Abstract
Epithelial-mesenchymal transition (EMT) in carcinoma cells enhances malignant progression by promoting invasion and survival. EMT is induced by microenvironmental factors including TGF-β and Wnt agonists, and by the E-box-binding transcription factors Twist, Snail and ZEB. Grainyhead-like-2 (GRHL2), a member of the mammalian Grainyhead family of wound healing regulatory transcription factors, suppresses EMT and restores sensitivity to anoikis by repressing ZEB1 expression and inhibiting TGF-β signaling. In this study, we elucidate the functional relationship between GRHL2 and ZEB1 in EMT/MET and tumor biology. At least three homeodomain proteins, Six1, LBX1, and HoxA5, transactivated the ZEB1 promoter, in the case of Six1, through direct protein-promoter interaction. GRHL2 altered the Six1-DNA complex, inhibiting this transactivation. Correspondingly, GRHL2 expression prevented tumor initiation in xenograft assays, sensitized breast cancer cells to paclitaxel and suppressed the emergence of CD44highCD24low cells (defining the cancer stem cell phenotype in the cell type studied). GRHL2 was down-regulated in recurrent mouse tumors that had evolved to an oncogene-independent, EMT-like state, supporting a role for GRHL2 down-regulation in this phenotypic transition, modeling disease recurrence. The combination of TGF-β and Wnt activation repressed GRHL2 expression by direct interaction of ZEB1 with the GRHL2 promoter, inducing EMT. Together, our observations indicate that a reciprocal feedback loop between GRHL2 and ZEB1 controls epithelial vs. mesenchymal phenotypes and EMT-driven tumor progression.
Keywords: Grainyhead-like-2, ZEB1, epithelial-mesenchymal transition
Introduction
The oncogenic epithelial-mesenchymal transition (EMT) contributes to tumor progression by enhancing tumor cell invasiveness and anoikis-resistance, which may enhance the early steps of metastasis (1–5). Chemoresistance and radioresistance also accompany EMT, confounding stable patient responses to treatment, especially in the claudin-low subclass of breast cancer, where a frank EMT-like pattern of genes is expressed (6–9). In certain contexts, EMT also promotes the transition of tumor cells to cancer stem cell/tumor initiating cells, although cancer stem cells also can arise independently of EMT (10–14).
The transcription factors ZEB1 and ZEB2 (SIP1) are pivotal activators of EMT. ZEB1 and, subsequently, ZEB2 were identified as the first repressors of the mammalian E-cadherin promoter (15, 16). They are now known to regulate cytoskeletal, cell polarity, cell adhesion and apoptosis-regulatory genes that collectively suffice for EMT induction (17, 18). ZEB1 is induced by transcription factors that include NF-kB, Twist/Snail (acting in concert), TCF4 and LBX1 (19–22). Conversely, ZEB1 expression is translationally attenuated by mir-200b/c, whose expression is, in turn, repressed by TGF-β signaling, explaining (in part) the engagement of stable autocrine TGF-β signaling in the maintenance of EMT (23).
Grainyhead-like-2 is a transcription factor that regulates the EMT/MET-related processes of wound healing, epidermal junction assembly, and neural tube closure (24–27). Previously, we reported that GRHL2 is a generalized suppressor of oncogenic EMT, through at least two mechanisms, direct repression of ZEB1 expression and inhibition of the TGF-β pathway (28). Here, we show that GRHL2 inhibited transactivation of the ZEB1 promoter mediated by the homeodomain proteins Six1, LBX1 and HoxA5. GRHL2 acted as a tumor suppressor gene, by the criterion of suppressed tumor initiation frequency. GRHL2 also sensitized tumor cells to paclitaxel and prevented the emergence of CD44highCD24low mammary epithelial cells. The combination of Wnt and TGF-β pathway activation up-regulated ZEB1 expression. ZEB1 reciprocally repressed GRHL2 expression through a direct interaction with the GRHL2 promoter. These observations reveal a novel GRHL2-ZEB1 reciprocal feedback loop that drives EMT vs. MET in response to extracellular signals.
Materials and Methods
Cell lines
HMLE and HMLE+twist-ER were kindly provided by R. Weinberg (Whitehead Institute); MDA-MB-231LN were from E. Pugacheva (West Virginia University). Cells were cultured and stable cell lines were generated via retroviral transduction as described previously (28) GRHL2 was expressed using either MSCV-IRES-puro or MSCV-IRES-GFP (pMIG, Addgene) and mixed populations or multiple clonal lines were used as indicated. The β-catenin S33Y mutant (provided by S.P.S. Monga, University of Pittsburgh) was subcloned into the pMXS-IRES-PURO retroviral vector (contributed by Russ Carstens, University of Pennsylvania in-frame with a C-terminal FLAG tag). The human Six1 sequence was also subcloned into the pMXS-IRES-PURO vector. ZEB1 shRNA (V3LHS_356186, Open Biosystems) and ZEB1 cDNA were expressed via the pTRIPZ and pLUT lentiviral vectors, respectively, as described previously (28). Human H-rasV12 in MSCV-IRES-Blast (Addgene) was used to express activated Ras in CD44low (flow-sorted) HMLE cells for tumor assays described below. HMLE+Ras cells were subjected to stable GRHL2 knockdown (Open Biosystems RHS4430-99291384) using pGIPZ, followed by selection for puromycin resistance and sorting for GFP.
Xenograft Assays
MDA-MB-231LN stably expressing either empty-MSCV-IRES-Puro or GRHL2-IRES-Puro were trypsinized and re-suspended in growth media and the indicated cell numbers in 0.1 ml were injected into the 4th inguinal mammary fat pad of female BALB/c nude mice (Charles River) at approximately 4 weeks of age. HMLE+Ras cells with GRHL2 or control shRNA (see above) were trypsinized and suspended in growth medium; the indicated cell numbers were injected in 0.1ml into the 4th inguinal mammary fat pad of female NOD/SCID (Charles River). Assays were carried out for ten weeks (HMLER) or for indicated times. Tumor volume was determined using the formula π(L1×L22)/6, where L1 is the long axis and L2 is the short axis. Tumor burden was monitored according to the WVU ACUC Tumor Burden Policy. Animal studies were approved by the West Virginia University Animal Care and Use Committee.
Western blotting
Electrophoresis, transfer, immunoblotting and antibodies used were described in (28). Other antibodies used here were: rabbit anti-vimentin (Cell Signaling), mouse anti-FLAG (M2; Sigma) and mouse anti-GAPDH (Thermo Scientific).
Chemosensitivity assays
MDA-MB-231 cells with stable GRHL2-MSCV-IRES-puro or empty MSCV-IRES-puro were plated in 96-well dishes at 5,000 cells per well and assayed in biological triplicate for cell viability after three days of incubation, with the indicated concentration of paclitaxel, using Presto-Blue (Invitrogen) according to the manufacturer’s protocol. For the chemo-induced marker analysis, the protocol was modeled after (11). HMLE cells expressing empty-MSCV-IRES-GFP or GRHL2–MSCV-IRES-GFP were treated with paclitaxel (10nM) for 4 days, followed by 7 days recovery and flow analysis for CD44 using a CD44-PE antibody (BD Biosciences). HMLE induced to undergo EMT by ectopic twist were used as a positive control for the CD44high phenotype.
qRT-PCR analysis of primary vs. recurrent mouse tumors
RNAs from primary and recurrent tetO-neuNT (n=5 for each group) (29) or tetO-Wnt1 (30)(n=4 for each group) were provided by L. Chodosh (University of Pennsylvania) or E. Gunther (Penn State University, Hershey) and analyzed for GRHL2 expression using a β2-microglobulin internal control as described previously (28), using the primers: mGRHL2-f: caaggacgaccagcgcagca; mGRHL2-r: ggccctccccctgcttctga; mB2M-f: tggtctttctggtgcttgtc; mB2M-r: gggtggaactgtgttacgtag.
BIO, TGF-β and BMP2 treatments
HMLE cells were treated with 5 μM 6-bromo-3-[(3E)-1,3-dihydro-3-(hydroxyimino)-2H-indol-2-ylidene]-1,3-dihydro-(3Z)-2H-indol-2-one (BIO;(31)) and/or TGF-β1 (5ng/ml) for 4 days. BIO was removed and the TGF-β was continued for another 6 days; samples were then analyzed by western blotting. HMLE+twist-ER were treated with either 50 nM 4-hydroxy-tamoxifen (4OHT), 0.5 μg/ml BMP-2 (R and D Biosystems) or both for 7–10 days and then analyzed by western blotting.
Reporter Assays
Co-transfections, luciferase and β-galactosidase assays as well as the GRHL2 expression vector and ZEB1-WT- promoter reporter were described previously (28) all values represent the average relative luciferase activity/beta-galactosidase activity of biological duplicates; error bars represent standard deviation from the mean. The transcription factors used in the co-transfection/reporter assays were obtained and cloned as follows: hLBX1 was obtained from D. Haber (Dana-Farber Cancer Center) in pBABE and cloned into pCDNA3.1. hHOXA5 was purchased from Addgene and subcloned from pET28B into pCDNA-3.1. mTwist (pBABE-mTWIST-ER) was purchased from Addgene and the twist cDNA was subcloned into pCDNA-3.1 (GC rich Phusion buffer). HA-Snail-pCDNA-3.1 was purchased from Addgene. The GRHL2 genomic sequence from −625 to +335 (relative to the transcription start site of the standard GRHL2 transcript) was subcloned into pGL4.14 using the following primers: Gra-prom-f3: CTCACCTGTTTGAAAATCTG and Gra-prom-r12: TGTTTGATCCAATGAACTTGC, using the Bacterial Artificial Chromosome clone 2058A8 (Invitrogen) as a template; these coordinates were based on the GRHL2 transcript NM_024915. The band resulting from this PCR was re-amplified with similar primers containing NheI (forward) and BglII (reverse) restriction sites, and subcloned into pGL4.14 (Promega).
The ZEB1 promoters containing a mutant downstream homeobox site or deleted upstream homeobox site or both were generated as follows. The downstream site was mutated by using the mutagenic primers Mut-f3: cgtgcctccctctccccaccagtccgtaggaaaacttttccc Mut-r3: gggaaaagttttcctacggactggtggggagagggaggcacg in the Quickchange-IIXL kit (Agilent) with the ZEB1 promoter/pGL3basic clone (provided by Antonio Garcia de Herreros, University of Barcelona) as a template. The deletion of the upstream site was generated by amplifying the same template in two separate PCR reactions, using Ant-f: TAATTTACGCGTCCTTAAGGTCCTGCACGGCG, Ant-r: TTTAAAAAGCTTCCGCCATGATCCTCTCGC (reaction 1) and Ant-f2: tatataGCGGCCGCAAGGGAAGGGAAGGGAGTCC, Ant-r2: TTTAAAGCGGCCGCTTCAATGAGATTGAACTTCA. Following restriction digestion and isolation from an agarose gel, fragments were three-way ligated into pGL3basic.
Chromatin Immunoprecipitation (CHIP) assays
The protocol for crosslinking, immunoprecipitation, de-crosslinking and qPCR analysis was as described (28). For Six1 protein interaction with the ZEB1 promoter, 5 dishes of HMLE+ empty vector (pMXS-IRES-puro) or FLAG-Six1 were immunoprecipitated with mouse anti-FLAG-M2-agarose or mouse IgG (Santa Cruz BioTechnology)-agarose in conjunction with DNA/protein-blocked protein A/G agarose (Pierce/Thermo). Primers to amplify the ZEB1 promoter were: f: gccgccgagcctccaacttt; r: tgctagggaccgggcggttt. For interaction of ZEB1 protein with the GRHL2 promoter, 5 dishes HMLE expressing ZEB1 (in pLUT vector) were processed. To pull down ZEB1 1 μg of the following antibodies (3 μg total) were used: ZEB1, rb, Sigma HPA004820; ZEB1, rb, Cell Signalling 3396S; ZEB1, rb, Santa Cruz Biotechnology SC-25388. Control antibody was rabbit IgG (3 μg) Santa Cruz Biotechnology. Primers to analyzed the ZEB1 chIP were: Positive control: Ecad-ZEB1-f: ggccggcaggtgaaccctca; Ecad-ZEB1-r: gggctggagtctgaactga; GRHL2 promoter set 1: f: cgccgctccactcgggtaaa ; r: tgcgcgatgattggctgggg; GRHL2 promoter set2: f: tcagctctcacaggctgccg; r: gaggcgcgctcaggtaaggc. Negative control primer set for all CHIP assays was from an intergenic region upstream of the GAPDH locus: GAPDH-f: atgggtgccactggggatct; GAPDH-r: tgccaaagcctaggggaaga.
Electrophoretic mobility shift assay
Purified recombinant Six1 protein was generated as described previously (32). Purified recombinant GRHL2 protein was obtained by expressing the GRHL2 coding sequence in E. coli BL21 using the pGEX-6P3 vector and purifying the fusion protein as described previously (15). After binding to the Glutathione Sepharose resin (GE Healthcare) the protein was cleaved free of GST sequence using Pre-Scission Protease, which was then dialyzed against EMSA buffer (see below) using Slide-A-Lyzer Mini-dialysis Unit (Thermo Scientific). 2x EMSA buffer was: 30mM HEPES pH 7.6, 20% Glycerol, 1mM EDTA, 200mM KCL, 0.5% NP40, 5mg/ml BSA (added fresh), 10mM DTT (added fresh). For 20ul binding reactions: 10ul 2x EMSA Buffer+1 μL poly dIdC (0.5 μg/μL)+2 μL annealed 100 nM oligonucleotide, and recombinant protein (as indicated). Oligonucleotides for EMSA were synthesized with Cy5 groups on both 5′ ends (Operon) and annealed at 10 mM Tris/1mM EDTA/100mM NaCl. Binding reactions were incubated at 4 degrees for twenty minutes, fifteen minutes at room temperature and analyzed on 6% polyacrylamide/TBE gels in 0.25x TBE buffer (pre-run for 1h) at 170V. The fluorescent signals were imaged using the LiCor Odyssey imaging system.
Oligo sequences(Forward strands)
ZEB1-WT-Promoter: (Cy5)-GGAAGGTGATGTCGTAAAGCCGGGAGTGTCGTAAACCAGGTGCGGTGGG
ZEB1-MT-Promoter: (Cy5)-GGAAGGTGATGTCGTCCCGCCGGGAGTGTCGTCCCCCAGGTGCGGTGGG
GRHL2-Pos-Con: (Cy5)-AACTATAAAACCGGTTTATCTAGTTGG
Six1-Pos-Con: (Cy5)-GGGGGCTCAGGTTTCTGTGGC
Negative Control (From E-cadherin promoter):
TGGCCGGCAGGTGAACCCTCAGCCAATCAGCGCTACGGGGGGCGGTGCTCCGGGGCTCACCTGGCTGCAG
Flow cytometry
Trypsinized cells (1×105) were stained with APC mouse anti-human CD44 (BD 560890) and PerCP-Cy5.5 mouse anti-human CD24 (BD 561647) in 100 uL of phosphate-buffered saline containing 2.5 mM EDTA, 10 mM HEPES and 2% horse serum, followed by washing and analysis using a FACS-Aria.
Statistical Methods
Error bars in graphs represent standard deviations, using a two-tailed t-test. P-values for tumor assays were calculated using the two-tailed Fisher’s exact test. Tumors were considered present when the tumor volume was reported to be greater than 100mm3. Odds ratios and p-values for tumor incidence were calculated using the Fisher Exact Test as implemented in R v2.15.1 (www.r-project.org).
Results
GRHL2 inhibits the activation of ZEB1 expression by homeodomain proteins
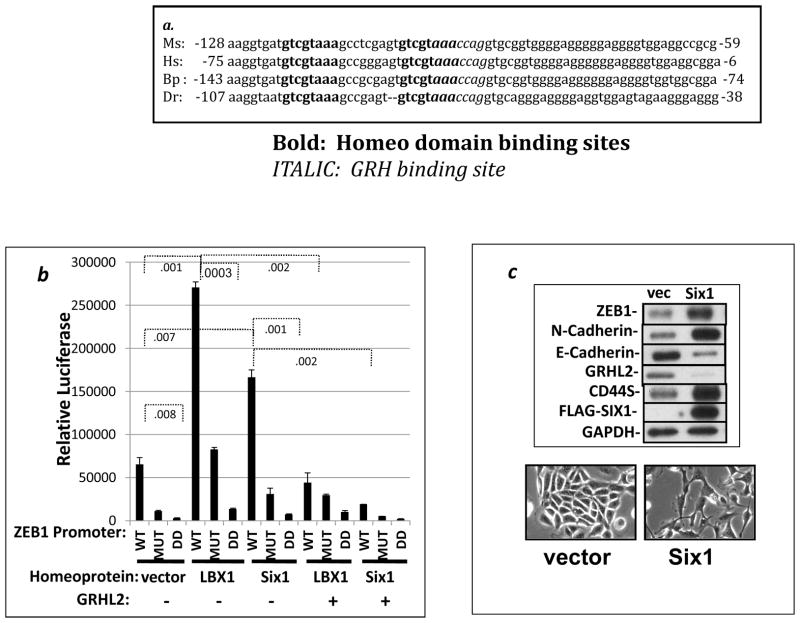
Previously, we reported that GRHL2 binds and represses the ZEB1 promoter so as to suppress EMT (28). To characterize this transcriptional regulation further, we analyzed the ZEB1 promoter using a transcription factor binding prediction program (Genomatix) and identified two tandem homeodomain consensus binding sites, overlapping with and just upstream of the putative GRHL2 site. Alignment of this sequence between several species revealed that both the putative GRHL2 binding site as well as these homeobox sites were highly conserved (Figure 1a). In this light, we tested the effect of two homeodomain factors that were previously shown to be involved in EMT induction in breast cancer cells, LBX1 and Six1, on the activity of the ZEB1 promoter (22, 33). These factors activated transcription of the ZEB1 promoter significantly; mutation of the 3′ homeodomain binding site substantially diminished transactivation by these factors. The double deletion totally eliminated it, and, interestingly, GRHL2 co-transfection inhibited the transactivation by both homeodomain factors (Figure 1b).
Figure 1. Homeodomain proteins LBX1 and Six1 activate ZEB1 expression and are inhibited by GRHL2 protein.
(a) The region of interest from the ZEB1 promoter is conserved between various species: Ms: Mus musculus, Hs: Homo sapiens, Bp: Bos primigenius, Dr: Danio rerio. Bold=homeobox consensus sites; Italics=grainyhead consensus sites. (b). LBX1 and Six1 activate transcription from the ZEB1 promoter in a GRHL2-sensitive manner. The ZEB1 promoter-luciferase reporter with wild-type sequence (WT), the 3′ homeobox site mutated (MUT) or both the mutation of the latter site plus deletion of the 5′ homeobox binding site (DD) were co-transfected into MSP cells together with the indicated transcription factors; values represent the average of biological duplicates from one representative of two experiments. (c) Six1 induces the endogenous ZEB1 gene and EMT in unfractionated HMLE cells. HMLE cells expressing vector or FLAG-Six1 were analyzed for the indicated EMT markers (upper panel); or morphologic changes (lower panel).
We then tested the effect of Six1 on endogenous ZEB1 expression. HMLE cells are a mixed population containing 5–10% mesenchymal cells in equilibrium with epithelial cells (11). HMLE cells were infected with Six1 retroviral expression constructs. Interestingly, Six1 by itself up-regulated ZEB1, down-regulated GRHL2 and induced EMT (figure 1c). Upon removal of the CD44high sub-population by flow sorting prior to infection with Six1 retrovirus, however, induction of ZEB1 by Six1 required the concomitant knockdown of GRHL2 through an shRNA (previously shown to have identical effects with other shRNAs targeting GRHL2), consistent with the antagonistic effect of GRHL2 on EMT that we observed previously (figure S1, (28)). Knockdown of the individual homeodomain proteins examined failed to block ZEB1 induction during EMT, indicating that multiple redundant factors, likely to include additional, unidentified factors, activate ZEB1 (data not shown). In this connection, HoxA5, another factor that was predicted to bind these two sites, also demonstrated robust, GRHL2-sensitive ZEB1 activation as well as the ability to induce mammosphere formation in HMLE cells, although frank EMT was not evident (figure S2, S3 and data not shown).
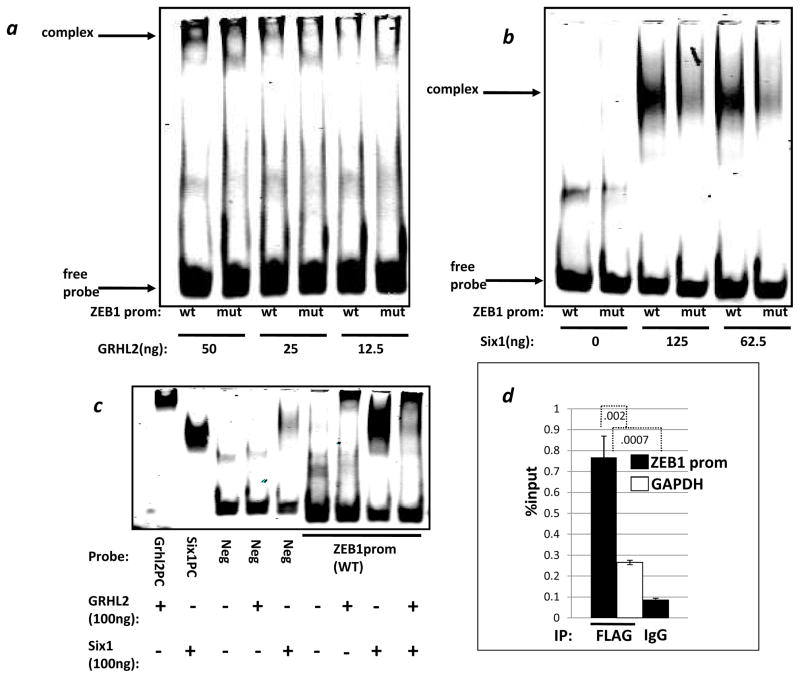
We tested Six1 for selective, direct binding to an oligonucleotide corresponding to the conserved sequence of the ZEB1 promoter. Six1 protein interacted preferentially with the wild-type but not the doubly mutated (transcriptionally inactive) sequence, as did GRHL2 protein (figure 2a, b). The addition of stoichiometric amounts of GRHL2 protein diminished the Six1-DNA complex dramatically, indicating either a competition for DNA binding or an inhibitory Six1-GRHL2 interaction, which the data do not allow us to distinguish (figure 2c). CHIP analysis revealed that Six1 was recruited to the endogenous ZEB1 promoter, indicating a direct Six1 protein-ZEB1 promoter interaction (figure 2d).
Figure 2. GRHL2 and Six1 bind specifically to sequences in the ZEB1 promoter, and GRHL2 alters this complex.
Wild-type (wt) or mutant (mut) oligonucleotides corresponding to the conserved sequence of interest from the ZEB1 promoter were used to analyze and demonstrate the specificity of GRHL2 protein (a) or Six1 (b) protein interactions. (c) GRHL2 positive control oligo (GRHL2PC), Six1 positive control oligo (Six1PC), oligo from the E-cadherin promoter which neither protein is predicted to bind (NC), and WT ZEB1 promoter oligo were used in an EMSA with the indicated combinations of GRHL2 and Six1 proteins. (d). Six1 protein interacts with the ZEB1 promoter in vivo. Chromatin from HMLE expressing either vector or FLAG-Six1 were used in a CHIP assay and analyzed by qRT-PCR using ZEB1 promoter primers or GAPDH control primers. Values are the average of biological duplicates from one representative of two experiments.
The combined data support a model under which GRHL2 is a general inhibitor of homeodomain protein-induced ZEB1 expression and EMT.
GRHL2 suppresses primary tumor growth and sensitizes tumor cells to chemotherapy-induced cytotoxicity
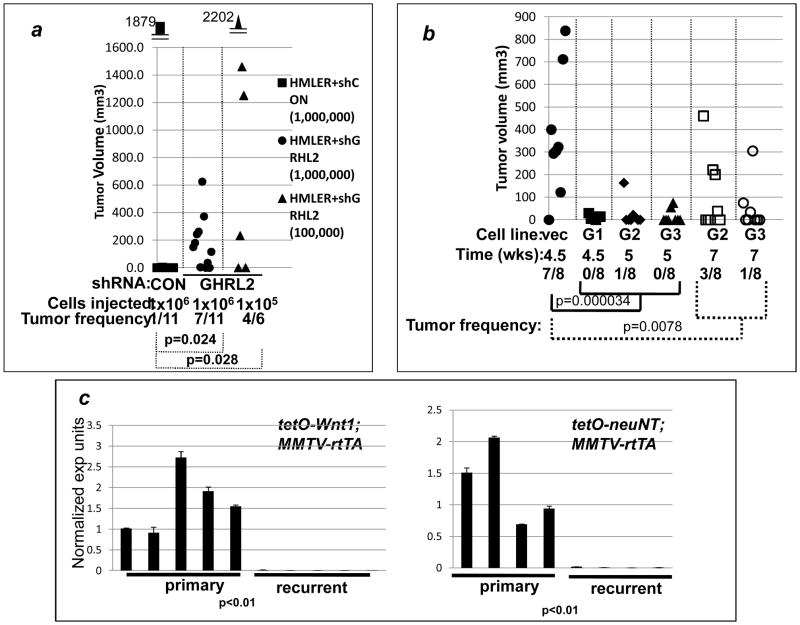
Previously, we reported that GRHL2 suppressed or reversed the oncogenic EMT, in part, through the repression of the ZEB1 gene (28). EMT can increase the frequency of breast cancer stem cells (CSC) in certain cell lines such as HMLE (12), and consistent with this, GRHL2 suppressed mammosphere generation (28). Here, we tested the effects of GRHL2 on tumor initiation frequency, a measure of CSC frequency. HMLER cells are immortalized mammary epithelial cells that express H-rasV12 ectopically and have not undergone EMT, but are induced to do so by stable GRHL2 knockdown using multiple, independently targeting shRNAs (28, 34). As reported previously, HMLER cells possessed a low tumor-initation frequency, but this frequency was increased significantly by the knockdown of the GRHL2 gene (figure 3a). Correspondingly, the knockdown of GRHL2 increased the frequency of CD44highCD24low cells (figure S4), which have been correlated with cancer stem cell phenotype in this cell line previously (12, 28). To extend this to another cell line, and use the converse approach, the triple-negative breast cancer line, MDA-MB-231LN was stably transduced with a GRHL2 expressing retrovirus or with or empty vector (28) and injected. GRHL2 suppressed the frequency of tumors significantly (figure 3b). These data were consistent with the ability of GRHL2 to suppress the tumor initiation-promoting effect of EMT (11, 12, 35). In conjunction with the functional assays and clinical correlations – downregulation of GRHL2 expression in EMT-like tumors -- that we published previously (28), these results identify GRHL2 as a tumor suppressor gene.
Figure 3. GRHL2 suppresses tumor initiation capacity and sensitizes breast cancer cells to chemotherapy-induced cytotoxicity.
(a) GRHL2 suppresses tumor initiation (knockdown approach). Immunodeficient mice were injected orthotopically with HMLER cells expressing control shRNA vs. GRHL2 shRNA (one of two GRHL2 shRNAs shown previously to suppress EMT in this cell line (22)); individual tumor volumes at ten weeks are shown. (b) GRHL2 suppresses tumor initiation (over-expression approach). MDA-MB-231LN cells expressing GRHL2 (three individual clones, labeled G1, G2, G3) or MDA-MB-231LN cells with empty vector (vec) were assayed at the indicated time points. (Mice bearing vector control tumors were sacrificed between 4.5–7 weeks due to signs of tumor burden-related illness.) Odds ratios for tumor initiation were 0.0108 and 0.0558 for the early and late time point data, respectively. (c). GRHL2 is down-regulated during the transition from primary to recurrent phenotype in tetO-Wnt1 and tetO-neuNT mouse models: qRT-PCR on RNAs from independent tumors, normalized against β2-microglobulin, conducted in technical duplicate.
In doxycycline-inducible mouse mammary tumor models, the expression of neuNT, Wnt1 or c-myc promotes primary tumors that regress upon the removal of doxycycline. The tumors recur frequently, and the recurrent tumor cells have undergone EMT, providing a model for evolution from oncogene/proliferation rate-driven tumors to oncogene-independent, EMT-driven recurrences (29, 30, 36). We analyzed RNA from the tetO-neuNT and tetO-Wnt1 mouse models for GRHL2 expression by qRT-PCR. In both cases, GRHL2 expression was greatly reduced in the recurrent tumor relative to primary (figure 3c). These findings indicate that GRHL2 is down-regulated during tumor evolution to an EMT phenotype, further supporting the tumor suppressor function of GRHL2, in a mouse model for disease recurrence.
EMT is associated with chemoresistance (37, 38). We reasoned that the mesenchymal to epithelial transition induced by GRHL2 would be associated with enhanced sensitivity to the microtubule-targeting chemotherapy drug, paclitaxel. Ectopic GRHL2 expression induced dose-dependent cell death in the otherwise refractory MDA-MB231LN tumor cell line (Figure S5). HMLE cells contain a subpopulation of CD44highCD24low cells that is enriched by paclitaxel treatment; this subpopulation has enhanced tumor initiation frequency (11, 38). We confirmed previous reports that, following paclitaxel treatment and recovery, this subpopulation predominated (figure S5). The paclitaxel-selected cells also expressed lower levels of GRHL2 than the original population, consistent with their previously reported EMT phenotype. Ectopic GRHL2 expression prevented the chemotherapy-induced emergence of CD44highCD24low cells, indicating that GRHL2-mediated suppression of this phenotype was resistant to the effects of challenge with a chemotherapy drug (figure S5 and (28)). The current data did not, however, allow us to exclude the possibility that paclitaxel generated a second, as yet uncharacterized, cancer stem cell subpopulation from HMLE that was not suppressed by GRHL2.
Based on the extensive data supporting an oncogenic role for EMT, the suppression of EMT by GRHL2, the effects of GRHL2 on tumorigenicity and chemo-sensitivity, and the down-regulation of GRHL2 during EMT in cell culture, patient and mouse models, we set out to identify pathways that down-regulate GRHL2 expression.
GRHL2 is down-regulated by Wnt and TGF-β
TGF-β is able to induce EMT in only restricted cell contexts (39). In HMLE cells, TGF-β induced EMT only in conjunction with Wnt pathway agonists or in cells where GRHL2 had been knocked down, providing insight into this restriction (11, 28).
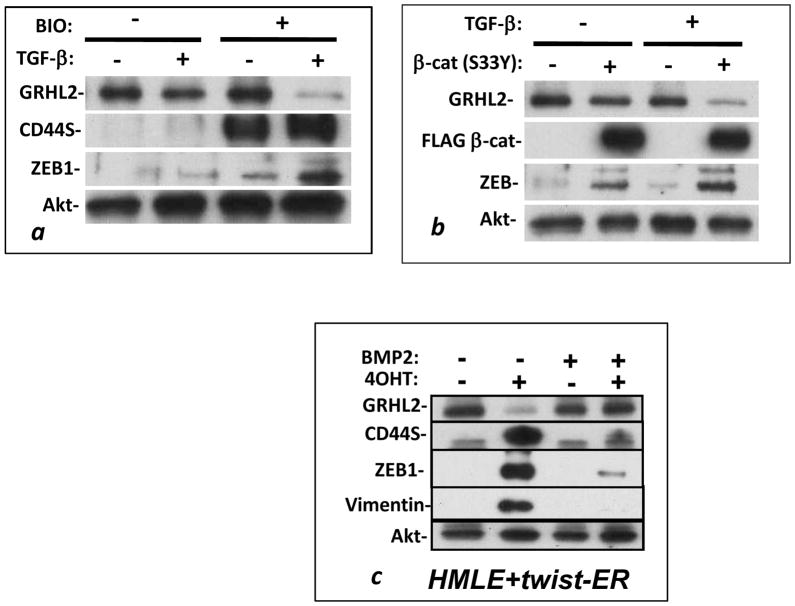
Activation of the Wnt signaling pathway by transient treatment with the GSK-3 inhibitor, 6-bromo-3-[(3E)-1,3-dihydro-3-(hydroxyimino)-2H-indol-2-ylidene]-1,3-dihydro-(3Z)-2H-indol-2-one (BIO;(31)) or, by stable expression of the degradation-resistant β-catenin S33Y mutant (40), down-regulated GRHL2 expression only modestly, as did extended treatment with TGF-β. Co-activation of both pathways, however, caused a dramatic down-regulation of GRHL2 expression (figure 4a, b), accompanied by up-regulation of the GRHL2-repressed target gene, ZEB1. BMP2, a functional antagonist of TGF-β signaling and Twist-induced EMT (11, 41), alleviated both GRHL2 down-regulation and ZEB1 induction in response to the activation of Twist-ER protein by 4-hydroxytamoxifen (figure 4c).
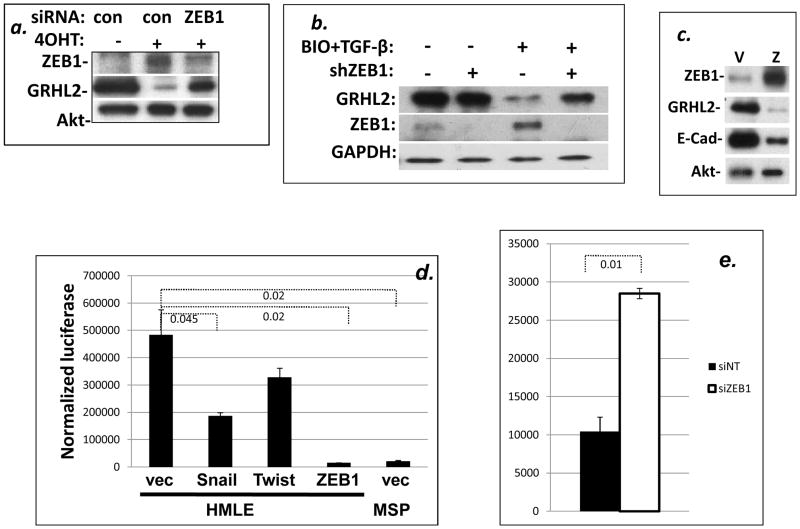
Figure 4. Wnt and TGF-β cooperate to down-regulate GRHL2.
HMLE cells treated with the canonical Wnt pathway agonist BIO (a) or stably expressing β-catenin S33Y (b) were assayed for GRHL2 down-regulation in response to TGF-β treatment by Western blotting; each is one representative of two duplicate experiments. (c) HMLE cells that stably expressed Twist-ER protein were treated with or without 4-OHT (to activate Twist-ER) in the presence or absence of the TGF-β pathway antagonist BMP2, and cell lysates were assayed for GRHL2 and other indicated protein expression.
These observations signified that Wnt and TGF-β pathways collaborated to down-regulate GRHL2 as an intermediate step in EMT, correlating with increased ZEB1 expression, which prompted us to investigate the potential role of ZEB1 in repressing GRHL2.
ZEB1 represses the GRHL2 promoter directly
To test the role of ZEB1 in the down-regulation of GRHL2, HMLE cells with an estrogen-inducible Twist fusion protein (HMLE+Twist-ER) were induced to undergo EMT by the addition of 4-hydroxytamoxifen. As we reported previously, this up-regulated ZEB1 and down-regulated GRHL2 (28). Suppression of ZEB1 with siRNA, however, largely prevented the GRHL2 down-regulation (figure 5a). To test this in a different context using a different knockdown reagent, HMLE cells with doxycycline-inducible ZEB1 shRNA expression or control cells were induced with the combination of BIO and TGF-β. Depletion of ZEB1 by GRHL2 shRNA alleviated the down-regulation of GRHL2, indicating that the Wnt+TGF-β mediated downregulation of GRHL2 is dependent on ZEB1 expression (figure 5b). Conversely, we expressed ZEB1 ectopically in HMLE cells using a doxycycline-inducible lentiviral vector. Induction of ZEB1 down-regulated GRHL2 expression as well as the previously characterized ZEB1 target, E-cadherin ((15) and figure 5c). These observations indicated that ZEB1 repressed GRHL2 expression.
Figure 5. ZEB1 down-regulates the GRHL2 promoter.
(a, b): si/shRNA mediated knockdown of ZEB1 partially alleviated GRHL2 down-regulation caused by 4-OHT-activated Twist-ER protein (a) or BIO plus TGF-β treatment (b). (c) HMLE stably expressing a doxycycline-inducible ZEB1 expression construct were induced with doxycycline and lysates were analyzed for ZEB1, GRHL2, E-cadherin and Akt by Western blotting. (d) The GRHL2 promoter in a luciferase reporter vector was co-transfected with Snail, Twist or ZEB1 expression vectors in HMLE cells or the promoter was transfected by itself into MSP cells; normalized luciferase activity values are shown. (e) HMLE-MSP transfected with control or ZEB1 siRNA, were transfected with the GRHL2 promoter-luciferase construct; normalized luciferase activity values are shown. Values are the average of biological duplicates from one representative of two experiments.
We then explored the possibility that ZEB1 protein might repress the GRHL2 gene through direct interaction with promoter sequences. As an initial test, we subcloned a genomic fragment containing the sequences from −625 to +335 (relative to the transcription start site of the standard GRHL2 transcript) into a luciferase reporter vector and assayed this for transcriptional activity. The transcriptional activity of the GRHL2 promoter clone was substantially greater in HMLE cells than in the mesenchymal subpopulation (MSP) cells derived from HMLE, consistent with the low expression of endogenous GRHL2 in the latter, validating this promoter clone ((28) and figure 5d). Co-transfected ZEB1 repressed the promoter, nearly to background level. By contrast, two other E-box binding EMT-related transcription factors, Snail and Twist, only modestly repressed the promoter. Conversely, the promoter activity assayed in MSP cells was stimulated by transfection of ZEB1 siRNA (figure 5e).
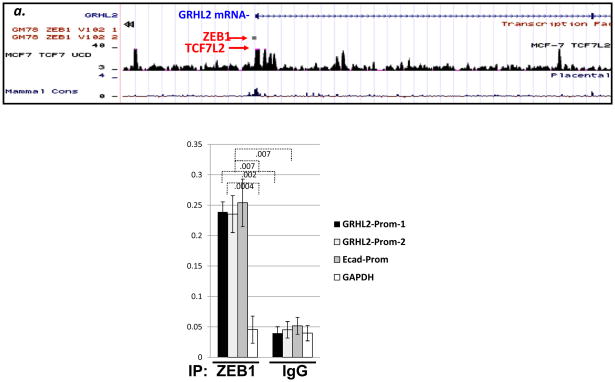
Inspection of published ZEB1 CHIP-seq data indicated binding sites positioned near the transcription start site of GRHL2 (Figure 6a). By CHIP analysis using anti-ZEB1 antibody, we detected an interaction of ZEB1 protein with GRHL2 sequences near the transcription start site, with efficiency comparable to that of the E-cadherin promoter and significantly stronger than a negative control sequence (the intergenic region upstream of the GAPDH gene; figure 6b). When combined, these data indicated that ZEB1 protein represses the GRHL2 promoter directly, establishing that a mutual antagonism between expression of ZEB1 and GRHL2 directs tumor cells toward epithelial or mesenchymal phenotypes.
Figure 6. ZEB1 protein interacts directly with the GRHL2 promoter.
(a): CHIP–seq data (Encode project;(55)) for ZEB1 reveal a potential binding site near the transcription start site. (b) Confirmation of CHIP-seq. Chromatin from HMLE expressing tet-ON ZEB1 were subjected to CHIP using ZEB1 antibody and analyzed by qRTPCR using the indicated primers. The average of biological duplicates from one representative of two experiments is shown.
Discussion
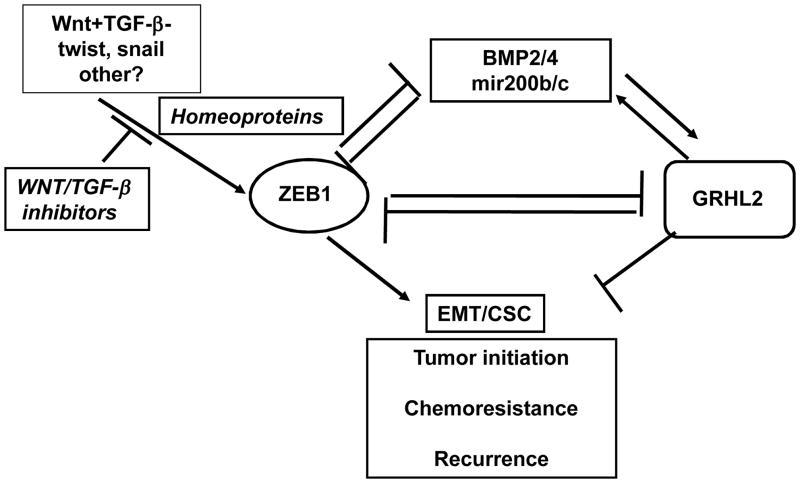
Our observations inform a model of reciprocal antagonism between ZEB1 and GRHL2 as a pivotal mechanism underlying EMT (figure 7). Under this model, GRHL2 and ZEB1 – each regulated by micro-environmental factors – repress each other’s transcription. GRHL2 represses ZEB1 by inhibiting at least three activators of the ZEB1 promoter: LBX1, Six1 and HoxA5.
Figure 7. A reciprocal feedback loop between Grainyhead-like-2 and ZEB1 controls EMT and tumor suppression.
Details are explained in the text.
We focused on these three transcription factors in light of the functional importance of homeodomain consensus sites for activity of the ZEB1 promoter (figure 1). Although numerous homeodomain proteins may have partially redundant functions, confounding efforts to show an effect of knockdown/knockout of an individual factor, their overall significance in diverse cancer types is established (reviewed in (42)). In particular, previous reports have implicated LBX1 and Six1 in breast cancer-related EMT (22, 33, 43). While Six1 was shown previously to induce ZEB1 expression, this was proposed to be post-transcriptional in colorectal cancer cell lines (44), in contrast with our evidence indicating a direct interaction of Six1 with the ZEB1 promoter.
Importantly, GRHL2 protein, the first direct transcriptional repressor of the ZEB1 gene to be reported (28), interfered with the transcriptional activation of the ZEB1 promoter by all three of these factors. While GRHL2 protein effectively abolished the Six1 protein-DNA complex on the ZEB1 promoter oligonucleotide that contained two tandem Six1 binding sites, there was no effect on Six1 protein binding to a positive control sequence containing only one binding site (data not shown), suggesting that GRHL2 selectively affects two Six1 protein molecules on DNA vs. a single Six1 molecule. We were unable to detect an interaction of GRHL2 with Six1 protein in co-transfection assays (data not shown), suggesting that they interact only on DNA.
The antagonistic effects of GRHL2 vs. ZEB1, mediated, in part, by homeodomain activators of ZEB1, may relate conceptually to previously reported developmental roles of these genes. GRHL2 is required for neural tube closure, the stage of development directly preceding delamination. Subsequently, anterior HOX genes (HoxA1, HoxB1) induce EMT during delamination of the neural crest, through activation of Snail and Msx1/2 transcription (45). Speculatively, the homeodomain factors may also activate EMT factors such as ZEB1, thereby repressing GRHL2 expression, which would indicate, analogously, functional opposition of these factors during development that mirrors their respective functions in regulating the oncogenic EMT.
We have now shown the other half of the reciprocal feedback loop (figure 7) as well: ZEB1 is also a direct repressor of GRHL2 (figures 4,5). When combined, this represents a switch that dictates cellular phenotype between epithelial and mesenchymal states, formally analogous to the relationship between ZEB1 and mir200 (46). Thus, one effect of inducing ZEB1 expression by micro-environmental factors is to repress GRHL2 expression, stabilizing ZEB1 expression. In this connection, we found that neither TGF-β nor canonical Wnt pathway stimulation was sufficient to induce ZEB1/down-regulate GRHL2; however, the two in conjunction tipped the balance in favor of ZEB1. This finding is precisely consistent with those showing that multiple micro-environmental signals are needed to regulate EMT or MET (10, 47) and may prove to be the underlying mechanism. Other combinations of factors induce ZEB1 and EMT, at least in specific cell contexts, including TGF-β+ FGF2, TGF-β+ TNF-alpha, TGF-β+EGF(R), TGF-β+mechanotransduction from extracellular matrix, or factors in the “senescent cell secretory phenotype” (SASP) (19, 48–51). It is also notable that Six1 expression activates autocrine TGF-β and Wnt signaling loops, suggesting that it activates ZEB1 both directly and indirectly (33).
Our results indicate that these combinations of factors may down-regulate GRHL2 as an intermediate step leading to EMT. Moreover, stable GRHL2 expression suppressed EMT, decreased tumor frequency and sensitized tumor cells to the effects of chemotherapeutic drugs. Thus, factors that down-regulate GRHL2 are potential drug targets for novel therapies acting to enhance chemotherapeuic responses and prevent disease recurrence. In this connection, it is interesting to note that the related gene, GRHL3, is a tumor suppressor in squamous cell carcinoma that up-regulates PTEN (52, 53). During development, GRHL2 and GRHL3 regulate some unique and other common genes (54), suggesting that the biological functions of GRHL2 should be investigated in the context of other Grainyhead family members for optimal translational benefit.
Cellular commitment to undergo an oncogenic EMT is formally analogous to cell cycle control in that both involve a transition from dependence on extracellular signals to an autonomous, stable state. For example, the cell cycle becomes autonomous past the mitogen-driven restriction point, activation of cyclinD-CDK4/6 complexes, and maintains that state by a number of mechanisms, including stable activation of cyclin E expression by free E2F. Similarly, oncogenic EMT is initiated by micro-environmental factors, for example, TGF-β and Wnt agonists. We propose that the transition to an autonomous state is defined, in part, by down-regulation of GRHL2, stabilizing autocrine signaling through ZEB1, TGF-β, Wnt, and other factors. Thus, GRHL2 down-regulation is an “oncogenic EMT restriction point.”
Supplementary Material
Acknowledgments
We wish to thank Sarah McLaughlin for technical assistance with mouse tumor assays, Kathy Brundage for flow cytometry, Philip Riley IV for assistance with molecular biology, James Coad and Ryan Livengood for analysis of tumor pathology, Daniel Haber, Stephen Jane, Paul Monga and Peter Stoilov for constructs and advice, and Lewis Chodosh for tumor RNA samples.
Grant Support
S.M. Frisch was supported by NIH grant R01CA123359. The flow cytometry core facility (Mary Babb Randolph Cancer Center) was supported by NIH grants RR020866 and P20 RR16440. J. Denvir was supported in part by NIH grants 2P20RR016477 and 8P20GM103434.
Footnotes
Conflict of interest statement: The authors have no financial interests to declare, in any category of employment, sponsored research or other financial arrangements listed in the AACR journals conflict of interest form.
References
- 1.Tiwari N, Gheldof A, Tatari M, Christofori G. EMT as the ultimate survival mechanism of cancer cells. Semin Cancer Biol. 2012 doi: 10.1016/j.semcancer.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 3.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Guadamillas MC, Cerezo A, Del Pozo MA. Overcoming anoikis--pathways to anchorage-independent growth in cancer. J Cell Sci. 124:3189–97. doi: 10.1242/jcs.072165. [DOI] [PubMed] [Google Scholar]
- 5.Frisch SM, Schaller M, Cieply B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J Cell Sci. 2013;126:21–9. doi: 10.1242/jcs.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito Y, Iwase T, Hatake K. Eradication of breast cancer cells in patients with distant metastasis: the finishing touches? Breast Cancer. 2011 doi: 10.1007/s12282-011-0266-5. [DOI] [PubMed] [Google Scholar]
- 7.Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A. 2007;104:618–23. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Usary JE, Zhao W, Darr D, et al. Predicting drug responsiveness in human cancers using genetically engineered mice. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-13-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis MJ, Perou CM. The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov. 2013;3:27–34. doi: 10.1158/2159-8290.CD-12-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheel C, Weinberg RA. Phenotypic plasticity and epithelial-mesenchymal transitions in cancer and normal stem cells? Int J Cancer. 2011;129:2310–4. doi: 10.1002/ijc.26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheel C, Eaton EN, Li SH, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell States in the breast. Cell. 2011;145:926–40. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brabletz T. EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell. 2013;22:699–701. doi: 10.1016/j.ccr.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Liu S, Clouthier SG, Wicha MS. Role of microRNAs in the regulation of breast cancer stem cells. J Mammary Gland Biol Neoplasia. 2012;17:15–21. doi: 10.1007/s10911-012-9242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grooteclaes ML, Frisch SM. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–8. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- 16.Comijn J, Berx G, Vermassen P, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–78. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 17.Browne G, Sayan AE, Tulchinsky E. ZEB proteins link cell motility with cell cycle control and cell survival in cancer. Cell Cycle. 2010;9:886–91. doi: 10.4161/cc.9.5.10839. [DOI] [PubMed] [Google Scholar]
- 18.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 2011;11:670–7. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–24. doi: 10.1038/sj.onc.1209808. [DOI] [PubMed] [Google Scholar]
- 20.Dave N, Guaita-Esteruelas S, Gutarra S, et al. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J Biol Chem. 2011;286:12024–32. doi: 10.1074/jbc.M110.168625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Tillo E, de Barrios O, Siles L, Cuatrecasas M, Castells A, Postigo A. beta-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci U S A. 2012;108:19204–9. doi: 10.1073/pnas.1108977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu M, Smolen GA, Zhang J, et al. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes Dev. 2009;23:1737–42. doi: 10.1101/gad.1809309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Samakovlis C. Grainy head and its target genes in epithelial morphogenesis and wound healing. Curr Top Dev Biol. 2012;98:35–63. doi: 10.1016/B978-0-12-386499-4.00002-1. [DOI] [PubMed] [Google Scholar]
- 25.Schafer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell Biol. 2008;9:628–38. doi: 10.1038/nrm2455. [DOI] [PubMed] [Google Scholar]
- 26.Pyrgaki C, Liu A, Niswander L. Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Dev Biol. 2011;353:38–49. doi: 10.1016/j.ydbio.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerosuo L, Bronner-Fraser M. What is bad in cancer is good in the embryo: importance of EMT in neural crest development. Semin Cell Dev Biol. 2012;23:320–32. doi: 10.1016/j.semcdb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cieply B, Riley Pt, Pifer PM, et al. Suppression of the Epithelial-Mesenchymal Transition by Grainyhead-like-2. Cancer Res. 2012;72:2440–53. doi: 10.1158/0008-5472.CAN-11-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moody SE, Perez D, Pan TC, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 30.Debies MT, Gestl SA, Mathers JL, et al. Tumor escape in a Wnt1-dependent mouse breast cancer model is enabled by p19Arf/p53 pathway lesions but not p16 Ink4a loss. J Clin Invest. 2008;118:51–63. doi: 10.1172/JCI33320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 32.Patrick AN, Schiemann BJ, Yang K, Zhao R, Ford HL. Biochemical and functional characterization of six SIX1 Branchio-oto-renal syndrome mutations. J Biol Chem. 2009;284:20781–90. doi: 10.1074/jbc.M109.016832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Micalizzi DS, Christensen KL, Jedlicka P, et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-beta signaling. J Clin Invest. 2009;119:2678–90. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elenbaas B, Spirio L, Koerner F, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–95. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 36.Leung JY, Andrechek ER, Cardiff RD, Nevins JR. Heterogeneity in MYC-induced mammary tumors contributes to escape from oncogene dependence. Oncogene. 2012 doi: 10.1038/onc.2011.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Creighton CJ, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–5. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–59. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown KA, Aakre ME, Gorska AE, et al. Induction by transforming growth factor-beta1 of epithelial to mesenchymal transition is a rare event in vitro. Breast Cancer Res. 2004;6:R215–31. doi: 10.1186/bcr778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yook JI, Li XY, Ota I, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 41.Candia AF, Watabe T, Hawley SH, et al. Cellular interpretation of multiple TGF-beta signals: intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development. 1997;124:4467–80. doi: 10.1242/dev.124.22.4467. [DOI] [PubMed] [Google Scholar]
- 42.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 10:361–71. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 43.McCoy EL, Iwanaga R, Jedlicka P, et al. Six1 expands the mouse mammary epithelial stem/progenitor cell pool and induces mammary tumors that undergo epithelial-mesenchymal transition. J Clin Invest. 2009;119:2663–77. doi: 10.1172/JCI37691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ono H, Imoto I, Kozaki K, et al. SIX1 promotes epithelial-mesenchymal transition in colorectal cancer through ZEB1 activation. Oncogene. 2012 doi: 10.1038/onc.2011.646. [DOI] [PubMed] [Google Scholar]
- 45.Gouti M, Briscoe J, Gavalas A. Anterior Hox genes interact with components of the neural crest specification network to induce neural crest fates. Stem Cells. 2011;29:858–70. doi: 10.1002/stem.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gregory PA, Bracken CP, Smith E, et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell. 2011;22:1686–98. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao D, Vahdat LT, Wong S, Chang JC, Mittal V. Microenvironmental regulation of epithelial-mesenchymal transitions in cancer. Cancer Res. 2013;72:4883–9. doi: 10.1158/0008-5472.CAN-12-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shirakihara T, Horiguchi K, Miyazawa K, et al. TGF-beta regulates isoform switching of FGF receptors and epithelial-mesenchymal transition. EMBO J. 2011;30:783–95. doi: 10.1038/emboj.2010.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohashi S, Natsuizaka M, Wong GS, et al. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res. 2010;70:4174–84. doi: 10.1158/0008-5472.CAN-09-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laberge RM, Awad P, Campisi J, Desprez PY. Epithelial-mesenchymal transition induced by senescent fibroblasts. Cancer Microenviron. 2011;5:39–44. doi: 10.1007/s12307-011-0069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gjorevski N, Boghaert E, Nelson CM. Regulation of Epithelial-Mesenchymal Transition by Transmission of Mechanical Stress through Epithelial Tissues. Cancer Microenviron. 2011;5:29–38. doi: 10.1007/s12307-011-0076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhandari A, Gordon W, Dizon D, et al. The Grainyhead transcription factor Grhl3/Get1 suppresses miR-21 expression and tumorigenesis in skin: modulation of the miR-21 target MSH2 by RNA-binding protein DND1. Oncogene. 2012 doi: 10.1038/onc.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darido C, Georgy SR, Wilanowski T, et al. Targeting of the Tumor Suppressor GRHL3 by a miR-21-Dependent Proto-Oncogenic Network Results in PTEN Loss and Tumorigenesis. Cancer Cell. 2012;20:635–48. doi: 10.1016/j.ccr.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 54.Boglev Y, Wilanowski T, Caddy J, et al. The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Dev Biol. 2011;349:512–22. doi: 10.1016/j.ydbio.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 55.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.