbon metabolism found to be essential for stem cell pluripotency and later observed to alter histone methylation status.
2011 – De novo serine and glycine metabolism is found to be necessary and sufficient for cell transformation and malignancy.
2012 – Glycine catabolism is found to be associated with rapid cell proliferation.
2012 - Choline C11 PET imaging is FDA-approved.
2013 – Serine and glycine dietary restriction found to inhibit tumor growth in preclinical studies
References
- 1.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Locasale JW, Cantley LC. Metabolic flux and the regulation of mammalian cell growth. Cell metabolism. 2011;14:443–451. doi: 10.1016/j.cmet.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature reviews. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 5.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 7.de Koning TJ, et al. L-serine in disease and development. The Biochemical journal. 2003;371:653–661. doi: 10.1042/BJ20021785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalhan SC, Hanson RW. Resurgence of serine: an often neglected but indispensable amino Acid. The Journal of biological chemistry. 2012;287:19786–19791. doi: 10.1074/jbc.R112.357194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shane B, Stokstad EL. Vitamin B12-folate interrelationships. Annual review of nutrition. 1985;5:115–141. doi: 10.1146/annurev.nu.05.070185.000555. [DOI] [PubMed] [Google Scholar]
- 10.Stipanuk MH. Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Annual review of nutrition. 2004;24:539–577. doi: 10.1146/annurev.nutr.24.012003.132418. [DOI] [PubMed] [Google Scholar]
- 11.Stover PJ. Physiology of folate and vitamin B12 in health and disease. Nutrition reviews. 2004;62:S3–12. doi: 10.1111/j.1753-4887.2004.tb00070.x. discussion S13. [DOI] [PubMed] [Google Scholar]
- 12.Farber S, et al. The Action of Pteroylglutamic Conjugates on Man. Science. 1947;106:619–621. doi: 10.1126/science.106.2764.619. [DOI] [PubMed] [Google Scholar]
- 13.Farber S, Diamond LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. The New England journal of medicine. 1948;238:787–793. doi: 10.1056/NEJM194806032382301. This pioneering study by Sydney Farber and colleagues provided proof of concept of cancer chemotherapy. They demonstrated that chemically targeting folate metabolism could induce remissions in leukemic patients. [DOI] [PubMed] [Google Scholar]
- 14.Chabner BA, Roberts TG., Jr Timeline: Chemotherapy and the war on cancer. Nature reviews. Cancer. 2005;5:65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- 15.Johnston PG, et al. Antimetabolites. Cancer chemotherapy and biological response modifiers. 1997;17:1–39. [PubMed] [Google Scholar]
- 16.Chabner BA, Myers CE, Coleman CN, Johns DG. The clinical pharmacology of antineoplastic agents (second of two parts) The New England journal of medicine. 1975;292:1159–1168. doi: 10.1056/NEJM197505292922206. [DOI] [PubMed] [Google Scholar]
- 17.Chabner BA, Myers CE, Coleman CN, Johns DG. The clinical pharmacology of antineoplastic agents (first of two parts) The New England journal of medicine. 1975;292:1107–1113. doi: 10.1056/NEJM197505222922107. [DOI] [PubMed] [Google Scholar]
- 18.McKnight SL. On getting there from here. Science. 2010;330:1338–1339. doi: 10.1126/science.1199908. [DOI] [PubMed] [Google Scholar]
- 19.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locasale JW. The consequences of enhanced cell-autonomous glucose metabolism. Trends in endocrinology and metabolism: TEM. 2012;23:545–551. doi: 10.1016/j.tem.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nature reviews. Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 22.DeBerardinis RJ. Serine metabolism: some tumors take the road less traveled. Cell metabolism. 2011;14:285–286. doi: 10.1016/j.cmet.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birsoy K, Sabatini DM, Possemato R. Untuning the tumor metabolic machine: Targeting cancer metabolism: a bedside lesson. Nature medicine. 2012;18:1022–1023. doi: 10.1038/nm.2870. [DOI] [PubMed] [Google Scholar]
- 24.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature reviews. Molecular cell biology. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallingford JB, Niswander LA, Shaw GM, Finnell RH. The continuing challenge of understanding, preventing, and treating neural tube defects. Science. 2013;339:1222002. doi: 10.1126/science.1222002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iskandar BJ, et al. Folate regulation of axonal regeneration in the rodent central nervous system through DNA methylation. The Journal of clinical investigation. 2010;120:1603–1616. doi: 10.1172/JCI40000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth C, et al. Folic acid supplements in pregnancy and severe language delay in children. JAMA : the journal of the American Medical Association. 2011;306:1566–1573. doi: 10.1001/jama.2011.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, et al. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vander Heiden MG, et al. Metabolic pathway alterations that support cell proliferation. Cold Spring Harbor symposia on quantitative biology. 2011;76:325–334. doi: 10.1101/sqb.2012.76.010900. [DOI] [PubMed] [Google Scholar]
- 30.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Current opinion in genetics & development. 2008;18 :54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finkelstein JD. Methionine metabolism in mammals. The Journal of nutritional biochemistry. 1990;1:228–237. doi: 10.1016/0955-2863(90)90070-2. [DOI] [PubMed] [Google Scholar]
- 32.Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Current opinion in genetics & development. 2009;19:32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gumaa KA, MacLeod RM, McLean P. The pentose phosphate pathway of glucose metabolism. Influence of a growth-hormone-secreting pituitary tumour on the oxidative and non-oxidative reactions of the cycle in liver. The Biochemical journal. 1969;113:215–220. doi: 10.1042/bj1130215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zatz M, Dudley PA, Kloog Y, Markey SP. Nonpolar lipid methylation. Biosynthesis of fatty acid methyl esters by rat lung membranes using S-adenosylmethionine. The Journal of biological chemistry. 1981;256:10028–10032. [PubMed] [Google Scholar]
- 35.Spector AA, Yorek MA. Membrane lipid composition and cellular function. Journal of lipid research. 1985;26:1015–1035. [PubMed] [Google Scholar]
- 36.Aveldano MI, Bazan NG. Molecular species of phosphatidylcholine, -ethanolamine, -serine, and -inositol in microsomal and photoreceptor membranes of bovine retina. Journal of lipid research. 1983;24:620–627. [PubMed] [Google Scholar]
- 37.Kinney AJ, Moore TS. Phosphatidylcholine Synthesis in Castor Bean Endosperm : I. Metabolism of l-Serine. Plant physiology. 1987;84:78–81. doi: 10.1104/pp.84.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hickman MJ, et al. Coordinated regulation of sulfur and phospholipid metabolism reflects the importance of methylation in the growth of yeast. Molecular biology of the cell. 2011;22:4192–4204. doi: 10.1091/mbc.E11-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rowe PB, Lewis GP. Mammalian folate metabolism. Regulation of folate interconversion enzymes. Biochemistry. 1973;12:1862–1869. doi: 10.1021/bi00734a020. [DOI] [PubMed] [Google Scholar]
- 40.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nature chemical biology. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nature protocols. 2008;3:1299–1311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paul BD, Snyder SH. H(2)S signalling through protein sulfhydration and beyond. Nature reviews. Molecular cell biology. 2012;13:499–507. doi: 10.1038/nrm3391. [DOI] [PubMed] [Google Scholar]
- 43.Yun J, Johnson JL, Hanigan CL, Locasale JW. Interactions between epigenetics and metabolism in cancers. Frontiers in oncology. 2012;2:163. doi: 10.3389/fonc.2012.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. The Journal of nutritional biochemistry. 2012;23:853–859. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Gluckman PD. Epigenetics and metabolism in 2011: Epigenetics, the life-course and metabolic disease. Nature reviews. Endocrinology. 2012;8:74–76. doi: 10.1038/nrendo.2011.226. [DOI] [PubMed] [Google Scholar]
- 47.Teperino R, Schoonjans K, Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell metabolism. 2010;12:321–327. doi: 10.1016/j.cmet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nature reviews. Molecular cell biology. 2011;12:629–642. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- 49.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nature reviews. Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 50.Chiang PK, et al. S-Adenosylmethionine and methylation. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1996;10:471–480. [PubMed] [Google Scholar]
- 51.Heby O, Persson L. Molecular genetics of polyamine synthesis in eukaryotic cells. Trends in biochemical sciences. 1990;15:153–158. doi: 10.1016/0968-0004(90)90216-x. [DOI] [PubMed] [Google Scholar]
- 52.Snyder SH, Russell DH. Polyamine synthesis in rapidly growing tissues. Federation proceedings. 1970;29:1575–1582. [PubMed] [Google Scholar]
- 53.Janne J, Raina A, Siimes M. Mechanism of stimulation of polyamine synthesis by growth hormone in rat liver. Biochimica et biophysica acta. 1968;166:419–426. doi: 10.1016/0005-2787(68)90230-x. [DOI] [PubMed] [Google Scholar]
- 54.Farber S. Some observations on the effect of folic acid antagonists on acute leukemia and other forms of incurable cancer. Blood. 1949;4:160–167. [PubMed] [Google Scholar]
- 55.Vogelzang NJ, et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2003;21:2636–2644. doi: 10.1200/JCO.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 56.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nature reviews. Drug discovery. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 57.Cheson BD. New antimetabolites in the treatment of human malignancies. Seminars in oncology. 1992;19:695–706. [PubMed] [Google Scholar]
- 58.Bertino JR New York Academy of Sciences. Folate antagonists as chemotherapeutic agents. New York Academy of Sciences; 1971. Section of Biological and Medical Sciences. [Google Scholar]
- 59.Sirotnak FM. Folate antagonists as therapeutic agents. Academic Press; 1984. [Google Scholar]
- 60.Baram J, Allegra CJ, Fine RL, Chabner BA. Effect of methotrexate on intracellular folate pools in purified myeloid precursor cells from normal human bone marrow. The Journal of clinical investigation. 1987;79:692–697. doi: 10.1172/JCI112872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allegra CJ, Fine RL, Drake JC, Chabner BA. The effect of methotrexate on intracellular folate pools in human MCF-7 breast cancer cells. Evidence for direct inhibition of purine synthesis. The Journal of biological chemistry. 1986;261:6478–6485. [PubMed] [Google Scholar]
- 62.Diasio RB, Harris BE. Clinical pharmacology of 5-fluorouracil. Clinical pharmacokinetics. 1989;16:215–237. doi: 10.2165/00003088-198916040-00002. [DOI] [PubMed] [Google Scholar]
- 63.Pinedo HM, Peters GF. Fluorouracil: biochemistry and pharmacology. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1988;6:1653–1664. doi: 10.1200/JCO.1988.6.10.1653. [DOI] [PubMed] [Google Scholar]
- 64.Spears CP, Shahinian AH, Moran RG, Heidelberger C, Corbett TH. In vivo kinetics of thymidylate synthetase inhibition of 5-fluorouracil-sensitive and -resistant murine colon adenocarcinomas. Cancer research. 1982;42:450–456. [PubMed] [Google Scholar]
- 65.Burris HA, 3rd, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 66.Plunkett W, et al. Gemcitabine: metabolism, mechanisms of action, and self-potentiation. Seminars in oncology. 1995;22:3–10. [PubMed] [Google Scholar]
- 67.Heinemann V, Schulz L, Issels RD, Plunkett W. Gemcitabine: a modulator of intracellular nucleotide and deoxynucleotide metabolism. Seminars in oncology. 1995;22:11–18. [PubMed] [Google Scholar]
- 68.Bouffard DY, Laliberte J, Momparler RL. Kinetic studies on 2′,2′-difluorodeoxycytidine (Gemcitabine) with purified human deoxycytidine kinase and cytidine deaminase. Biochemical pharmacology. 1993;45:1857–1861. doi: 10.1016/0006-2952(93)90444-2. [DOI] [PubMed] [Google Scholar]
- 69.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nature reviews. Drug discovery. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 71.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 72.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nature medicine. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 73.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nature reviews. Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer research. 2006;66:2794–2800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 75.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. Journal of the National Cancer Institute. 2005;97:1498–1506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 76.Spannhoff A, Hauser AT, Heinke R, Sippl W, Jung M. The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem. 2009;4:1568–1582. doi: 10.1002/cmdc.200900301. [DOI] [PubMed] [Google Scholar]
- 77.Tsai HC, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wagner T, Jung M. New lysine methyltransferase drug targets in cancer. Nature biotechnology. 2012;30:622–623. doi: 10.1038/nbt.2300. [DOI] [PubMed] [Google Scholar]
- 79.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes & development. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 81.Brueckner B, Kuck D, Lyko F. DNA methyltransferase inhibitors for cancer therapy. Cancer J. 2007;13:17–22. doi: 10.1097/PPO.0b013e31803c7245. [DOI] [PubMed] [Google Scholar]
- 82.Yao Y, et al. Selective inhibitors of histone methyltransferase DOT1L: design, synthesis, and crystallographic studies. Journal of the American Chemical Society. 2011;133:16746–16749. doi: 10.1021/ja206312b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nature reviews. Drug discovery. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 84.Snell K. Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Advances in enzyme regulation. 1984;22:325–400. doi: 10.1016/0065-2571(84)90021-9. [DOI] [PubMed] [Google Scholar]
- 85.Fell DA, Snell K. Control analysis of mammalian serine biosynthesis. Feedback inhibition on the final step. The Biochemical journal. 1988;256:97–101. doi: 10.1042/bj2560097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Snell K, Natsumeda Y, Weber G. The modulation of serine metabolism in hepatoma 3924A during different phases of cellular proliferation in culture. The Biochemical journal. 1987;245:609–612. doi: 10.1042/bj2450609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Snell K, Weber G. Enzymic imbalance in serine metabolism in rat hepatomas. The Biochemical journal. 1986;233:617–620. doi: 10.1042/bj2330617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kit S. The biosynthesis of free glycine and serine by tumors. Cancer research. 1955;15:715–718. [PubMed] [Google Scholar]
- 89.Locasale JW, Cantley LC. Genetic selection for enhanced serine metabolism in cancer development. Cell Cycle. 2011;10:3812–3813. doi: 10.4161/cc.10.22.18224. [DOI] [PubMed] [Google Scholar]
- 90.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature genetics. 2011;43:869–874. doi: 10.1038/ng.890. These studies demonstrated that de novo serine biosynthesis through Phosphoglycerate dehydrogenase (PHGDH) was dramatically enhanced in a subset of breast cancers and melanomas. Those with copy number gains selectively depended on PHGDH for proliferation supporting the hypothesis that PHGDH is a candidate oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. These studies demonstrated that de novo serine biosynthesis through Phosphoglycerate dehydrogenase (PHGDH) was dramatically enhanced in a subset of breast cancers and melanomas. Those with copy number gains selectively depended on PHGDH for proliferation supporting the hypothesis that PHGDH is a candidate oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pollari S, et al. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast cancer research and treatment. 2011;125:421–430. doi: 10.1007/s10549-010-0848-5. [DOI] [PubMed] [Google Scholar]
- 93.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chaneton B, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hitosugi T, et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer cell. 2012;22:585–600. doi: 10.1016/j.ccr.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kung C, et al. Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chemistry & biology. 2012;19:1187–1198. doi: 10.1016/j.chembiol.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ye J, et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6904–6909. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maddocks OD, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. This study demonstrated the dietary requirements of serine and glycine for in vivo tumor growth as well as the p53 dependency of cancer cells to adapt to serine and glycine deprivation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma L, et al. Control of Nutrient Stress-Induced Metabolic Reprogramming by PKCzeta in Tumorigenesis. Cell. 2013;152:599–611. doi: 10.1016/j.cell.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang WC, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. This study demonstrated that increased Glycine Decarboxylase (GLDC) activity was found in a subset of tumor initiating cells and that increased GLDC activity was sufficient to induce tumor growth in a mouse xenograft. [DOI] [PubMed] [Google Scholar]
- 101.Wang J, Alexander P, McKnight SL. Metabolic specialization of mouse embryonic stem cells. Cold Spring Harbor symposia on quantitative biology. 2011;76:183–193. doi: 10.1101/sqb.2011.76.010835. [DOI] [PubMed] [Google Scholar]
- 102.Alexander PB, Wang J, McKnight SL. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15828–15833. doi: 10.1073/pnas.1111312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shyh-Chang N, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. This study demonstrated the ability of changes in the levels of metabolites in one-carbon metabolism to regulate the levels of histone methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ramm Sander P, et al. Stem cell metabolic and spectroscopic profiling. Trends in biotechnology. 2013 doi: 10.1016/j.tibtech.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 105.Sassone-Corsi P. Physiology. When metabolism and epigenetics converge. Science. 2013;339:148–150. doi: 10.1126/science.1233423. [DOI] [PubMed] [Google Scholar]
- 106.Jain M, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. This study carried out a metabolic flux analysis across the panel of NCI-60 cell lines. It was found that glycine uptake was the flux that most strongly correlated correlated with cell proliferation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sreekumar A, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. This study carried out a metabolomics analysis of urine from a large cohort of prostate cancer patients. The studies identified glycine and sarcosine as predictors of metastatic prostate cancer. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 108.Scuoppo C, et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature. 2012;487:244–248. doi: 10.1038/nature11126. This study identified genes involved in polyamine metabolism as having a tumor-suppressive role in oncogenesis. Polyamines are derived from S-adenosylmethione. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Igarashi K, Kashiwagi K. Modulation of cellular function by polyamines. The international journal of biochemistry & cell biology. 2010;42:39–51. doi: 10.1016/j.biocel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 110.Park MH, Nishimura K, Zanelli CF, Valentini SR. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee SB, et al. The effect of hypusine modification on the intracellular localization of eIF5A. Biochemical and biophysical research communications. 2009;383:497–502. doi: 10.1016/j.bbrc.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) Journal of biochemistry. 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nature reviews. Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 115.Hoeijmakers JH. Genome maintenance mechanisms are critical for preventing cancer as well as other aging-associated diseases. Mechanisms of ageing and development. 2007;128:460–462. doi: 10.1016/j.mad.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 116.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 117.Hu CM, et al. Tumor cells require thymidylate kinase to prevent dUTP incorporation during DNA repair. Cancer cell. 2012;22:36–50. doi: 10.1016/j.ccr.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 118.Wilson PM, LaBonte MJ, Lenz HJ, Mack PC, Ladner RD. Inhibition of dUTPase induces synthetic lethality with thymidylate synthase-targeted therapies in non-small cell lung cancer. Molecular cancer therapeutics. 2012;11:616–628. doi: 10.1158/1535-7163.MCT-11-0781. [DOI] [PubMed] [Google Scholar]
- 119.Stover PJ, Weiss RS. Sensitizing cancer cells: is it really all about U? Cancer cell. 2012;22:3–4. doi: 10.1016/j.ccr.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bester AC, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. This study showed that changes in the levels of nucleotide intermediates that are derived from one carbon metabolism are sufficient to induce tumorigenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ohta M, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. This study identified a tumor suppressor gene FHIT encoding a metabolic enzyme involved in nucleotide metabolism as being recurrently deleted in multiple cancers including colorectal cancer. [DOI] [PubMed] [Google Scholar]
- 122.Sozzi G, et al. The FHIT gene 3p14.2 is abnormal in lung cancer. Cell. 1996;85:17–26. doi: 10.1016/s0092-8674(00)81078-8. [DOI] [PubMed] [Google Scholar]
- 123.Pekarsky Y, Zanesi N, Palamarchuk A, Huebner K, Croce CM. FHIT: from gene discovery to cancer treatment and prevention. The lancet oncology. 2002;3:748–754. doi: 10.1016/s1470-2045(02)00931-2. [DOI] [PubMed] [Google Scholar]
- 124.Saldivar JC, et al. Initiation of genome instability and preneoplastic processes through loss of Fhit expression. PLoS genetics. 2012;8:e1003077. doi: 10.1371/journal.pgen.1003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Siprashvili Z, et al. Replacement of Fhit in cancer cells suppresses tumorigenicity. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13771–13776. doi: 10.1073/pnas.94.25.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rahman L, et al. Thymidylate synthase as an oncogene: a novel role for an essential DNA synthesis enzyme. Cancer cell. 2004;5:341–351. doi: 10.1016/s1535-6108(04)00080-7. This study provided evidence that thymidylate synthase can function as a metabolic oncogene. [DOI] [PubMed] [Google Scholar]
- 127.Fukushima M, Fujioka A, Uchida J, Nakagawa F, Takechi T. Thymidylate synthase (TS) and ribonucleotide reductase (RNR) may be involved in acquired resistance to 5-fluorouracil (5-FU) in human cancer xenografts in vivo. Eur J Cancer. 2001;37:1681–1687. doi: 10.1016/s0959-8049(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 128.Takezawa K, et al. Identification of thymidylate synthase as a potential therapeutic target for lung cancer. British journal of cancer. 2010;103:354–361. doi: 10.1038/sj.bjc.6605793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xu X, et al. Broad overexpression of ribonucleotide reductase genes in mice specifically induces lung neoplasms. Cancer research. 2008;68:2652–2660. doi: 10.1158/0008-5472.CAN-07-5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bertino JR, Banerjee D. Thymidylate synthase as an oncogene? Cancer cell. 2004;5:301–302. doi: 10.1016/s1535-6108(04)00086-8. [DOI] [PubMed] [Google Scholar]
- 131.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 132.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nature reviews. Molecular cell biology. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 134.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. The New England journal of medicine. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 135.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. The New England journal of medicine. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ceol CJ, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zee BM, et al. In vivo residue-specific histone methylation dynamics. The Journal of biological chemistry. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Barth TK, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends in biochemical sciences. 2010;35:618–626. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 142.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends in molecular medicine. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 143.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 144.Shaw RJ, Cantley LC. Decoding key nodes in the metabolism of cancer cells: sugar & spice and all things nice. F1000 biology reports. 2012;4:2. doi: 10.3410/B4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nature biotechnology. 2012;30:671–678. doi: 10.1038/nbt.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Teicher BA, Linehan WM, Helman LJ. Targeting cancer metabolism. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:5537–5545. doi: 10.1158/1078-0432.CCR-12-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dann SG, Abraham RT. Serine biosynthesis: fuel for the melanoma cell growth engine. Pigment cell & melanoma research. 2011;24:875–877. doi: 10.1111/j.1755-148x.2011.00894.x. [DOI] [PubMed] [Google Scholar]
- 148.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nature reviews. Drug discovery. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 149.Inzucchi SE, et al. Efficacy and metabolic effects of metformin and troglitazone in type II diabetes mellitus. The New England journal of medicine. 1998;338:867–872. doi: 10.1056/NEJM199803263381303. [DOI] [PubMed] [Google Scholar]
- 150.Cabreiro F, et al. Metformin Retards Aging in C. elegans by Altering Microbial Folate and Methionine Metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jalving M, et al. Metformin: taking away the candy for cancer? Eur J Cancer. 2010;46:2369–2380. doi: 10.1016/j.ejca.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 152.Pierotti MA, et al. Targeting metabolism for cancer treatment and prevention: metformin, an old drug with multi-faceted effects. Oncogene. 2013;32:1475–1487. doi: 10.1038/onc.2012.181. [DOI] [PubMed] [Google Scholar]
- 153.Chong CR, Chabner BA. Mysterious metformin. The oncologist. 2009;14:1178–1181. doi: 10.1634/theoncologist.2009-0286. [DOI] [PubMed] [Google Scholar]
- 154.Corominas-Faja B, et al. Metabolomic fingerprint reveals that metformin impairs one-carbon metabolism in a manner similar to the antifolate class of chemotherapy drugs. Aging. 2012;4:480–498. doi: 10.18632/aging.100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fedirko V, et al. Glycemic index, glycemic load, dietary carbohydrate, and dietary fiber intake and risk of liver and biliary tract cancers in Western Europeans. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2013;24:543–553. doi: 10.1093/annonc/mds434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nagle CM, et al. Carbohydrate intake, glycemic load, glycemic index, and risk of ovarian cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2011;22:1332–1338. doi: 10.1093/annonc/mdq595. [DOI] [PubMed] [Google Scholar]
- 157.Patel AV, et al. Glycemic load, glycemic index, and carbohydrate intake in relation to pancreatic cancer risk in a large US cohort. Cancer causes & control : CCC. 2007;18:287–294. doi: 10.1007/s10552-006-0081-z. [DOI] [PubMed] [Google Scholar]
- 158.Oh K, Willett WC, Fuchs CS, Giovannucci EL. Glycemic index, glycemic load, and carbohydrate intake in relation to risk of distal colorectal adenoma in women. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2004;13:1192–1198. [PubMed] [Google Scholar]
- 159.Ho VW, et al. A low carbohydrate, high protein diet slows tumor growth and prevents cancer initiation. Cancer research. 2011;71:4484–4493. doi: 10.1158/0008-5472.CAN-10-3973. [DOI] [PubMed] [Google Scholar]
- 160.Myers AP, Cantley LC. Sugar free, cancer free? Nutrition. 2012;28:1036. doi: 10.1016/j.nut.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fine EJ, et al. Targeting insulin inhibition as a metabolic therapy in advanced cancer: a pilot safety and feasibility dietary trial in 10 patients. Nutrition. 2012;28:1028–1035. doi: 10.1016/j.nut.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 162.Tavana O, Gu W. The Hunger Games: p53 Regulates Metabolism upon Serine Starvation. Cell metabolism. 2013;17:159–161. doi: 10.1016/j.cmet.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nature reviews. Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lowering blood homocysteine with folic acid based supplements: meta-analysis of randomised trials. Homocysteine Lowering Trialists’ Collaboration. BMJ. 1998;316:894–898. [PMC free article] [PubMed] [Google Scholar]
- 165.Shirota Y, et al. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2001;19:4298–4304. doi: 10.1200/JCO.2001.19.23.4298. [DOI] [PubMed] [Google Scholar]
- 166.Metzger R, et al. ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1998;16:309–316. doi: 10.1200/JCO.1998.16.1.309. [DOI] [PubMed] [Google Scholar]
- 167.Edler D, et al. Thymidylate synthase expression in colorectal cancer: a prognostic and predictive marker of benefit from adjuvant fluorouracil-based chemotherapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2002;20:1721–1728. doi: 10.1200/JCO.2002.07.039. [DOI] [PubMed] [Google Scholar]
- 168.Popat S, Matakidou A, Houlston RS. Thymidylate synthase expression and prognosis in colorectal cancer: a systematic review and meta-analysis. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22:529–536. doi: 10.1200/JCO.2004.05.064. [DOI] [PubMed] [Google Scholar]
- 169.Walther A, et al. Genetic prognostic and predictive markers in colorectal cancer. Nature reviews. Cancer. 2009;9:489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 170.Dervieux T, Greenstein N, Kremer J. Pharmacogenomic and metabolic biomarkers in the folate pathway and their association with methotrexate effects during dosage escalation in rheumatoid arthritis. Arthritis and rheumatism. 2006;54:3095–3103. doi: 10.1002/art.22129. [DOI] [PubMed] [Google Scholar]
- 171.Valik D, Radina M, Sterba J, Vojtesek B. Homocysteine: exploring its potential as a pharmacodynamic biomarker of antifolate chemotherapy. Pharmacogenomics. 2004;5:1151–1162. doi: 10.1517/14622416.5.8.1151. [DOI] [PubMed] [Google Scholar]
- 172.Vazquez A, Tedeschi PM, Bertino JR. Overexpression of the mitochondrial folate and glycine-serine pathway: a new determinant of methotrexate selectivity in tumors. Cancer research. 2013;73:478–482. doi: 10.1158/0008-5472.CAN-12-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jentzmik F, et al. Sarcosine in urine after digital rectal examination fails as a marker in prostate cancer detection and identification of aggressive tumours. European urology. 2010;58:12–18. doi: 10.1016/j.eururo.2010.01.035. discussion 20–11. [DOI] [PubMed] [Google Scholar]
- 174.Ploussard G, de la Taille A. Urine biomarkers in prostate cancer. Nature reviews. Urology. 2010;7:101–109. doi: 10.1038/nrurol.2009.261. [DOI] [PubMed] [Google Scholar]
- 175.Couzin J. Biomarkers. Metabolite in urine may point to high-risk prostate cancer. Science. 2009;323:865. doi: 10.1126/science.323.5916.865a. [DOI] [PubMed] [Google Scholar]
- 176.Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nature reviews. Cancer. 2011;11:835–848. doi: 10.1038/nrc3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer research. 1999;59:80–84. [PubMed] [Google Scholar]
- 178.Hara T, Kosaka N, Kishi H. PET imaging of prostate cancer using carbon-11-choline. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 1998;39:990–995. [PubMed] [Google Scholar]
- 179.Reske SN, et al. Imaging prostate cancer with 11C-choline PET/CT. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2006;47:1249–1254. [PubMed] [Google Scholar]
- 180.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. The New England journal of medicine. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 181.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Losman JA, Kaelin WG., Jr What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes & development. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wu K, et al. A prospective study on folate, B12, and pyridoxal 5′-phosphate (B6) and breast cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1999;8:209–217. [PubMed] [Google Scholar]
- 185.Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv Nutr. 2012;3:21–38. doi: 10.3945/an.111.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Guerreiro CS, et al. Risk of colorectal cancer associated with the C677T polymorphism in 5,10-methylenetetrahydrofolate reductase in Portuguese patients depends on the intake of methyl-donor nutrients. The American journal of clinical nutrition. 2008;88:1413–1418. doi: 10.3945/ajcn.2008.25877. [DOI] [PubMed] [Google Scholar]
- 187.Ma J, et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer research. 1997;57:1098–1102. [PubMed] [Google Scholar]
- 188.Giovannucci E, et al. Folate, methionine, and alcohol intake and risk of colorectal adenoma. Journal of the National Cancer Institute. 1993;85:875–884. doi: 10.1093/jnci/85.11.875. [DOI] [PubMed] [Google Scholar]
- 189.van Engeland M, et al. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer research. 2003;63:3133–3137. [PubMed] [Google Scholar]
- 190.Figueiredo JC, et al. Folic acid and prevention of colorectal adenomas: a combined analysis of randomized clinical trials. International journal of cancer. Journal international du cancer. 2011;129:192–203. doi: 10.1002/ijc.25872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ibrahim EM, Zekri JM. Folic acid supplementation for the prevention of recurrence of colorectal adenomas: metaanalysis of interventional trials. Med Oncol. 2010;27:915–918. doi: 10.1007/s12032-009-9306-9. [DOI] [PubMed] [Google Scholar]
- 192.Logan RF, Grainge MJ, Shepherd VC, Armitage NC, Muir KR. Aspirin and folic acid for the prevention of recurrent colorectal adenomas. Gastroenterology. 2008;134:29–38. doi: 10.1053/j.gastro.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 193.Chae YK, Yun JH. Folic acid and prevention of colorectal adenomas. JAMA : the journal of the American Medical Association. 2007;298:1397. doi: 10.1001/jama.298.12.1397-a. author reply 1397. [DOI] [PubMed] [Google Scholar]
- 194.Cole BF, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA : the journal of the American Medical Association. 2007;297:2351–2359. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- 195.Christensen BC, et al. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS genetics. 2010;6:e1001043. doi: 10.1371/journal.pgen.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhang S, et al. A prospective study of folate intake and the risk of breast cancer. JAMA : the journal of the American Medical Association. 1999;281:1632–1637. doi: 10.1001/jama.281.17.1632. [DOI] [PubMed] [Google Scholar]