Abstract
Fluorescent proteins (FPs) have been powerful tools for cell biologists for over 15 years. The large variety of FPs available rarely comes with an instruction manual or a warning label. The potential pitfalls of the use of FPs in cellular organelles represent a significant concern for investigators. FPs generally did not evolve in the often distinctive physicochemical environments of subcellular organelles. In organelles, FPs can misfold, go dark, and even distort organelle morphology. In this minireview, we describe the issues associated with FPs in organelles and provide solutions to enable investigators to better exploit FP technology in cells.
Introduction
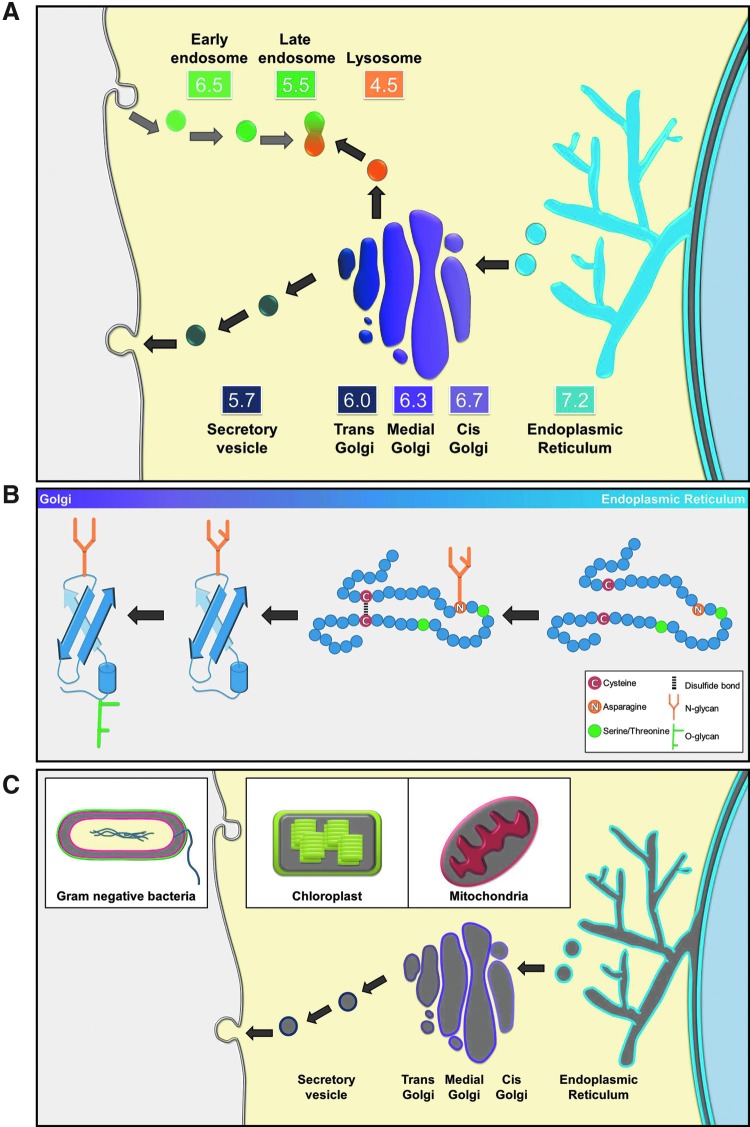
Eukaryotic cells contain membrane-bound compartments, which often have chemically distinct environments to perform specialized tasks. Different organelles within the secretory pathway have lumina with increasingly acidic pH values (Fig. 1A) (Paroutis et al., 2004). In addition, proteins can form disulfide bonds in the endoplasmic reticulum (ER), the intermembrane space of mitochondria, and the periplasm of bacteria (Fig. 1B, C). Secretory proteins containing consensus glycosylation sequences (Hebert et al., 2005; Schwarz and Aebi, 2011; Hanover et al., 2012) can be glycosylated on asparagines and serines/threonines (Fig. 1B). Regardless of whether a protein is native to an organelle, the protein will be susceptible to these modifications.
FIG. 1.
The diverse chemical environments within cellular organelles pose undesirable consequences for fluorescent proteins (FPs). (A) The organelles of the eukaryotic secretory pathway maintain distinct luminal pH values. FPs targeted to the secretory or endosomal pathways will encounter pH values that range from 7.2 to 5.7 or 6.5 to 4.5, respectively. Therefore, it is important to consider the pKa values of FPs in these environments. (B) Secretory proteins with cysteine(s) and/or glycosylation consensus sequences are modified as they traverse through the secretory pathway. In the endoplasmic reticulum (ER), N-linked sugars are added covalently, and as proteins within the secretory pathway continue to the Golgi complex, N-linked sugars are modified and potential O-linked glycosylation can be added as well. Cysteines have the potential to form disulfide bonds, primarily within the ER, although the entire secretory pathway is effectively oxidizing. (C) Eukaryotic (secretory pathway and the intermembrane space of chloroplasts and mitochondria) and prokaryotic (gram-negative bacteria periplasm) cellular environments with oxidizing environments, depicted with gray coloring.
Fluorescent proteins (FPs) were first identified in the cytoplasm of jellyfish (Shimomura et al., 1962). The ability of the investigators to express the green fluorescent protein (GFP) in non-native species (bacteria, Caenorhabditis elegans, human cells, etc.) ushered in a new era of cell biology in which investigators could genetically label cellular proteins and compartments with fluorophores (Chalfie et al., 1994). However, FPs appear to work more or less well in different cell compartments. In some cases, FPs work (or are obviously fluorescent), but as shown below, the fraction of successfully folded and fluorescent molecules may be quite low. Worse, FPs can even distort cellular organelle structures. How can FPs be made truly inert and fully exploited in all cellular compartments?
FP Essentials
All FPs share the common properties of forming a β-barrel and formation of the barrel is necessary before an autocatalytic reaction converts a tripeptide into a fluorophore (Tsien, 1998). Different FPs form fluorophores at rates ranging from minutes to several hours (Shaner et al., 2007). Relevant to this minireview, perturbation of the β-barrel or failure to form the barrel will result in a nonfluorescent protein. Once the protein achieves a barrel formation and produces a fluorophore, the fluorophore properties can be modified by the environment. Most commonly in cells, the major environmental change that impacts fluorescence is pH. This is true for fluorescent dyes, such as FITC, as well. Fortunately, most FPs have been extensively characterized and the pH range that impacts a particular FP is described as the FP's pKa (Kneen et al., 1998). Specifically, the pKa defines the pH at which the brightness of a fluorophore decreases by one half. The fluorophore will continue to become darker at decreasing pH values. Thus, in the progressively more acidic endocytic pathway of a mammalian cell (Fig. 1A), an FP may be fluorescent at the cell surface, dimmer in an early endosome (pH 5.5), and dark in a late endosome or lysosome (pH 4.5) (Paroutis et al., 2004). Therefore, it is important to choose an FP that will be suitable for studies in a particular cellular pH.
FPs can be used by themselves as inert probes or reporters (Dayel et al., 1999; Nehls et al., 2000; Snapp et al., 2006) in biosensor fluorescence resonance energy transfer reporters (Zadran et al., 2012), or as fusions with a protein of interest. In the latter case, placement of the FP relative to the protein of interest is essential. For example, proteins in the secretory pathway contain sequences critical for targeting to the ER (i.e., the signal sequence) and sequences involved in retrieval or retention of a protein in a compartment (i.e., a KDEL sequence). Both types of sequence are positional and must be at the absolute NH2 or COOH terminus of a protein. Placement of the FP before or after the start or end of the protein will result in significant mistargeting. Issues of FP placement in fusion proteins are discussed more extensively elsewhere (Snapp, 2005, 2009).
Assuming one has proper placement within a fusion protein of an FP appropriate to an organelle's pH, what other issues might an investigator face? Surprisingly, there are at least three major issues that can dramatically impact the outcome and interpretation of FP-fusion experiments in cells: (1) disulfide bond formation, (2) glycosylation, and (3) oligomerization.
Oxidizing Environments and Disulfide Bond Formation
Multiple cellular compartments contain proteins with covalently disulfide-bonded cysteines, even the cytoplasm (Cumming et al., 2004). Disulfide bonds form inter- and intrachain between cysteines in proteins exposed to oxidizers, especially in environments with strong oxidizing potentials, including the ER, the intermembrane space of chloroplasts and mitochondria, and the periplasm of gram-negative bacteria (Fig. 1C). Proteins native to these environments have evolved sequences and folds that promote the formation of specific disulfide bonds and this process is further guided by chaperones such as protein disulfide isomerase family members. However, any protein with cysteines potentially can be a substrate for disulfide bond formation machinery. The original FP molecules and their derivatives all evolved in the relatively nonoxidizing environment of a cell and the bacterial cytoplasm. In such an environment, there was little selection against the accumulation of cysteines. GFP and its derivatives [BFP (Heim and Griesbeck, 2004), CFP (Ormo et al., 1996), cerulean (Rizzo et al., 2004), EGFP (Yang et al., 1996), emerald (Cubitt et al., 1999), YFP (Tsien, 1998), Venus (Nagai et al., 2002), Citrine (Griesbeck et al., 2001)] all contain two cysteines. When expressed in the secretory pathway of eukaryotic cells, a significant population of the FPs forms interchain disulfide bonds, as evidenced by the presence of a ladder pattern on nonreducing SDS-PAGE immunoblots (Fig. 2A) (Jain et al., 2001; Aronson et al., 2011). The cysteines of EGFP normally face the inside of the β-barrel, which means that interchain disulfide bond formation must occur before a β-barrel forms. Equally curious is the implication that despite all of the different proteins in oxidizing environments that EGFP could form a random disulfide with, there appears to be a strong tendency for the non-β-barrel form of GFP to oligomerize (Fig. 2A).
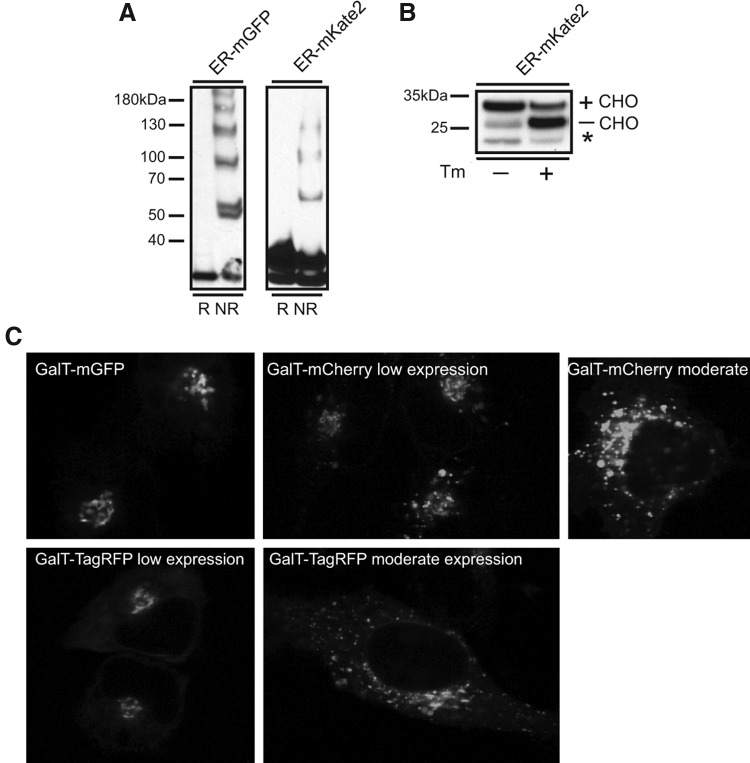
FIG. 2.
FPs localized to the secretory pathway can form disulfide bonds, become N-glycosylated, and oligomerize. ER localized mGFP and mKate2 (ER-mGFP and ER-mKate2, respectively) form inappropriate disulfide bonds. Representative immunoblots (A) reveal the presence of higher molecular weight oligomers in nonreducing (NR) conditions. Oligomers form due to non-native interchain disulfide bond formation. mKate2 also contains a potential N-glycosylation consensus sequence. (B) When localized to the ER, ER-mKate2 is N-glycosylated (+CHO). Treatment with the N-glycosylation inhibitor, tunicamycin (Tm), shifts the majority of ER-mKate2 to a lower molecular weight band, indicating the nonglycosylated population of FP (-CHO). Asterisk (*) denotes band corresponding to a cleavage product due to sample preparation. (C) Different FPs fused with the integral membrane Golgi complex marker Galactosyltransferase (GalT) can distort Golgi complex morphology. Representative images of cells expressing GalT-mGFP, -mCherry, or -TagRFP reveal fusion proteins, GalT-mCherry and -TagRFP expressed at moderate levels significantly disrupt the normal organelle structure. Compared to the low expression levels, GalT fusions retain typical compact Golgi complex morphology.
Despite the absence of an obvious structural role for the two cysteines, mutation of either causes the GFP to dim considerably and mutation of both to relatively conservative serines makes the mutant FP dark (Jain et al., 2001). Obviously, such an FP is useless for imaging. Even the visible FP is suboptimal as a significant fraction (in some cases greater than 50% of the total FP population) is misfolded, dark, and of unclear functionality (Fig. 2A). To maximize the fluorescent signal, minimize the amount of an FP fusion that needs to be expressed to visually detect it, and to minimize the formation of misfolded proteins, investigators need to use FPs resistant to disulfide bond formation.
Many other FPs have three or more cysteines. The recently developed FusionRed (Shemiakina et al., 2012) contains four cysteines. Kusabira orange derivatives even have a cysteine that forms part of the fluorophore (Karasawa et al., 2004). At $400 or more for some FP expression plasmids, selection of an FP unsuitable for a cellular environment of interest can be a costly mistake. Fortunately, alternatives exist and we describe these at the end of this minireview.
Glycosylation
Within the secretory pathway of eukaryotic cells, two types of sugars, asparagine linked (N-linked) and serine/threonine linked (O-linked) can be attached to proteins. N-linked are added in the ER as nascent proteins emerge from the translocon channel if the proteins contain an N-X-S/T consensus sequence (where X is any amino acid except proline) (Fig. 1B). O-linked sugars are added to serines and threonines in the Golgi complex by N-acetylgalactosaminyl-transferases (Fig. 1B). In contrast to N-linked glycosylation, O-linked consensus sites remain poorly defined, although flanking residues are often enriched in proline, serine, threonine, and alanine (Potter et al., 2006). There are additional forms of O-linked glycosylation, but these tend to be highly specialized for a small subset of proteins and are unlikely to be a significant concern for FPs.
The consequences of sugar additions can be substantial. First, N-linked sugars are added to the primary sequence of the protein still in the translocation channel, often before significant folding has occurred. If the sugar is placed on a residue that faces the inside of the β-barrel, then the ∼1.3 nm sugar could potentially interfere with β-barrel folding and/or fluorophore formation. On the outside of the β-barrel, the sugar will dramatically increase the size of the ∼4.6 nm diameter molecule. The size increase could cause steric interference blocking interactions of the fusion protein with a binding partner. More problematic on a global scale, the sugar marks the FP as a client for the glycosylation quality control machinery, including the lectin chaperones calnexin and calreticulin, UDP-glucose glycoprotein:glucosyltransferase, and the ER-associated degradation pathway (Hebert and Molinari, 2012). The overexpressed glycosylated FP will potentially titrate the glycosylation quality control machinery and could upset ER homeostasis or the half-life of the fusion protein could be shortened. If one is performing biophysical measurements (i.e., fluorescence recovery after photobleaching to determine a protein's diffusion coefficient in a cell) of a reporter FP or a fusion protein, then the sugar can substantially alter the FP's physical properties (Costantini et al., 2013).
Many of the newer evolved FPs contain N-linked glycosylation consensus sites. TagRFP contains one, TagBFP contains three, and mKate2 contains one. Figure 2B illustrates that, when localized to the ER, mKate2 is N-glycosylated. The GFP family members and the DsRed family members typically lack N-linked glycosylation consensus sites.
FP Oligomerization
Some FPs, such as DsRed, are obligate oligomers. FP-mediated protein oligomerization is generally highly undesirable for fusion proteins, as oligomerization can lead to activation of the fusion protein, inappropriate localization of the fusion protein, or can even alter organelle architecture. For example, standard EGFP is a weak dimer (Kd=0.11 mM) (Zacharias et al., 2002). In the context of a fusion protein in a confined or constrained space, this modest affinity becomes more than sufficient to promote interactions with the same protein on an apposing ER membrane, which in turn leads to membrane stacking and whorl formation and global reorganization of the ER (Snapp et al., 2003). Constraint of EGFP to a membrane protein restricts the tumbling movement of EGFP in a solution to a two-dimensional plane and increases the effective concentration, leading to an increasing probability of membrane stacking. Thus, it is critical to avoid oligomerizing FPs. Unfortunately, this has proven difficult because there is no agreed upon standard in the field for distinguishing oligomeric from monomeric FPs. The definitions of monomeric range from quantitative to experimental, although the criteria for monomeric FPs vary between laboratories. With the goal of establishing a quantitative standard for monomeric FPs, we developed a visual assay that reports on the tendency of an FP to oligomerize under physiologic conditions, when attached to an integral membrane protein (Costantini et al., 2012). Using this tool, we found that TagRFP is not, as previously reported (Shaner et al., 2008) monomeric. In fact, TagRFP is the FP most prone to oligomerizing that we have identified to date. The CyTERM construct is freely available from our laboratory and can be used to test any FP for oligomerizing tendencies.
In addition to oligomerization, FPs can fold correctly, but be sticky and aggregate into bright puncta (Yanushevich et al., 2002; Snapp, 2005). This behavior can be so severe that it can disrupt organelles in the secretory pathway. For example, the integral membrane Golgi complex marker Galactosyltransferase (GalT) can be fused to mGFP and localizes to a typical compact Golgi complex structure in a live cell (Fig. 2C). In contrast, fusion to TagRFP disrupts the Golgi complex into a fragmented dispersed structure (Fig. 2C). Whereas red FPs have been successfully used for a number of studies, it is important to recognize the potential of these (and any FP) to distort organelles. Therefore, it remains critical to compare an FP fusion protein's localization pattern to the native endogenous protein (preferably) or at least to the distribution of an epitope tagged fusion (i.e., myc or HA tagged). Note that the latter option is suboptimal for uncharacterized proteins as the tag may mask position-dependent targeting sequences.
Solutions
Taken together, this sampling of post-translational modifications and environmental differences illustrates the potential of cellular organelle environments to complicate and compromise the use of FPs. Therefore, it is critical for investigators to recognize the potential caveats of using FPs evolved for the cytoplasm. Table 1 summarizes properties of commonly used FPs and, thus, illuminates their potential utilities in different cellular environments. Inertness of FPs should always be a primary goal when selecting an FP for experiments.
Table 1.
Properties of Commonly Used Fluorescent Proteins
| FP | Monomeric | pKa | N-linked consensus seq. | Disulfide forming |
|---|---|---|---|---|
| EGFPa | No | 6.0 | No | Yes |
| mGFPa–c | Yes | 6.0 | No | Yes |
| mVenusd | Yes | 6.0 | No | Yese |
| mCitrinef | Yes | 5.7 | No | Predicted |
| Ceruleanc,g | Yes | 3.2 | No | Yese |
| TagBFPh,i | Yes | 2.7 | Yes | Yes |
| TagRFPa | No | <4.0 | Yes | Predicted |
| mCherrya | Yes, but sticky | <4.5 | No | No |
| sfGFPc,j | Yes | 5.5 | No | No |
Shaner et al. (2007).
Snapp et al. (2003).
Aronson et al. (2011).
Nagai et al. (2002).
Our unpublished results.
Griesbeck et al. (2001).
Markwardt et al. (2011).
Costantini et al. (2013).
Subach et al. (2008).
Pedelacq et al. (2006).
FP, fluorescent protein; sfGFP, superfolder green fluorescent protein.
Fortunately, solutions are available. First, the monomerized derivatives of DsRed (i.e., mRFP and mCherry) naturally lack cysteines and readily fluoresce in the oxidizing bacterial periplasm and ER (Campbell et al., 2002; Shaner et al., 2004; Snapp et al., 2006). In addition, DsRed and its derivatives do not contain consensus sequences for N-linked glycosylation. Therefore, these proteins are reasonable choices, especially for fusions with soluble monomeric proteins. Other FPs should be used with integral membrane protein fusions or with naturally oligomerizing proteins due to the stickiness of monomeric DsRed family members.
What about truly inert FPs? Through educated guesses and directed mutagenesis, it has been possible to mutate the cysteines of TagBFP and a GFP variant and maintain much of the original fluorescence brightness of the original proteins (Suzuki et al., 2012; Costantini et al., 2013). Curiously, at least one FP with cysteines, superfolder GFP (sfGFP) (Pedelacq et al., 2006), is resistant to disulfide bond formation in multiple environments (Aronson et al., 2011). It is truly monomeric (Pedelacq et al., 2006; Costantini et al., 2012) and contains no N-linked glycosylation consensus sites. In our hands, sfGFP appears to be the most inert FP and we strongly recommend sfGFP for use in oxidizing environments. We anticipate that, as more investigators become aware of the problems of environmental impacts on FP functionality, improved inert FPs of a wider palette will be developed.
Acknowledgments
Work leading to this review was supported by grants from the National Institute of General Medical Sciences (NIGMS) (R01GM086530-01) (E.L.S.), the Marion Bessin Liver Center Imaging and Cell Structure Core supported by the National Institute of Diabetes and Digestive and Kidney Diseases NIDDK (2P30DK041296) (E.L.S), and the NIH Training Program in Cellular and Molecular Biology and Genetics Grant T32 GM007491 (L.M.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the NIH.
Disclosure Statement
No competing financial interests exist.
References
- Aronson D.E. Costantini L.M. Snapp E.L. Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon. Traffic. 2011;12:543–548. doi: 10.1111/j.1600-0854.2011.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R.E. Tour O. Palmer A.E. Steinbach P.A. Baird G.S. Zacharias D.A. Tsien R.Y. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M. Tu Y. Euskirchen G. Ward W.W. Prasher D.C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Costantini L.M. Fossati M. Francolini M. Snapp E.L. Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions. Traffic. 2012;13:643–649. doi: 10.1111/j.1600-0854.2012.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini L.M. Subach O.M. Jaureguiberry-Bravo M. Verkhusha V.V. Snapp E.L. Cysteineless non-glycosylated monomeric blue fluorescent protein, secBFP2, for studies in the eukaryotic secretory pathway. Biochem Biophys Res Commun. 2013;430:1114–1119. doi: 10.1016/j.bbrc.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubitt A.B. Woollenweber L.A. Heim R. Understanding structure-function relationships in the Aequorea victoria green fluorescent protein. Methods Cell Biol. 1999;58:19–30. doi: 10.1016/s0091-679x(08)61946-9. [DOI] [PubMed] [Google Scholar]
- Cumming R.C. Andon N.L. Haynes P.A. Park M. Fischer W.H. Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem. 2004;279:21749–21758. doi: 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- Dayel M.J. Hom E.F. Verkman A.S. Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophys J. 1999;76:2843–2851. doi: 10.1016/S0006-3495(99)77438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck O. Baird G.S. Campbell R.E. Zacharias D.A. Tsien R.Y. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- Hanover J.A. Krause M.W. Love D.C. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol. 2012;13:312–321. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- Hebert D.N. Garman S.C. Molinari M. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 2005;15:364–370. doi: 10.1016/j.tcb.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Hebert D.N. Molinari M. Flagging and docking: dual roles for N-glycans in protein quality control and cellular proteostasis. Trends Biochem Sci. 2012;37:404–410. doi: 10.1016/j.tibs.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim N. Griesbeck O. Genetically encoded indicators of cellular calcium dynamics based on troponin C and green fluorescent protein. J Biol Chem. 2004;279:14280–14286. doi: 10.1074/jbc.M312751200. [DOI] [PubMed] [Google Scholar]
- Jain R.K. Joyce P.B. Molinete M. Halban P.A. Gorr S.U. Oligomerization of green fluorescent protein in the secretory pathway of endocrine cells. Biochem J. 2001;360:645–649. doi: 10.1042/0264-6021:3600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasawa S. Araki T. Nagai T. Mizuno H. Miyawaki A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneen M. Farinas J. Li Y. Verkman A.S. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys J. 1998;74:1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwardt M.L. Kremers G.J. Kraft C.A. Ray K. Cranfill P.J. Wilson K.A. Day R.N. Wachter R.M. Davidson M.W. Rizzo M.A. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One. 2011;6:e17896. doi: 10.1371/journal.pone.0017896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T. Ibata K. Park E.S. Kubota M. Mikoshiba K. Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nehls S. Snapp E.L. Cole N.B. Zaal K.J. Kenworthy A.K. Roberts T.H. Ellenberg J. Presley J.F. Siggia E. Lippincott-Schwartz J. Dynamics and retention of misfolded proteins in native ER membranes. Nat Cell Biol. 2000;2:288–295. doi: 10.1038/35010558. [DOI] [PubMed] [Google Scholar]
- Ormo M. Cubitt A.B. Kallio K. Gross L.A. Tsien R.Y. Remington S.J. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- Paroutis P. Touret N. Grinstein S. The pH of the secretory pathway: measurement, determinants, and regulation. Physiology (Bethesda) 2004;19:207–215. doi: 10.1152/physiol.00005.2004. [DOI] [PubMed] [Google Scholar]
- Pedelacq J.D. Cabantous S. Tran T. Terwilliger T.C. Waldo G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Potter B.A. Hughey R.P. Weisz O.A. Role of N- and O-glycans in polarized biosynthetic sorting. Am J Physiol Cell Physiol. 2006;290:C1–C10. doi: 10.1152/ajpcell.00333.2005. [DOI] [PubMed] [Google Scholar]
- Rizzo M.A. Springer G.H. Granada B. Piston D.W. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Schwarz F. Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21:576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Shaner N.C. Campbell R.E. Steinbach P.A. Giepmans B.N. Palmer A.E. Tsien R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shaner N.C. Lin M.Z. McKeown M.R. Steinbach P.A. Hazelwood K.L. Davidson M.W. Tsien R.Y. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nature Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C. Patterson G.H. Davidson M.W. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- Shemiakina II. Ermakova G.V. Cranfill P.J. Baird M.A. Evans R.A. Souslova E.A. Staroverov D.B. Gorokhovatsky A.Y. Putintseva E.V. Gorodnicheva T.V., et al. A monomeric red fluorescent protein with low cytotoxicity. Nat Commun. 2012;3:1204. doi: 10.1038/ncomms2208. [DOI] [PubMed] [Google Scholar]
- Shimomura O. Johnson F.H. Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- Snapp E. Design and use of fluorescent fusion proteins in cell biology. In: Bonafacino J.S., editor; Dasso M., editor; Harford J., editor; Lippincott-Schwartz J., editor; Yamada K., editor. In Current Protocols in Cell Biology. John Wiley & Sons, Inc.; New York: 2005. p. 21.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E.L. Fluorescent proteins: a cell biologist's user guide. Trends Cell Biol. 2009;19:649–655. doi: 10.1016/j.tcb.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E.L. Hegde R.S. Francolini M. Lombardo F. Colombo S. Pedrazzini E. Borgese N. Lippincott-Schwartz J. Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E.L. Sharma A. Lippincott-Schwartz J. Hegde R.S. Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc Natl Acad Sci USA. 2006;103:6536–6541. doi: 10.1073/pnas.0510657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach O.M. Gundorov I.S. Yoshimura M. Subach F.V. Zhang J. Gruenwald D. Souslova E.A. Chudakov D.M. Verkhusha V.V. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T. Arai S. Takeuchi M. Sakurai C. Ebana H. Higashi T. Hashimoto H. Hatsuzawa K. Wada I. Development of cysteine-free fluorescent proteins for the oxidative environment. PLoS One. 2012;7:e37551. doi: 10.1371/journal.pone.0037551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R.Y. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Yang T.T. Cheng L. Kain S.R. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Res. 1996;24:4592–4593. doi: 10.1093/nar/24.22.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanushevich Y.G. Staroverov D.B. Savitsky A.P. Fradkov A.F. Gurskaya N.G. Bulina M.E. Lukyanov K.A. Lukyanov S.A. A strategy for the generation of non-aggregating mutants of Anthozoa fluorescent proteins. FEBS Lett. 2002;511:11–14. doi: 10.1016/s0014-5793(01)03263-x. [DOI] [PubMed] [Google Scholar]
- Zacharias D.A. Violin J.D. Newton A.C. Tsien R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zadran S. Standley S. Wong K. Otiniano E. Amighi A. Baudry M. Fluorescence resonance energy transfer (FRET)-based biosensors: visualizing cellular dynamics and bioenergetics. Appl Microbiol Biotechnol. 2012;96:895–902. doi: 10.1007/s00253-012-4449-6. [DOI] [PubMed] [Google Scholar]