SUMMARY
Gametes are among the most highly specialized cells produced during development. Although gametogenesis culminates in transcriptional quiescence in plants and animals, regulatory mechanisms controlling this are unknown. Here, we confirm that gamete differentiation in the single-celled yeast Saccharomyces cerevisiae is accompanied by global transcriptional shutoff following the completion of meiosis. We show that Jhd2, a highly conserved JARID1-family histone H3K4 demethylase, activates protein-coding gene transcription in opposition to this programmed transcriptional shutoff, sustaining the period of productive transcription during spore differentiation. Moreover, using genome-wide nucleosome, H3K4me, and transcript mapping experiments, we demonstrate that JHD2 globally represses intergenic noncoding transcription during this period. The widespread transcriptional defects of JHD2 mutants are associated with precocious differentiation and the production of stress-sensitive spores, demonstrating that Jhd2 regulation of the global postmeiotic transcriptional program is critical for the production of healthy meiotic progeny.
INTRODUCTION
Sporulation is the gametogenesis phase of the ascomycete life cycle, and couples meiosis of diploid cells with the subsequent differentiation of the meiotic products into gametes called spores. These spores are stored inside the ascus, the remnant of the mother cell within which sporulation occurs. Pioneering studies of Saccharomyces cerevisiae by Hopper et al. (1974), later confirmed by Brengues et al. (2002), first described spore dormancy by demonstrating dramatic reductions in mitochondrial respiration, transcription, and translation during sporulation. In spores, like in animal gametes, transcriptional dormancy, a phenomenon often referred to as “quiescence,” is counter-balanced by a dramatic increase in mRNA stability (Brengues et al., 2002; Hamatani et al., 2004; Schultz, 2002; Tadros and Lipshitz, 2009). Although the mechanisms underlying the transition to transcriptional quiescence in gametes remain unknown, given the global nature of the phenomenon, mechanisms that act throughout the genome irrespective of DNA sequence seem likely to be involved.
Much of gene regulation involves histone modifications. Methylation of histone H3 on lysine-4 (H3K4) likely represents the most comprehensively studied histone modification. Like all other known histone lysine methylations, H3K4 exhibits mono, di, and tri-methylated forms (H3K4me1, H3K4me2, H3K4me3). These modification states can have distinctive regulatory outputs reflecting the recruitment of histone-methyl-specific binding proteins (Kim and Buratowski, 2009; Santos-Rosa et al., 2003; Taverna et al., 2006). Studies in effectively all eukaryotes have demonstrated a strong correlation between transcriptional activity and H3K4me deposition downstream of transcription start sites (TSSs), supporting a general view of H3K4me as a transcriptionally activating epigenetic mark. Mechanistic insights into how H3K4me globally impacts transcription during development and differentiation, however, are surprisingly limited.
Recent studies have revealed that eukaryotic genomes are subject to widespread transcriptional activity beyond the boundaries of protein coding genes. Indeed, TSSs of protein coding genes are not only reliably marked by high levels of H3K4me3, but these regions have more recently been shown to be associated with abundant noncoding transcription in organisms ranging from yeast to human (van Bakel et al., 2010; Xu et al., 2009). Similarly, TTSs also exhibit significant levels of noncoding transcription (van Bakel et al., 2010; Yadon et al., 2010). Although elegant regulatory functions for select noncoding transcription events have been described in yeast, the general relevance and significance of noncoding transcription remains a topic of debate (van Bakel et al., 2010).
The regulatory mechanisms controlling noncoding transcription are even less well understood. In human embryonic stem cells, global inhibition of intergenic noncoding transcription requires JARID1B, a highly conserved H3K4 demethylase of the Jumonji super-family (Xie et al., 2011). Biologically, JARID family genes control a balance between pluripotency and differentiation of mammalian stem cells, and confer drug resistance and slow cell cycle timing properties to distinct subpopulations of tumor cells when hyperactivated (Benevolenskaya et al., 2005; Dey et al., 2008; Landeira et al., 2010; Lopez-Bigas et al., 2008; Pasini et al., 2010; Peng et al., 2009; Roesch et al., 2010; Sharma et al., 2010; Shen et al., 2009; Xie et al., 2011). The yeast genome encodes five Jumonji proteins including two orthologs of the JARID subfamily, JHD2 and ECM5 (Klose et al., 2006). Although Ecm5 seems unlikely to possess histone demethylase activity owing to a degraded catalytic domain (Klose et al., 2006), Jhd2, like all JARID1-family proteins, is a biochemically verified histone demethylase with specificity for H3K4me (Ingvarsdottir et al., 2007; Liang et al., 2007).
We confirm here that yeast attains transcriptional quiescence in postmeiotic cells undergoing spore differentiation, and find that Jhd2 functions globally to promote gene transcription during this period. Using high-resolution genomic approaches, we show that JHD2 enhances transcription of protein coding genes while repressing the transcription of their associated intergenic noncoding regions in postmeiotic cells globally. For ribosomal protein encoding and Rap1-bound genes, major classes of coregulated genes, we describe evidence that Jhd2 positively regulates their transcription by negatively regulating inhibitory noncoding transcription mechanisms. These studies provide mechanistic insight into how JARID H3K4 demethylases impact global transcription during differentiation. More fundamentally, our findings build upon yeast sporulation as a powerful model to study programmed metabolic quiescence underlying gametogenesis.
RESULTS AND DISCUSSION
JHD2 Promotes Global Demethylation of H3K4me, mRNA Accumulation, and Spore Fitness during Sporulation
Consistent with a previous report (Lenstra et al., 2011), we were unable to identify any impact of JHD2 on transcript patterns in vegetative cells (Figure S1A and Table S1 available online). Because JHD2 mRNA accumulates dramatically during sporulation (Chu et al., 1998; Lardenois et al., 2011), we investigated JHD2 function during this process. We constructed isogenic diploid WT and jhd2 mutant strains in the SK1 strain background, which exhibits highly efficient and synchronous meiosis. All of the experiments described here utilize this strain background. We first determined that jhd2Δ did not perturb meiotic progression or spore formation (Figure 1A and data not shown).
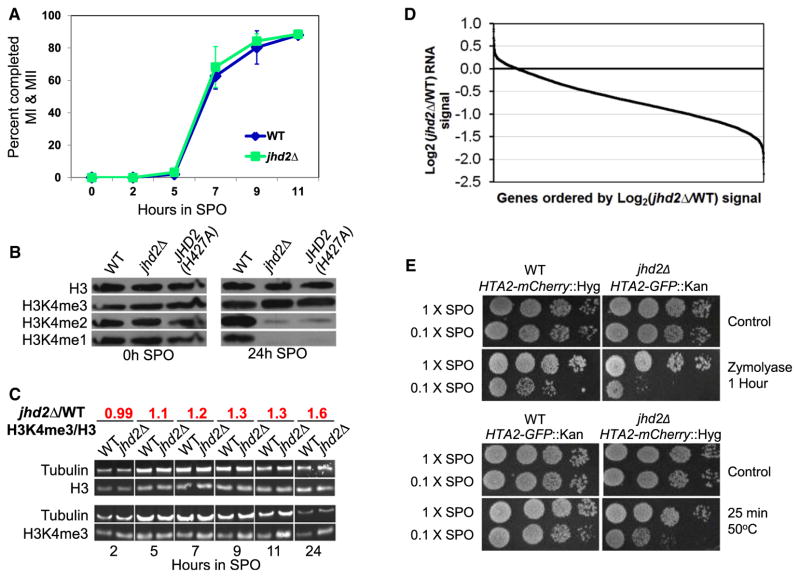
Figure 1. JHD2 Functions during Sporulation to Control Global Transcript Accumulation, H3K4 Demethylation, and Gamete Fitness.
(A) Meiotic progression of WT and jhd2Δ cultures was quantified as percent of cells that have completed meiosis I and II. Shown are averages of three independent experiments. Error bars indicate 1 SD.
(B) Western blotting was used to detect abundance of H3 and H3K4me in WT and jhd2Δ strains at the indicated time points of sporulation. Membranes were stripped and reprobed with the indicated antibodies.
(C) Quantitative western blotting was performed on aliquots from sporulating WT and jhd2Δ cultures using the indicated primary antibodies. After normalization to tubulin signal, the H3-3meK4/total-H3 ratio of jhd2Δ compared with WT was calculated and is shown. See also Figure S1.
(D) Affymetrix tiling arrays were used to measure RNA abundance from WT and jhd2Δ spores (20 hr) at a resolution of 4 bp. Normalized intensities for protein coding genes were plotted for jhd2Δ cells relative to WT and ordered by decreasing fold change for all annotated genes. For comparison, see Figure S1A and Table S1 for relative WT and jhd2Δ RNA abundance data in vegetative cells.
(E) WT and jhd2Δ mutants of the indicated genotypes were cosporulated in 1× or 0.1× SPO medium. Sporulated cultures were treated with zymolyase or 55°C heat and were assayed for survival by monitoring postgermination growth.
Using western blots with H3K4me-specific antibodies, we found that bulk H3K4me3 levels are reduced ~1.7-fold following sporulation (Figure S1B), consistent with a previous report (Zhang et al., 2011). Interestingly, not only did jhd2Δ mutant spores contain higher levels of H3K4me3 compared with WT, they also contained substantially reduced quantities of H3K4me1/2 (Figures 1B, S1C, and S1D). A time course analysis revealed that jhd2Δ cells exhibited a progressive increase in bulk H3K4me3 levels compared with WT cells as they advanced through sporulation (Figure 1C). The amount of H3K4me3 was ~1.6-fold higher in jhd2Δ spores compared to WT spores (Figures 1C and S1C). Western blot analysis of biological replicates of cells at the 10 hr time point confirmed these findings (Figures S1C and S1D), and revealed that this increase was accompanied by a decreased level of H3K4me1/2 in sporulating cells (Figure S1D). As there is very little H3K4me1/2 in terminal jhd2Δ spores, we infer that sustained H3K4 methylation in the absence of Jhd2-mediated demethylation eventually converts bulk H3K4me1/2 to H3K4me3. A strain that is homozygous for the jhd2(H427A) allele, which encodes a mutated protein with a disrupted Fe2+-binding pocket that is essential for the enzymatic activity of the Jumonji family (Ingvarsdottir et al., 2007; Liang et al., 2007), exhibited bulk H3K4me defects in spores that were indistinguishable from those in jhd2Δ spores (Figure 1B). These results strongly suggest that Jhd2 demethylation activity toward H3K4 controls the balance of H3K4me levels during sporulation.
The jhd2 mutant defects in H3K4me abundance during sporulation suggested that they might also show transcriptional defects. To test this, we used whole-genome Affymetrix tiling microarray profiling to measure the relative abundance of transcribed RNA in WT and jhd2Δ spores at high resolution. In stark contrast to jhd2Δ in vegetative cells (Figure S1A; Lenstra et al., 2011), we found that jhd2Δ mutant spores exhibited a pronounced and global reduction in the abundance of protein coding mRNAs (Figure 1D). Surprisingly, despite these changes, jhd2Δ mutant spores did not exhibit any detectable fitness defects.
We have found that robust sporulation occurs in the presence of greatly reduced environmental carbon compared with during optimized laboratory protocols (Eastwood et al., 2012). Under such low-carbon conditions, which are presumably more relevant to the natural ecological situation (Eastwood et al., 2012), each sporulating cell differentiates only a portion of its haploid meiotic products into spores, producing mostly “dyads” (2-spored asci); in contrast, sporulation in the standard laboratory sporulation medium generates mostly tetrads (Davidow et al., 1980; Eastwood et al., 2012). To determine if the gene expression defects of jhd2Δ mutants caused spore fitness defects under such conditions, we evaluated the fitness of jhd2Δ mutant spores produced during reduced carbon sporulations. In order to eliminate flask-to-flask variability in the experiment, we cosporulated WT and jhd2Δ mutant strains that expressed differing dominant drug resistance markers and fluorescently tagged histone proteins in the same flasks under high and low carbon conditions. Inspection using fluorescent microscopy confirmed that these strains sporulated with essentially identical efficiencies, with low carbon causing an ~50% reduction in the number of spores formed for each genotype (data not shown). jhd2Δ spores produced under low carbon conditions, however, exhibited increased sensitivity to stresses such as heat or to treatment with zymolyase, an enzyme that digests yeast cell walls (Figure 1E). Stress resistance tests on paired cocultures sporulated under high carbon conditions confirmed that these phenotypes were only exhibited in jhd2Δ mutant spores produced during low carbon sporulation (Figure 1E). Thus, JHD2 is critical for the production of robust gametes under ecologically relevant conditions.
JHD2 Delays the Onset of Global Transcriptional Quiescence in Postmeiotic Cells
Because we found that jhd2Δ cells completed meiosis with the same kinetics and efficiency as WT cells, but that jhd2Δ spores exhibited a global reduction in the abundance of mRNAs, we hypothesized that Jhd2 protects sporulating cells from becoming quiescent prior to terminal differentiation. The first descriptions of spore quiescence were obtained using strain backgrounds that exhibit moderately reduced sporulation efficiency and kinetics (Brengues et al., 2002; Hopper et al., 1974). To confirm that SK1 spores attain metabolic quiescence, we measured dissolved O2 consumption rates of sporulating cells and found that terminally differentiated spores (36 hr post-induction) consume oxygen at ~4% the rate of cells at time 0 of time courses, demonstrating that mitochondrial respiration is dramatically downregulated during sporulation (Figure 2A). Measurement of O2 consumption rates throughout sporulation showed that cells began showing reduced mitochondrial activity during meiosis and attained a terminal metabolic rate by the 16 hr time point (Figure 2A).
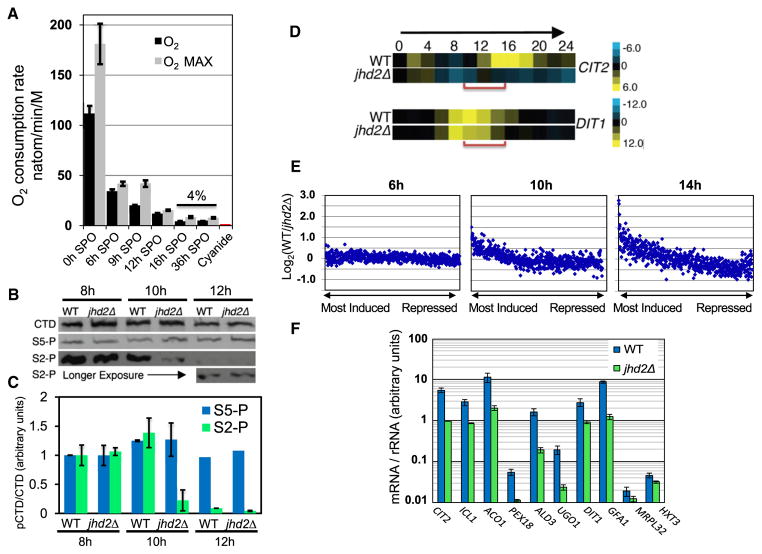
Figure 2. JHD2 Promotes Global Transcription in Opposition to Programmed Quiescence during Sporulation.
(A) Mitochondrial respiration rates of sporulating cells were assessed by measurement of rate of dissolved oxygen consumption. Error bars represent one standard deviation of technical replicates. Maximal respiration rate (O2 MAX) was measured after addition of the respiratory chain uncoupler carbonyl cyanide m-chlorophenyl hydrazone. Cyanide was added to inhibit the mitochondrial electron transport chain (cytochrome C oxidase).
(B) Quantitative western blots of bulk RNA Pol II CTD (CTD) and RNA Pol II CTD phosphorylated on serine-2 (S2-P) or serine 5 (S5-P) from WT and jhd2Δ cells collected at the 8 hr, 10 hr, and 12 hr time points of sporulation.
(C) Quantification of four technical replicates of western blot data described in (B), normalized to the 8 hr time points. Error bars represent 1 SD. See also Figure S2.
(D) Relative transcript abundance of CIT2 and DIT1 from 24 hr sporulation time course microarray profiles are shown.
(E) mRNA signals were ranked in WT cells at 6 hr, 10 hr, and 14 hr into the sporulation program. The differences in mRNA signal from jhd2Δ was calculated, averaged over bins of 10 genes, and displayed as scatter plots. See also Figures S2C, S2D, and Table S2.
(F) RT-qPCR was used to measure transcript abundance of the indicated genes at the 10 hr time points of independent sporulation time courses. Error bars represent 1 SD of technical replicates.
Before investigating further the role of Jhd2 in regulating mRNA accumulation during sporulation, we assessed transcription in sporulating cells in greater detail by measuring phosphorylations in the heptad repeats of the C-terminal domain of the largest subunit of RNA polymerase II (RNA Pol II CTD) that are diagnostic of transcriptional activity. Phosphorylations of the RNA Pol II CTD on serines-2 or -5 (CTD-pS2, CTD-pS5) are mechanistically coupled with transcriptional elongation and initiation respectively (Kim et al., 2010). During WT sporulation, we found by western blot analysis that the abundance of CTD-pS2 dropped sharply between the 8 and 12 hr time points, to ~5% of CTD-pS2 levels at time 0 (Figure S2A). Interestingly, CTD-pS5 levels remained relatively constant, suggesting that transcription may be selectively affected at the elongation stage in postmeiotic cells, similarly to “stationary phase” haploid cells that become transcriptionally quiescent upon media exhaustion (Radonjic et al., 2005). As our time course data show that meiosis was completed between 7–9 hr, these results suggest that cells sustain global transcriptional activity for a period of ~2 hr following the completion of meiosis.
We next compared CTD-pS2/5 levels in paired WT and jhd2Δ sporulating cultures at the 8, 10, and 12 hr postmeiotic time points. These experiments showed that although jhd2Δ mutants contained indistinguishable quantities of bulk CTD-pS2/5 at the 8 hr time point, jhd2Δ caused a marked defect in the abundance of bulk CTD-pS2 starting at the 10 hr time point (Figures 2B, 2C, and S2B). Analysis of the same extracts did not reveal any defects in jhd2Δ cells for the accumulation of CTD-pS5 (Figures 2B, 2C, and S2B) implying that JHD2 function is required for bulk transcription as cells advance into the postmeiotic stages of sporulation.
Much, if not all, of the transcript accumulation defects of jhd2Δ spores may be a consequence of the failure of JHD2 mutants to sustain global transcription in opposition to programmed transcriptional quiescence in postmeiotic cells. To test this, we carried out whole-genome transcriptional profiling of RNA prepared from WT and jhd2Δ cells throughout 24 hr sporulation time courses with samples taken every 2 hr (Table S2). The application of hierarchical clustering to our data showed that the global dynamics of gene expression were broadly similar between WT and jhd2Δ (Figure S2C), and corresponded well with the known induction and repression events occurring during the first 11 hr of sporulation (Chu et al., 1998). Further analysis of our clustered data, however, revealed two classes of jhd2Δ defects in transcript accumulation (Figure S2D). CIT2 and DIT1 represent gene-specific examples of these classes (Figure 2D). Interestingly, although CIT2 and DIT1 show differing kinetics of expression during WT sporulation, transcript accumulation for each gene was most obviously reduced in postmeiotic jhd2Δcells, supporting our conclusion that jhd2Δ cells are defective in postmeiotic transcription.
To globally evaluate the jhd2Δ mRNA accumulation defect, we ranked genes from the microarray time course data set by the magnitude of their transcript induction at three time points in the WT program; midmeiosis (6 hr), early spore differentiation (10 hr), and late spore differentiation (14 hr). We then expressed the difference in magnitude of induction of these genes between WT and jhd2Δ on scatter plots, with each point representing the average impact of JHD2 deletion on 10 adjacently ranked genes (Figure 2E). At 6 hr into sporulation, the data points lie along a straight line centered on 0, showing equivalent levels of global transcript accumulation between WT and jhd2Δ cells. In striking contrast, at 10 and 14 hr, there is a pronounced and progressively increased skewing of the relative transcript abundance rankings for induced genes away from the 0-line of the y axis. This analysis shows that jhd2Δ cells have a general defect in accumulation of transcripts from those genes that are induced to high levels in postmeiotic wild-type cells. We selected 20 genes for RT-qPCR analysis to confirm these microarray studies, and found that all 20 showed reduced mRNA abundance in jhd2Δ cells compared with WT cells at the 10 hr time point of sporulation (Figures 2F and S2E).
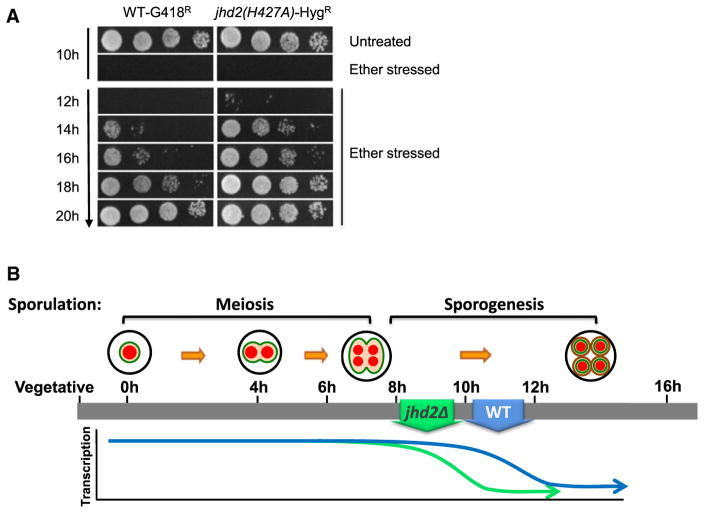
Our studies show that global transcription is turned down precociously in sporulating jhd2Δ cells. We hypothesized that these cells might also carry out spore differentiation precociously. A hallmark of the completion of yeast spore differentiation is acquisition of resistance to a variety of environmental stresses. One such stress resistance, exposure to diethyl ether, provided an acutely acting treatment with which to monitor spore differentiation (Dawes and Hardie, 1974). We cosporulated WT and jhd2 mutant strains marked with differing dominant drug resistance markers in the same flasks, and assayed the cells for relative resistance to ether exposure at time points throughout spore differentiation by titering the cosporulated cultures onto drug plates after ether stress. jhd2 mutants attained precocious resistance to ether treatment compared with wild-type (Figure 3A, jhd2(H427A) is shown). Equivalent experiments were performed using differing permutations of genotype/drug-marker (e.g., WT-HygR, jhd2Δ-G418R, etc.) with identical results (data not shown).
Figure 3. JHD2 Specifies the Duration of the Spore Differentiation Program via Promoting Its Extension.
(A) WT-G418R and jhd2(H427A)-HygR cultures were cosporulated in the same flask. Aliquots were removed at the indicated times and treated with diethyl ether and relative survival of the two strains was determined by plating onto G418 or hygromycin containing plates.
(B) WT spores attain developmentally programmed transcriptional quiescence within a few hours following the completion of meiosis. JHD2 delays both the onset of global transcriptional quiescence, and the terminal differentiation of spores.
See also Figure S3.
Deletion of the histone methyl transferase SET1 causes a sporulation defect that is essentially the opposite of jhd2Δ mutants: set1Δ causes a delay in meiotic progression (Sollier et al., 2004). We found that set1Δ is epistatic to jhd2Δ with respect to meiotic progression (data not shown). Moreover, mutation of H3K4 to the unmethylatable residue alanine also exhibited delayed meiosis that is epistatic to jhd2Δ (Figure S3). Although these phenotypes are consistent with Jhd2 acting through H3K4me, because these mutations cause delays in meiosis, reliable analyses of SET1 or H3K4A epistasis to JHD2 during the time when JHD2 acts (following meiosis) are not technically feasible.
Our data support the model that Jhd2 demethylation of H3K4 functions to specify the duration of the program of spore differentiation by sustaining the postmeiotic gene expression program globally (Figure 3B). Although JHD2 function did not impact spore fitness under optimized laboratory sporulation conditions, jhd2Δ caused defects in spore fitness when sporulation is performed under more ecologically relevant conditions. Under these more challenging conditions, we hypothesize that the normal, JHD2-specified duration of the spore differentiation program is critical for production of robust gametes. Intriguingly, JHD2 function during sporulation is reminiscent of the function of mammalian JARID family proteins, which also control differentiation kinetics via their impact on transcription. Although differentiation in most mammalian cells, unlike during yeast sporulation, does not typically involve global transcriptional repression, both events nevertheless involve substantial transcriptional remodeling. The mechanisms of JHD2 function during spore differentiation— described further below—may therefore be relevant to how JARIDs control differentiation and oncogenesis in humans.
JHD2 Globally Represses Noncoding Transcription during Sporulation
Because we first detected global transcriptional defects in jhd2Δ mutants at the 10 hr time point of sporulation, it seemed likely that the mechanisms by which Jhd2-demethylation impinges upon transcription during sporulation would be best assessed then. We therefore measured H3K4me3 and H3K4me2 levels across genes at the 10 hr time point of sporulation using ChIP-chip and a high-density tiling array that covers yeast chromosome III (Figures 4A and 4B; Table S3) (Yuan et al., 2005). As expected, we detected a prominent enrichment of H3K4me3 abundance immediately downstream of TSSs in WT and jhd2Δ postmeiotic cells (Figure 4A). In agreement with our western blot data, jhd2Δ mutants showed significantly increased amounts of H3K4me3 spanning the regions from −500 to +100 base pair (bp) with respect to TSSs (Figure 4A). Strikingly, jhd2Δ mutants also accumulated significantly increased amounts of H3K4me3 between ~300 bp upstream and downstream of TTSs (right panel of Figure 4A). H3K4me2 showed essentially identical distributions as H3K4me3 except for within the bodies of ORFs, where jhd2Δ exhibited a decrease in the abundance of H3K4me2, consistent with a globally reduced rate of transcription coupled H3K4 di-methylation within transcription units (Figure 4B). To rule out the possibility that the accumulation of TTS-proximal H3K4me in jhd2Δ was due to overlap with TSSs of downstream tandemly transcribed genes as opposed to bona fide TTS-associated signal, we confirmed that H3K4me2/3 also accumulate at TTSs of convergently transcribed genes in jhd2Δ cells (Figures S4A and S4B).
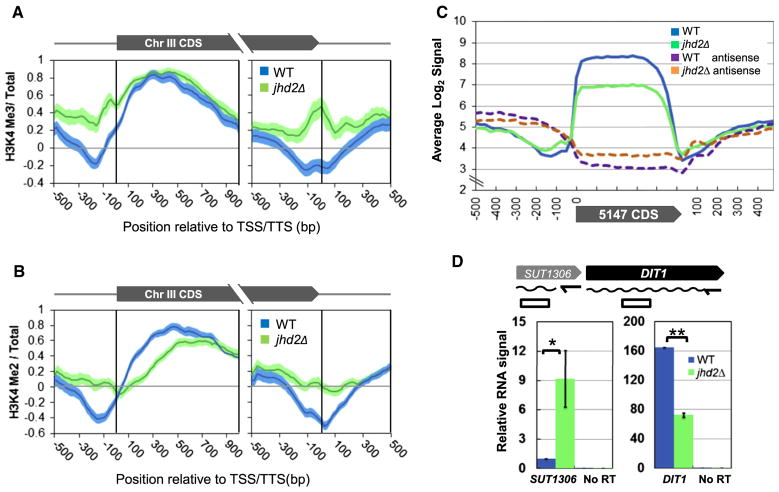
Figure 4. JHD2 Globally Represses Noncoding Transcription near the TSS and TTS Regions during Sporulation.
(A and B) Chromatin immunoprecipitation followed by high-resolution microarray hybridization (ChIP-chip) was performed using chromatin prepared from WT and jhd2Δ cells at the 10 hr sporulation time point. Shown are average gene profiles of H3K4me3 (A) and H3K4me2 (B) ChIP signal of 119 genes on Ch III in WT and jhd2Δ cells. Average profiles were centered at the TSSs (left panels) or TTSs (right panels). Error-“clouds” represent 1 SE. See also Table S3.
(C) Affymetrix RNA expression data from Figure 1E were reassembled. Normalized signals from 5,147 protein coding genes were scaled into 20 bins for each coding region and averaged. Shown are the binned average transcript and antisense signals and an additional 500 bp of flanking signals on either side.
(D) RT q-PCR was performed with transcript-specific primers to measure levels of SUT1306 and DIT1 RNA in WT and jhd2Δ cells at the 10 hr sporulation time point. RNA signals were normalized to input RNA quantity and shown as relative abundance compared to WT SUT1306 transcript levels.
Error bars represent 1 SD. *p < 0.05. **p < 0.005. See also Figure S4.
Because H3K4me deposition is dependent on transcription (Hampsey and Reinberg, 2003), the hyper accumulation of H3K4me 5′ of TSSs and surrounding TTSs in sporulating jhd2Δ cells suggested that JHD2 might repress noncoding transcription within these regions. Our microarray data presented in Figure 1D measured RNA produced from both DNA strands across the entire genome at high resolution. Although this data measured RNA from terminally sporulated cells at the 20 hr time point, as mRNA stability is dramatically upregulated during sporulation (Brengues et al., 2002), it nevertheless afforded us a means to evaluate this hypothesis. When we reassembled this data to construct a globally averaged protein coding gene transcript profile, we found, as expected, that the abundance of transcripts from this averaged transcript unit was reduced in jhd2Δ mutants, (Figure 4C). However, when we inspected the regions upstream and downstream of the averaged TSS and TTS, we found that jhd2Δ cells exhibited an increased accumulation of sense strand and antisense strand RNA, respectively. Indeed, we detected increased antisense RNA in jhd2Δ cells throughout the entire body of the averaged protein coding gene (Figure 4C).
Our results demonstrate that JHD2 functions globally to promote protein coding gene transcription and to repress the transcription of associated noncoding regions on both the sense and antisense strands in postmeiotic cells. Although such anti-correlated transcription events may be coupled in a nonregulatory manner when the TSSs of the coding and noncoding transcripts are in close proximity (Xu et al., 2009), noncoding transcription can negatively impact the transcription of associated protein coding genes (Shearwin et al., 2005). Such inhibition of protein coding gene transcription is often caused by an inhibitory noncoding transcription unit situated 5′ to the target gene and expressed on the same DNA strand, as has been described previously at the SER3, ADH1, ADH3, and FLO11 loci in yeast (Bird et al., 2006; Bumgarner et al., 2009; Martens et al., 2004). We noticed that the well-studied sporulation-specific gene DIT1 is flanked on its 5′ end by an intergenic “stable untranslated transcript” (SUT) called SUT1036 that is transcribed in the sense orientation with respect to DIT1 (Lardenois et al., 2011). We used RT-qPCR to measure the abundance of SUT1306 RNA at the 10 hr time point of sporulation. In jhd2Δ, we found that SUT1306 transcript levels were significantly increased, whereas DIT1 transcript levels were significantly decreased, consistent with a JHD2-repressed mechanism negatively impacting DIT1 transcription via the modulation of SUT1306 (Figures 4D and S4C).
JHD2 Represses a Program of Globally Increased Nucleosome Accumulation at TSSs during Sporulation
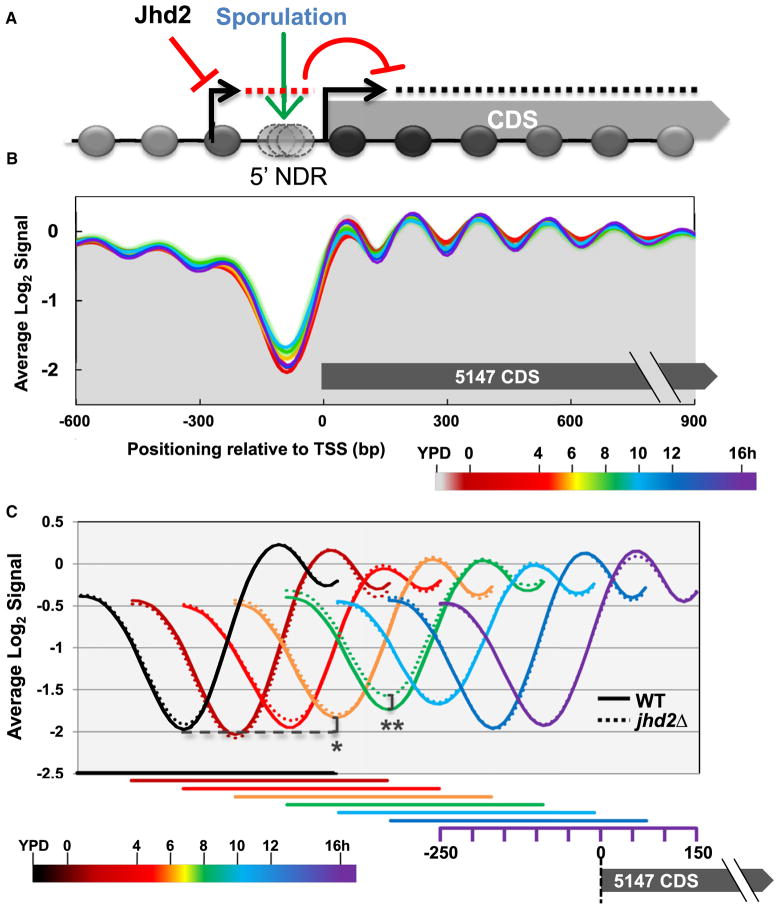
TSSs emanate from nucleosome depleted regions (NDRs) that are flanked on their 5′ and 3′ sides by well-positioned nucleosomes. Increased nucleosomal occupancies spanning these TSS regions inhibit transcription, presumably by interfering with the recruitment of activating factors (Badis et al., 2008; Bai et al., 2010; Hartley and Madhani, 2009). As nucleosome deposition occurs in the wake of elongating RNA Pol II during both coding and noncoding transcription (Belotserkovskaya et al., 2003; Hughes et al., 2012; Pruneski and Martens, 2011), we considered the model that the high levels of intergenic noncoding transcription observed in jhd2Δ cells might be accompanied by an increase in global nucleosome occupancy near these TSSs, which could account for at least some of the mechanisms by which JHD2 acts on protein coding genes (Figure 5A). In light of this potential mechanism, sporulation may comprise an unexplored biological context in yeast that involves globally acting nucleosomal occlusion near TSSs (Figure 5A).
Figure 5. JHD2 Represses a Program of Increasing Nucleosome Binding to TSSs during Sporulation.
(A) Working model: Jhd2 promotes protein coding gene expression by negatively regulating interfering noncoding transcription.
(B) Genome-wide high-resolution nucleosome occupancy data was collected from sporulating WT and jhd2Δ cultures. TSS centered and log2 transformed mean occupancy signal of 5147 coding genes is shown with the time point of sporulation represented by color, as indicated in the legend.
(C) Globally averaged regions from −250 to +150 with respect to the TSS are displayed. For clarity, data from different sporulation time points are shown offset from each other, with the x axes for each time point displayed beneath. *p value = 4 × 10−8, **p value = 4 × 10−10.
See also Figure S5.
To test this model, we measured nucleosome occupancies across the genomes of sporulating cells using high-resolution microarray hybridizations of mono-nucleosomal DNA isolated from these cells. Although a recent study found that WT cells progressing through meiosis (0–7 hr sporulation time points) exhibited surprisingly limited global dynamics in nucleosomal occupancies (Zhang et al., 2011), our data suggested a role for JHD2 in global transcriptional control in postmeiotic cells. We therefore evaluated nucleosomal occupancies throughout the 0–16 hr time points of sporulation. From this data, we extracted genomic regions flanking 5,147 protein coding gene TSSs and evaluated the averaged nucleosome occupancy at positions between 600 bp upstream and 900 bp downstream of these TSSs (Figure 5B). As expected, and in agreement with Zhang et al. (2011), we observed a prominent NDR directly 5′ of TSSs, with well-positioned nucleosomes moving into the bodies of transcript units in both vegetative cells and throughout sporulation (Figure 5B).
When we next displayed the TSS-proximal nucleosomal occupancies in temporal order throughout sporulation, we found that the depth of the globally averaged NDR decreased significantly as WT cells progressed through the program (Figure 5C). Beginning at the 6 hr time point and continuing through the 10 hr time point, confidence interval analysis (Bonn et al., 2012) and two-tailed t tests showed that this effect was both highly statistically significant and progressively increasing in magnitude (Figures 5C and S5A). Intriguingly, at the 12 hr time point (and continuing until our last examined time point at 16 hr), the averaged NDR depth reverted to the state found in vegetative cells (Figure 5C). We next compared the WT nucleosome occupancy properties during sporulation with those from experimentally paired jhd2Δ cells. Although the globally averaged nucleosome profiles of WT and jhd2Δ were nearly indistinguishable throughout most of sporulation, at the 8 hr time point, confidence interval analyses and two-tailed t tests showed that jhd2Δ cells exhibited a highly significant increase in nucleosome occupancy at the globally averaged TSS (Figures 5C and S5B).
Our results show that sporulation is accompanied by a global increase in nucleosomal occupancy at TSS-associated sites. This program appears to initiate at the 6 hr mark of sporulation, peak at the 10 hr time point, and revert to the “baseline” state at 12 hr coincident with the acquisition of transcriptional quiescence. Interestingly, JHD2 represses this program, with deletion of JHD2 resulting in a further increase in nucleosomal occupancy at TSSs at the 8 hr time point. Consistent with the hypothesis that the programmed increase in nucleosomal occupancy near TSSs is a consequence of noncoding transcription coupled nucleosome deposition, the reversion to the “baseline” nucleosome occupancy state occurs concomitant with dramatically reduced global transcription rates. Nucleosomes presumably return to their more energetically favorable states following this period (Kaplan et al., 2009). As this period coincides with when JHD2 promotes the transcription of protein coding genes and represses noncoding transcription near the TSSs of these genes, these results support our model (Figure 5A).
JHD2 Acts Broadly at Ribosomal Protein Encoding and Rap1-Bound Genes in Postmeiotic Cells
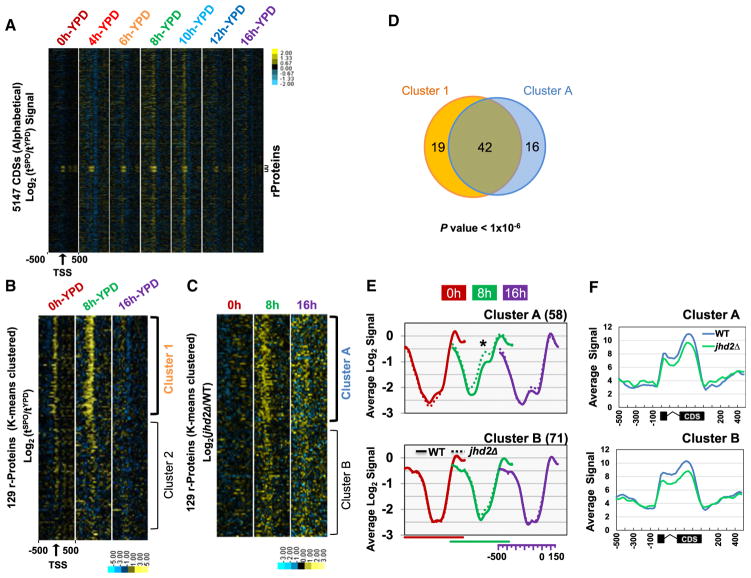
To further test our model for JHD2 function, we examined specific classes of genes. During sporulation, ribosomal protein (RP) gene transcription is repressed throughout meiosis and de-repressed following the completion of meiosis, presumably to provide spores with a “maternal” load of RP mRNAs and/or ribosomes to be utilized upon germination (Chu et al., 1998; Pearson and Haber, 1980). In yeast, coordinated transcription of the 137 RP genes represents a substantial energetic investment, accounting for ~50% of all RNA Pol II transcription events (Warner, 1999). Given the role of JHD2 in promoting postmeiotic transcription (Figure 3B), this postmeiotic de-repression phase of RP genes therefore seemed likely to be of significance. After first ordering the protein coding genes alphabetically so that all of the RP genes (named “RPL” and “RPS”) were adjacent to each other, we visualized the sporulation dynamics in nucleosome occupancies surrounding TSSs using heat maps (Figure 6A). This analysis showed that RP genes exhibited a robust ‘filling in” of TSS-associated NDRs during sporulation that stood out from the rest of the genome (Figure 6A).
Figure 6. JHD2 Acts at Ribosomal Protein Encoding Genes in Postmeiotic Cells.
(A) Differential analysis of global nucleosome positioning changes was performed by calculating the change in Log2 signal at each time point in sporulation compared to the signal in vegetative cells (Log2 (tSPO/tYPD)). Each row depicts 1 kb of signal from a TSS-centered gene. Genes are ordered alphabetically by their standard name. Data from each time point of sporulation is labeled. Small brackets demarcate the ribosomal protein (RP) encoding genes (RPL/RPS).
(B) Log2 (tSPO/tYPD) signal from 129 RP genes was K-means clustered into two groups (Clusters 1 and 2) using a Euclidian distance matrix.
(C) Log2(jhd2Δ/WT) signal of 129 RP genes were independently K-means clustered into two groups (Clusters A and B).
(D) There is significant overlap between Cluster 1 and Cluster A. The p value from the hypergeometric distribution test is shown.
(E) The average nucleosome occupancy signals from −500 to +150 with respect to the TSS of the RPs in Clusters A and B. For clarity, data from different sporulation time points are shown offset from each other, with the x axes for each time point shown underneath. *p value < 0.05.
(F) The average normalized Affymetrix RNA transcript signal at 20 hr of sporulation for clusters in (E) are shown. Signal associated with coding region are scaled into 20 bins. An additional 500 bp of flanking signals are shown on each side.
See also Figure S6 for analysis of Rap1-bound genes.
We identified two classes of RP genes using k-means clustering (Cluster 3.0): one that exhibited dramatic increases in nucleosomal occupancies near TSSs at the 8 hr sporulation time point, and one that did not (Figure 6B, clusters 1 and 2). We next identified two classes of RP genes in jhd2Δ mutants: one that hyper accumulated nucleosomes near TSSs at the 8 hr time point of sporulation compared with WT, and one that did not (Figure 6C, clusters A and B). The overlap between cluster 1 and cluster A was very significant, suggesting that JHD2 opposes a sporulation pathway that promotes the accumulation of nucleosomes near the TSSs of a large class (almost half) of RP genes (Figure 6D). Confidence interval analysis and two-tailed t tests confirmed that the differences in nucleosome occupancy between WT and jhd2Δ were highly significant for cluster A, but not for cluster B (Figure 6E and data not shown). If the RP genes in cluster A accumulated nucleosomes as a consequence of increased noncoding transcription, then they should also exhibit increased noncoding transcription 5′ of their TSSs in jhd2Δ compared with cluster B genes. In agreement with this, we found that cluster A was associated with increased noncoding RNA in jhd2Δ compared with cluster B (Figure 6F).
RP gene transcriptional control involves the general regulator factor Rap1 (Lieb et al., 2001). To extend our analysis of RP genes, we interrogated the nucleosomal occupancies of the 421 known Rap1-bound genes (Rhee and Pugh, 2011) after first removing all of the RP genes from this list. We again identified a cluster of Rap1-bound genes that exhibited increased nucleosomal binding at the 8 hr time point of sporulation in WT cells and one that did not (Figure S6A, clusters 1 and 2). In striking recapitulation of our RP gene analysis, jhd2Δ caused a highly significant hyper accumulation of nucleosomes in a cluster of genes that overlapped significantly with cluster 1 (Figures S6B–S6D). Like we observed with the RP genes, this cluster was associated with increased JHD2-repressed noncoding transcription 5′ of their TSSs (Figure S6E).
One commonality of the genes that exhibit strong evidence of JHD2-repressed nucleosomal occlusion is their significantly larger NDR regions (compare clusters A with clusters B in Figures 6E and S6D). As these NDRs possess more exposed DNA to accommodate both nucleosomal accumulation and noncoding transcription, they stand out dramatically in our analysis of JHD2-mediated nucleosome occupancy changes and may therefore be regarded as diagnostic of more broadly acting mechanisms. This interpretation is supported by the fact that both clusters A and clusters B from the RP and Rap1-bound genes exhibit equivalent defects in protein coding mRNA expression (Figure 6F and S6E). Perhaps relatedly, repressive noncoding transcription can act through a variety of mechanisms in addition to nucleosome occlusion, such as the accumulation of stalled polymerases, or the dislodgement of transcription factors and preinitiation complexes (Palmer et al., 2011).
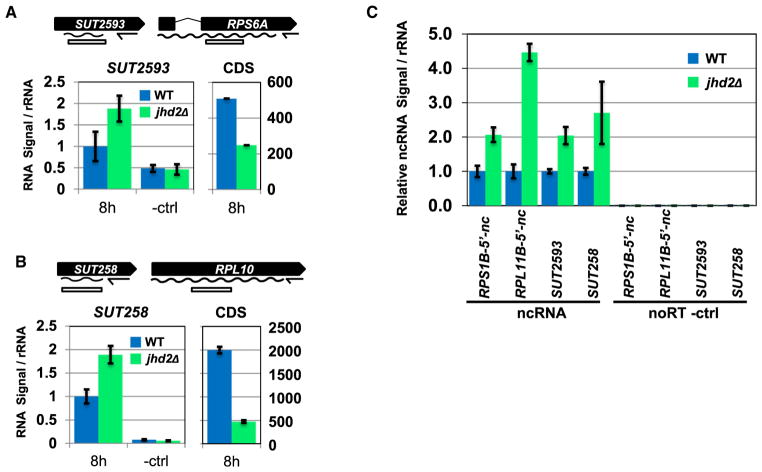
The cluster A and B RP genes RPS6A and RPL10 are flanked on their immediate 5′ ends by the annotated noncoding transcripts SUT2593 and SUT258 (Lardenois et al., 2011). In agreement with the conclusion that RP genes from both clusters are generally targeted by JHD2-repressed noncoding transcription, both SUT2593 and SUT258 were upregulated in jhd2Δ whereas their associated RP genes were downregulated at the 8 hr sporulation time point (Figures 7A and 7B). Although most RP genes do not have annotated noncoding transcripts in their 5′ regions (only 17 RPs have annotated 5′ sense ncRNAs), our analysis suggested that noncoding transcripts are widespread 5′ of RP genes. We used RT-qPCR to interrogate the 5′ region directly upstream of two such genes from cluster A and cluster B for the existence of JHD2-regulated noncoding transcripts on the sense strand. In addition to confirming our observations of SUT258 and SUT2593 in this biological replicate, we found that RPS1B and RPL11B are both flanked on their 5′ ends by previously unannotated JHD2-repressed noncoding transcripts (Figure 7C). RT-qPCR analysis of these regions in RNA prepared from vegetative cells grown in dextrose did not show any differences in the abundance of these transcripts between WT and jhd2Δ (data not shown). As a recent study has shown that noncoding transcription plays prominent roles in mediating transcriptional regulation in vegetative cells adapting to alternative carbon utilization (Kim et al., 2012), it will be of great interest to elucidate the global function of JHD2 under these conditions.
Figure 7. Jhd2 Negatively Regulates Abundance of Noncoding Transcripts during Sporulation.
Transcript-specific RT qPCR was performed to measure levels of noncoding transcripts associated with coding genes. Signals were normalized to RDN25-1 rRNA levels, and expressed as fold difference from WT ncRNA signal. Error bars represent 1 SD from technical replicates.
(A) Relative levels of SUT2593 and RPS6A transcript are shown.
(B) Relative levels of SUT258 and RPL10 transcript are shown.
(C) Relative levels of ncRNA were measured in cultures at 10 hr of sporulation for WT and jhd2Δ.
CONCLUSIONS
We have shown here that yeast sporulation is accompanied by a programmed onset of global transcriptional quiescence following the completion of meiosis, and that the highly conserved H3K4 demethylase JHD2 promotes protein coding gene transcription in opposition to this transcriptional quiescence (Figure 3B). Our studies also show that JHD2 represses 5′ sense-strand oriented intergenic noncoding transcription in postmeiotic cells and an associated program of globally increased nucleosomal occupancies near TSSs, suggesting an explanation for at least part of JHD2s mechanism of action (Figure 5A). Although we have not investigated their mechanism of action here, we report that JHD2 globally represses antisense RNA that overlaps with gene bodies and appears to originate from the TTSs of genes. It seems likely that this accumulation of antisense RNA may act to repress gene transcription and/or mRNA stability of their associated protein coding genes in postmeiotic cells. In addition to setting the time interval of postmeiotic transcriptional activity, JHD2 correspondingly defines the time period of spore differentiation. As JARID1A/B acts antagonistically to terminal differentiation in mammalian stem cells, the developmental role of Jhd2 behaves in an analogous manner to that seen for members of the mammalian JARID family.
Our analysis of spore fitness suggests that Jhd2 may modulate the final stages of spore differentiation in response to carbon metabolism. Because Jhd2 enzymatic activity, and indeed the activity of the entire Jumonji gene family, requires the TCA cycle metabolite alpha-ketoglutarate (Klose et al., 2006; Tsukada et al., 2006), it is tempting to hypothesize that this gene family could function as sensors of carbon metabolism which fine-tune transcriptional outputs in response to nutrient availability cues. More speculatively, given the extraordinary conservation of both H3K4 methylation and JARID1 family H3K4 demethylases in eukaryotes, our results suggest that metabolic control of JARID function may constitute a general mechanism to modulate transcriptional programs during differentiation.
EXPERIMENTAL PROCEDURES
Yeast Genetics
Yeast strains used in this study are listed in Supplemental Experimental Procedures. Standard yeast genetic methods were used for construction of all strains. For construction of the chromosomal JHD2(H427A) alleles, the delitto perfetto method was used (Storici and Resnick, 2003). Histone shuffle plasmids were obtained from the SHIMA collection (Nakanishi et al., 2008).
Sporulation
Strains were grown overnight to saturation in standard YPD media and then diluted into YPA (1% yeast extract, 2% bacto peptone, 1% potassium acetate) at an optical density (OD)600 of 0.3. Cultures were grown for 12–14 hr at 30°C with shaking at 200 RPM to an OD of ~1.5. Cells were pelleted, washed with ddH2O and resuspended into SPO media (1% potassium acetate, 0.02% raffinose) and sporulated at 30°C with shaking at 200 RPM. Meiotic progression was quantified microscopically using DAPI staining of fixed cells.
Western Blotting
Protein extracts were prepared from MMY718 (wild-type) and MMY1879 (jhd2Δ) at the indicated sporulation time points as described (Kushnirov, 2000) and normalized for total protein concentration. Equal amounts of protein were electrophoresed on SDS-PAGE gels, and transferred onto Amersham Hybond-P membranes. Primary antibodies for histone experiments were Abcam ab1791 (pan-H3), ab8580 (H3K4me3), ab7766 (H3K4me2), ab8895 (H3K4me1), and ab6161 (Rat mAb against tubulin). For quantitative western blotting presented in Figure 2B, the Li-Cor Odyssey system was used according to the manufacturer’s specifications. Primary antibodies to probe for RNA Pol II CTD phosphorylation states were Millipore 05-623 (pan-CTD clone 4H8), 04-1571 (CTD-S2P clone 3E10), and 04-1572 (CTD-S5P clone 3E8). Band intensities were quantified using ImageQuant and plotted as values of each phosphorylation modification normalized to bulk CTD from each protein extract.
Transcript Profiling Experiments
MMY718 and MMY1879 were sporulated and aliquots of culture were harvested at time points between 0 and 24 hr and flash-frozen in liquid nitrogen. RNA was prepared by bead beating cell pellets (Mini Bead beater, Biospec Products) for 4 × 2.5 min in Trizol (Invitrogen) with 5 min incubations on ice between cycles. After addition of chloroform and phase separation, RNA was further purified by extraction with acidic phenol at 65°C for 30 min. For ORF profiling time courses, microarrays were spotted with commercially available 60-mer oligonucleotides corresponding to yeast ORFs, and hybridizations, data extraction, and normalization were performed as described (Meneghini et al., 2003). For a hybridization reference, a pool combining equal amounts of cDNAs from all the WT and jhd2Δ time points was generated and coupled with Cy3.
The normalized expression values are presented in Table S2.
Affymetrix RNA hybridizations, normalization, and data analysis were performed as described (Badis et al., 2008). The complete Affymetrix transcript microarray data set (at 20 hr of sporulation) can be downloaded at NCBI Gene expression omnibus under the accession number GSE41002.
RT-qPCR Measurement of Coding and Noncoding RNAs
RNA was prepared as described above, precipitated, and resuspended in RNase-free water. Six micrograms of total RNA was reverse-transcribed using either random nonamers (Figures 2 and S2), or site-specific primers (Figure 7) and Superscript III RT (Invitrogen) according to the manufacturer’s instructions. The resulting cDNA preparations were used at a dilution of 1/200 for qPCR using 2× SYBR Green/PCR buffer (Aparicio et al., 2004) with the MyIQ thermocycler (BioRad). Reactions were quantified relative to a common genomic DNA sample and transcript abundance normalized to a qPCR of the RDN25-1 rRNA locus. Oligonucleotides are detailed in Supplemental Experimental Procedures.
Mononucleosome Isolation, ChIP-Chip Hybridization, and Analysis
For H3K4me ChIP-chip experiments, MMY718 (wild-type) and MMY1879 (jhd2Δ) were sporulated as described above. At the 10 hr time point, quadruplicate replicates of sporulating cells were harvested. Chromatin IP, array hybridizations, data extraction, and analysis of these replicates were performed as described previously using tiling microarrays that span chromosome III (Bernstein et al., 2002; Hartley and Madhani, 2009; Liu et al., 2005). For each gene, the values for probes representing the 2 kb region between −500 and +1,500 base pairs with respect to their transcription start sites (TSS) were extracted. The signals for all genes were aligned based on the positions of the TSS in wild-type cells, which were determined using published data (Xu et al., 2009). The aligned data were averaged to obtain a histone modification profile of the average gene on chromosome III. The complete data set is presented in Table S3.
Genome-wide Nucleosome Occupancy Studies
Chromatin preparations from YPD grown cells and sporulation time points corresponding to, 0, 4, 6, 8, 10, 12, and 16 hr were prepared as described above with modifications (Albert et al., 2007). Crosslinked cell pellets were lysed by sonication using a Diagenode Bioruptor followed by bead beating for 7 cycles of 1 min with 2 min rests on ice. Mononucleosomal DNA was prepared from these extracts and processed for Affymetrix microarray hybridization as described with some modifications (Lee et al., 2007). Data extraction and normalization was performed as described (Lee et al., 2007). The raw data as well as normalized TSS/TTS centered nucleosome occupancy data have been uploaded to NCBI Gene Expression Omnibus under the accession number GSE40874.
Ether Treatment Assays
MMY1662 and MMY1657 were cosporulated in the same flask as described above. At the indicated time points, aliquots were removed, briefly sonicated, and treated with 33% di-ethyl ether in nutating microcentrifuge tubes at room temperature. At 2 min intervals between 2–10 min, aliquots were removed, serial diluted in H2O, and spotted onto YPD plates containing G418 or hygromycin. Spotted sporulated cells were allowed to germinate and form colonies over 2 days at 30°C, and photographed.
Spore Fitness Assays
MMY1898/MMY1996 and MMY1995/MMY3032 were cosporulated as described above. Two paired sporulations, one in 1× SPO medium, and the other in 0.1× SPO medium, were sporulated for 18 hr and assayed for spore number and spore fitness as follows. Sporulation efficiency, spore number, and genotype were quantified using bright field and fluorescence microscopy utilizing the differential fluorescence of each cosporulated strain. Aliquots were harvested, sonicated, serial diluted, and spotted onto YPD plates containing G418 or hygromycin. The same aliquots were subjected to treatment with 0.6 mg/ml zymolyase for 1 hr at 37°C followed by resuspension in 0.5% Triton X-100, or to incubation at 50°C for 25 min. Following these treatments, relative spore survival was determined following spotting of serial dilutions onto YPD plates containing G418 or hygromycin.
Oxygen Consumption Assays
Oxygen consumption of sporulating cells in suspension was measured using an oxygen electrode (Strathkelvin 782 oxygen meter). For basal oxygen consumption measurements, ethanol (1% v/v) was added as a carbon source. For maximal oxygen consumption measurements, mitochondrial membrane potential was decoupled with addition of carbonyl cyanide m-chlorophenyl hydrazone (CCCP) to 8 μM. To confirm that mitochondrial respiration was responsible for the oxygen consumption that we observed, cyanide was used to inhibit electron transport chain function at a final concentration of 10 μM. Oxygen consumption rate was measured in units of nmol atoms (natoms) of oxygen per minute per one million cells (natom/min/M).
Supplementary Material
Acknowledgments
We thank Changtai Cui and Nguyen Nguyen for technical support. Special thanks to Jacqueline Segall and Julie Claycomb for critical reading of the manuscript, to Emma McCullagh for assistance with figure construction, and to Desiree Tillo for advice concerning nucleosome positioning data analysis. This work was supported by CIHR grant MOP-89996 to M.D.M., CIHR grant MOP-86705 to C.N., and NIH grant (GM071801) and a Leukemia and Lymphoma Society Scholar’s Award to H.D.M. M.D.M. holds a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
Footnotes
ACCESSION NUMBERS
Transcript profiling and nucleosome occupancy microarray data are available from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) with the accession numbers GSE41002 and GSE40874, respectively.
Supplemental Information includes six figures, three tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2012.10.005.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Aparicio O, Geisberg JV, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Cell Biol. 2004;Chapter 17(Unit 17.7) doi: 10.1002/0471143030.cb1707s23. [DOI] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Charvin G, Siggia ED, Cross FR. Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle. Dev Cell. 2010;18:544–555. doi: 10.1016/j.devcel.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG., Jr Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell. 2005;18:623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AJ, Gordon M, Eide DJ, Winge DR. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 2006;25:5726–5734. doi: 10.1038/sj.emboj.7601453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczyński B, Riddell A, Furlong EE. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Brengues M, Pintard L, Lapeyre B. mRNA decay is rapidly induced after spore germination of Saccharomyces cerevisiae. J Biol Chem. 2002;277:40505–40512. doi: 10.1074/jbc.M206700200. [DOI] [PubMed] [Google Scholar]
- Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci USA. 2009;106:18321–18326. doi: 10.1073/pnas.0909641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Davidow LS, Goetsch L, Byers B. Preferential occurrence of nonsister spores in two-spored asci of Saccharomyces cerevisiae: evidence for regulation of spore-wall formation by the spindle pole body. Genetics. 1980;94:581–595. doi: 10.1093/genetics/94.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes IW, Hardie ID. Selective killing of vegetative cells in sporulated yeast cultures by exposure to diethyl ether. Mol Gen Genet. 1974;131:281–289. doi: 10.1007/BF00264859. [DOI] [PubMed] [Google Scholar]
- Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor-Papidimitriou J, Wynder C. The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol Cell Biol. 2008;28:5312–5327. doi: 10.1128/MCB.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood MD, Cheung SW, Lee KY, Moffat J, Meneghini MD. Developmentally programmed nuclear destruction during yeast gametogenesis. Dev Cell. 2012;23:35–44. doi: 10.1016/j.devcel.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Magee PT, Welch SK, Friedman M, Hall BD. Macromolecule synthesis and breakdown in relation to sporulation and meiosis in yeast. J Bacteriol. 1974;119:619–628. doi: 10.1128/jb.119.2.619-628.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Jin Y, Rando OJ, Struhl K. A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.07.003. Published online August 8, 2012. http://dx.doi.org/10.1016/j.molcel.2012.07.003. [DOI] [PMC free article] [PubMed]
- Ingvarsdottir K, Edwards C, Lee MG, Lee JS, Schultz DC, Shilatifard A, Shiekhattar R, Berger SL. Histone H3 K4 demethylation during activation and attenuation of GAL1 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:7856–7864. doi: 10.1128/MCB.00801-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. Set3 HDAC Mediates Effects of Overlapping Noncoding Transcription on Gene Induction Kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jørgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardenois A, Liu Y, Walther T, Chalmel F, Evrard B, Granovskaia M, Chu A, Davis RW, Steinmetz LM, Primig M. Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc Natl Acad Sci USA. 2011;108:1058–1063. doi: 10.1073/pnas.1016459108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Klose RJ, Gardner KE, Zhang Y. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat Struct Mol Biol. 2007;14:243–245. doi: 10.1038/nsmb1204. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, Dewaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling- Hampton K, Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AC, Egan JB, Shearwin KE. Transcriptional interference by RNA polymerase pausing and dislodgement of transcription factors. Transcription. 2011;2:9–14. doi: 10.4161/trns.2.1.13511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Pearson NJ, Haber JE. Changes in regulation of ribosomal protein synthesis during vegetative growth and sporulation of Saccharomyces cerevisiae. J Bacteriol. 1980;143:1411–1419. doi: 10.1128/jb.143.3.1411-1419.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneski JA, Martens JA. Transcription of intergenic DNA deposits nucleosomes on promoter to silence gene expression. Cell Cycle. 2011;10:1021–1022. doi: 10.4161/cc.10.7.15167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bernstein BE, Karabetsou N, Morillon A, Weise C, Schreiber SL, Mellor J, Kouzarides T. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol Cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8:323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin- mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference— a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Lin W, Soustelle C, Suhre K, Nicolas A, Géli V, de La Roche Saint-André C. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J. 2004;23:1957–1967. doi: 10.1038/sj.emboj.7600204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Resnick MA. Delitto perfetto targeted mutagenesis in yeast with oligonucleotides. Genet Eng (N Y) 2003;25:189–207. [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011;30:1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadon AN, Van de Mark D, Basom R, Delrow J, Whitehouse I, Tsukiyama T. Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Mol Cell Biol. 2010;30:5110–5122. doi: 10.1128/MCB.00602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ma H, Pugh BF. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res. 2011;21:875–884. doi: 10.1101/gr.117465.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.