Abstract
Objective
The NOX2 NADPH oxidase complex produces reactive oxygen species and plays a critical role in the killing of microbes by phagocytes. Genetic mutations in genes encoding components of the complex result in both X-linked and autosomal recessive forms of chronic granulomatous disease (CGD). Patients with CGD often develop intestinal inflammation that is histologically similar to Crohn's colitis, suggesting a common aetiology for both diseases. The aim of this study is to determine if polymorphisms in NOX2 NADPH oxidase complex genes that do not cause CGD are associated with the development of inflammatory bowel disease (IBD).
Methods
Direct sequencing and candidate gene approaches were used to identify susceptibility loci in NADPH oxidase complex genes. Functional studies were carried out on identified variants. Novel findings were replicated in independent cohorts.
Results
Sequence analysis identified a novel missense variant in the neutrophil cytosolic factor 2 (NCF2) gene that is associated with very early onset IBD (VEO-IBD) and subsequently found in 4% of patients with VEO-IBD compared with 0.2% of controls (p=1.3×10−5, OR 23.8 (95% CI 3.9 to 142.5); Fisher exact test). This variant reduced binding of the NCF2 gene product p67phox to RAC2. This study found a novel genetic association of RAC2 with Crohn's disease (CD) and replicated the previously reported association of NCF4 with ileal CD.
Conclusion
These studies suggest that the rare novel p67phox variant results in partial inhibition of oxidase function and are associated with CD in a subgroup of patients with VEO-IBD; and suggest that components of the NADPH oxidase complex are associated with CD.
Introduction
Inflammatory bowel diseases (IBD) are hypothesised to occur in genetically susceptible individuals as a result of dysregulated immune responses to gut flora after exposure to an as yet unidentified environmental stimulus.1,2 Investigation of diseases that present with intestinal inflammation that are similar to IBD may provide an important insight into the pathogenesis of IBD. For example, chronic granulomatous disease (CGD) is a rare genetic disorder with a prevalence of 1/200 000 to 1/250 000 caused by X-linked and autosomal recessive mutations in genes encoding components of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex (also referred to as NOX2 NADPH oxidase or phagocyte oxidase).3 Patients with CGD have severe and recurrent infections as a result of the inability of phagocytes to mount sufficient respiratory burst to kill invading pathogens.4 Interestingly, up to 40% of patients with CGD develop a form of colitis that is endoscopically and pathologically very similar to the colitis observed in Crohn's disease (CD).5–7
Defective neutrophil respiratory burst has been observed in patients with IBD,8–10 and a genome-wide association study (GWAS) identified the NCF4 (encoding the p40phox subunit of the NOX2 NADPH oxidase) region as an ileal CD-specific susceptibility gene. This association did not meet genome-wide significance in the original published GWAS11,12 and was not replicated in a recent GWAS meta-analysis, perhaps due to the fact that the GWAS analysis focused on CD independent of disease location.13 We have therefore undertaken studies to determine if components of the NADPH oxidase complex play a role in the development of IBD. Here we report a novel missense variant in NCF2 in patients with very early onset IBD (VEO-IBD) that results in neutrophil dysfunction and susceptibility to CD. We also describe novel associations of the NADPH oxidase complex gene RAC2 with CD and replicate the previously described association of NCF4 with ileal CD. Together, these results demonstrate that the NADPH oxidase complex genes play a role in the pathogenesis of CD.
Materials and Methods
RT-PCR
RNA was isolated from the whole blood by PAX gene blood RNA kit (Qiagen, USA) according to the manufacturer's instructions. cDNA was synthesised using SuperScript III Reverse Transcriptase (Life Technologies, Carlsbad, California, USA). Primers for full length NCF2, NCF4 and RAC2 were designed and synthesised at CDI (see table 1 in online supplement). PCR was performed according to a standard protocol and the purified PCR product was cloned into pJET cloning vector (Fermentas, Hanover, Maryland, USA) and sequenced by ABI 3730 DNA analyser (Applied Biosystems, Melbourne, Australia).
NCF2 genotyping
In order to determine if the NCF2 variant c.113 G→A R38Q was found in other patients with IBD, we designed a custom Taqman probe (tcagtgccgtccaggacccccactccc(g/a) gatttgcttcaacattggctgc). Four hundred and eighty patients and 480 controls and a second cohort of 119 patients with VEO-IBD (Toronto samples described below) were genotyped using Taqman at the Centre for Applied Genomics (TCAG), Hospital for Sick Children, Toronto (see online supplement for binding studies).
Single nucleotide polymorphism analysis and genotyping
International HapMap project14 (http://www.hapmap.org) Caucasian (CEU) phase II data Release 23a were used to select tag single nucleotide polymorphisms (SNPs) (minor allelic frequency (MAF) >1%) that span the NADPH oxidase complex genes and flanking regions through the ‘Tagger’ software program (r2 >0.8).15 Twenty-one tag SNPs covering the NCF4 region (chromosome 22, 35 581 544 to 35 598 557), 15 tag SNPs covering the NCF2 region (chromosome 1, 180 256 354 to 180 291 372), 19 tag SNPs covering the RAC2 region (chromosome 22, 35 945 811 to 35 964 804), five tag SNPs covering the CYBA region (chromosome 16, 87 237 198 to 87 244 957) and six tag SNPs covering the NOX2/CYBB (X-chromosome, 37 395 536 to 37 428 930) were used to capture all SNPs with r2>0.8 to the tag SNPs (see table 1 for list of NADPH oxidase SNPs). As NCF1 is located in tandem with two nearly identical pseudogenes, analysis of this region was not carried out. Genotyping of samples was performed using the Illumina Goldengate Custom Chip genotyping system (Toronto discovery) and Taqman (North America and Scotland Replication) at the Centre for Applied Genomics, Hospital for Sick Children, Toronto and the University of Edinburgh. Sixty-two SNPs in the NADPH oxidase complex (RAC2, CYBA, CYBB, NCF2 and NCF4) passed quality control.
Table 1. Association of NADPH oxidase genes NCF4 and RAC2 with CD in the discovery cohort.
| Chromosome | SNP | Gene | Position | A1 | A2 | MAF | FA | FU | p Value | OR | L95 | U95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | rs6000448 | NCF4 | 35 580 717 | 1 | 3 | 0.108045 | 0.1176 | 0.1007 | 0.1839 | 1.184 | 0.9228 | 1.52 |
| 22 | rs10854693 | NCF4 | 35 581 956 | 3 | 1 | 0.405648 | 0.378 | 0.427 | 0.003823 | 0.7925 | 0.6769 | 0.9278 |
| 22 | rs1883113 | NCF4 | 35 582 630 | 3 | 2 | 0.0677741 | 0.07546 | 0.06184 | 0.01325 | 1.472 | 1.084 | 2 |
| 22 | rs4821542 | NCF4 | 35 582 864 | 1 | 3 | 0.165449 | 0.1776 | 0.1561 | 0.1857 | 1.149 | 0.9354 | 1.411 |
| 22 | rs7287350 | NCF4 | 35 583 185 | 1 | 3 | 0.0548537 | 0.06031 | 0.05065 | 0.02581 | 1.47 | 1.048 | 2.063 |
| 22 | rs1883112 | NCF4 | 35 586 792 | 1 | 3 | 0.422821 | 0.3941 | 0.4451 | 0.001388 | 0.7732 | 0.6605 | 0.9053 |
| 22 | rs4821544 | NCF4 | 35 588 449 | 3 | 1 | 0.344518 | 0.3735 | 0.3221 | 0.005118 | 1.257 | 1.071 | 1.475 |
| 22 | rs741997 | NCF4 | 35 588 759 | 3 | 1 | 0.0843854 | 0.09299 | 0.07774 | 0.121 | 1.243 | 0.9442 | 1.636 |
| 22 | rs746713 | NCF4 | 35 589 305 | 3 | 1 | 0.306977 | 0.3011 | 0.3115 | 0.7173 | 0.9696 | 0.8202 | 1.146 |
| 22 | rs909484 | NCF4 | 35 590 547 | 1 | 3 | 0.171761 | 0.17 | 0.1731 | 0.8553 | 0.9808 | 0.7968 | 1.207 |
| 22 | rs729749 | NCF4 | 35 593 792 | 1 | 3 | 0.217158 | 0.2184 | 0.2162 | 0.7703 | 1.028 | 0.853 | 1.239 |
| 22 | rs2075938 | NCF4 | 35 596 268 | 1 | 3 | 0.279255 | 0.2679 | 0.288 | 0.3198 | 0.915 | 0.7682 | 1.09 |
| 22 | rs8137456 | NCF4 | 35 604 389 | 2 | 3 | 0.0687708 | 0.07774 | 0.06184 | 0.1693 | 1.24 | 0.9125 | 1.685 |
| 22 | rs8137602 | NCF4 | 35 604 595 | 3 | 2 | 0.0916944 | 0.08155 | 0.09953 | 0.07419 | 0.7759 | 0.5872 | 1.025 |
| 22 | rs3959633 | NCF4 | 35 605 501 | 1 | 3 | 0.0249169 | 0.03125 | 0.02002 | 0.07318 | 1.56 | 0.9591 | 2.537 |
| 22 | rs5756379 | NCF4 | 35 605 850 | 3 | 1 | 0.334551 | 0.3285 | 0.3392 | 0.5302 | 0.9483 | 0.8033 | 1.119 |
| 22 | rs4821554 | NCF4 | 35 606 030 | 1 | 3 | 0.257475 | 0.2424 | 0.2691 | 0.1157 | 0.8646 | 0.7213 | 1.036 |
| 22 | rs5750326 | NCF4 | 35 606 231 | 1 | 2 | 0.496678 | 0.5023 | 0.4923 | 0.9018 | 1.01 | 0.8663 | 1.177 |
| 22 | rs5756564 | RAC2 | 35 942 049 | 3 | 1 | 0.397674 | 0.3598 | 0.427 | 0.0006546 | 0.7562 | 0.6439 | 0.888 |
| 22 | rs4820272 | RAC2 | 35 945 560 | 1 | 3 | 0.162791 | 0.189 | 0.1425 | 0.001566 | 1.401 | 1.137 | 1.727 |
| 22 | rs933222 | RAC2 | 35 948 583 | 1 | 3 | 0.281395 | 0.295 | 0.2709 | 0.2225 | 1.117 | 0.9348 | 1.336 |
| 22 | rs12166968 | RAC2 | 35 951 304 | 2 | 3 | 0.0671096 | 0.0686 | 0.06596 | 0.9971 | 1.001 | 0.7332 | 1.365 |
| 22 | rs6572 | RAC2 | 35 951 391 | 3 | 2 | 0.453488 | 0.4947 | 0.4217 | 0.00038 | 1.337 | 1.139 | 1.57 |
| 22 | rs739041 | RAC2 | 35 954 945 | 3 | 1 | 0.434492 | 0.4331 | 0.4356 | 0.8488 | 1.015 | 0.8678 | 1.188 |
| 22 | rs9607431 | RAC2 | 35 959 884 | 2 | 1 | 0.135548 | 0.1677 | 0.1107 | 0.00008594 | 1.577 | 1.256 | 1.98 |
| 22 | rs1476002 | RAC2 | 35 960 734 | 1 | 3 | 0.12766 | 0.1321 | 0.1243 | 0.6058 | 1.065 | 0.8383 | 1.353 |
| 22 | rs13058338 | RAC2 | 35 962 716 | 4 | 1 | 0.256981 | 0.2294 | 0.2783 | 0.006391 | 0.7773 | 0.6486 | 0.9316 |
| 22 | rs5756573 | RAC2 | 35 963 721 | 1 | 3 | 0.145376 | 0.1437 | 0.1466 | 0.7453 | 1.037 | 0.8329 | 1.291 |
| 22 | rs2284038 | RAC2 | 35 965 001 | 3 | 1 | 0.398007 | 0.404 | 0.3934 | 0.6625 | 1.037 | 0.8821 | 1.218 |
| 22 | rs2239774 | RAC2 | 35 967 599 | 2 | 3 | 0.164452 | 0.1974 | 0.139 | 0.0001514 | 1.495 | 1.214 | 1.84 |
| 22 | rs2239775 | RAC2 | 35 967 787 | 1 | 2 | 0.134884 | 0.1319 | 0.1372 | 0.4163 | 0.9086 | 0.7212 | 1.145 |
| 22 | rs2239773 | RAC2 | 35 968 235 | 1 | 3 | 0.277002 | 0.247 | 0.2996 | 0.004515 | 0.7696 | 0.6424 | 0.9221 |
| 22 | rs2213430 | RAC2 | 35 968 906 | 1 | 3 | 0.434219 | 0.436 | 0.4329 | 0.6503 | 1.038 | 0.8845 | 1.217 |
| 22 | rs6000632 | RAC2 | 35 974 061 | 1 | 3 | 0.245183 | 0.237 | 0.2515 | 0.4481 | 0.9314 | 0.7751 | 1.119 |
| 22 | rs4821615 | RAC2 | 35 976 077 | 2 | 3 | 0.447841 | 0.4512 | 0.4452 | 0.8961 | 1.01 | 0.8644 | 1.181 |
| 22 | rs12484031 | RAC2 | 35 979 216 | 3 | 1 | 0.100664 | 0.09299 | 0.1066 | 0.2213 | 0.8521 | 0.6593 | 1.101 |
| 22 | rs7288979 | RAC2 | 35 979 440 | 3 | 1 | 0.176412 | 0.1608 | 0.1885 | 0.06473 | 0.8266 | 0.6754 | 1.012 |
The discovery cohort consisted of 2049 subjects (656 with Crohn's disease, 544 with ulcerative colitis and 849 controls).
p Values are presented as uncorrected. Bonferroni correction threshold for 62 SNPs and IBD/UC/CD, α=2.0×10−4.
A1/A2, allele 1/2; CD, Crohn's disease; MAF, minor allelic frequency; FA, frequency affected; FU, frequency unaffected; IBD, inflammatory bowel disease; L95 and U95, lower and upper 95th confidence interval; SNP, single nucleotide polymorphism.; UC, ulcerative colitis.
Subjects, quality control and population stratification
All subjects in this study were of European descent by self-reporting of ethnic heritage. All probands had a confirmed diagnosis of IBD and fulfilled standard diagnostic criteria. Phenotypic characterisation was based on the Montreal Classification.16 Perianal disease (Montreal Classification ‘p’) included only those patients with perianal abscess and/or fistulae. Phenotypic information and DNA samples were obtained from study subjects with approval of the institutional review ethics board for IBD genetic studies at the Hospital for Sick Children and Mount Sinai Hospital in Toronto. Replication cohorts had review ethics board approval for genetic and phenotypic studies at the individual institutions. Written informed consent was obtained from all participants.
The discovery cohort included patients recruited from the Hospital for Sick Children (22%) and Mount Sinai Hospital (78%) in Toronto and local and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) control individuals. A total of 2049 subjects (656 with CD, 544 with ulcerative colitis (UC) and 849 controls; see table 2A in online supplement for demographic and phenotypic description) in the discovery analysis were used in the final analysis. The first replication cohort consisted of 1836 Caucasian individuals from North America including 443 patients with CD, 477 with UC and 916 controls (see table 2B in online supplement for demographic and phenotypic description); NIDDK patients recruited from Chicago and Pittsburgh with North America Caucasians of European origin control individuals obtained from the Centre for Applied Genomics (Ontario Population Genomics Platform (plates used: 1–4, 6, 9–12, 14); a complete description of this control population can be found at http://www.tcag.ca/cyto_population_control_DNA.html). The second cohort consisted of 2449 individuals exclusively recruited from Scotland including 691 patients with CD, 615 with UC and 1143 controls (see table 2C in online supplement for demographic and phenotypic description). All patients and control individuals were Caucasian and all related individuals were excluded by checking the estimated identity by descent for each pair of samples. Part of these cohorts has been used in previous GWAS including all the NIDDK patients in the North American replication cohort11,17 and 374 individuals from Scotland in the Paediatric IBD GWAS.18 None of the replication cohort control individuals were genotyped in IBD GWAS.
Preliminary analysis
HAPLOVIEW19 was used to obtain LD patterns, obtaining descriptive statistics for the SNPs. PLINK version 1.0620 was used to test for Hardy-Weinberg equilibrium (HWE) for each marker based on the Pearson χ2 test. Descriptive statistics of demographic variables were generated using SAS V.9.2 (SAS Institute). The Wilcoxon rank sum test and χ2 test were used to identify differences in demographic variables between subgroups.
Association analysis
The analysis was applied in stages. In stage 1, association analyses were applied to detect the associations with the 62 SNPs in five genes involved in the NADPH oxidase complex (RAC2, CYBA, CYBB (NOX2), NCF2 and NCF4) and IBD, CD and UC versus healthy controls. Logistic regression models were applied to an additive genetic model and Pearson χ2 tests were applied for dominant and recessive genetic models. We used an additive genetic model for primary analysis.21 For the CYBB analysis we used the chromosome-counting approach22 employed by the Wellcome Trust Case-Control Consortium.23 Throughout the report the p values are the additive model p value. ORs and 95% CIs were estimated for the disease group compared with the control group. The association, adjusting for selected principal component vectors from the Eigenstrat analysis, was tested using conditional logistic regression (SAS V.9.2). In stage 2 the four RAC2 SNPs identified from the discovery cohort were genotyped in a replication cohort (North America) and an independent validation cohort (Scotland). Independent analysis was applied to the replication and validation cohorts. Combined effect estimates from all three IBD cohorts were estimated using a logistic regression model including cohorts as adjusted covariates. All p values are two-sided.
Subphenotype analysis
In addition to comparing patients with IBD, CD and UC with healthy controls, we applied subphenotype analysis to evaluate the genetic effect on the disease risk of the IBD subpopulation according to the Montreal Classification system. The subpopulation comparisons were applied for each of the genetic markers on ileal only (L1), colonic only (L2), ileocolonic (L3), ileal any (L1/L3), colon only (L2 plus UC), colon any (L2/L3 plus UC), perianal (p) and early diagnosis patients (diagnosis age <16 years) versus healthy controls. The allelic model was used to test single marker associations between each of the subgroups. The analyses were applied to the discovery cohort, the North American replication cohort, the Scottish replication cohort and the pooled samples separately. Since the subphenotype analysis is exploratory and hypothesis-generating, p<0.05 was used to define nominal signals.
Results
Novel NCF2 variant associated with very early onset IBD
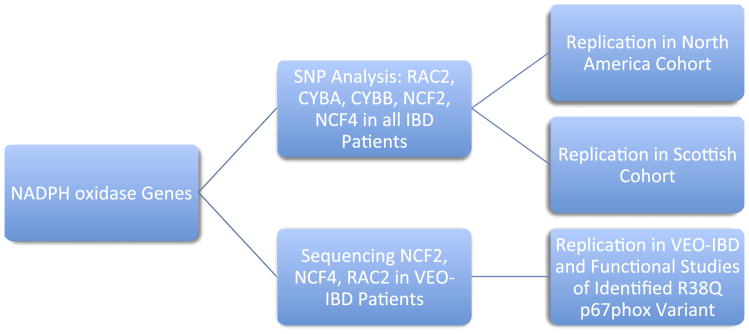
Many patients with VEO-IBD (defined as onset of disease before 10 years of age based on the Paris IBD Classification24) present with clinical intestinal features, including perianal disease and pancolitis, similar to those observed in CGD. We therefore sequenced coding regions of NADPH oxidase complex genes (including NCF2, NCF4 and RAC2) in 10 patients with VEO-IBD with pancolitis and perianal disease without evidence of immunodeficiency, as determined by commercial genetic testing for known CGD mutations, and a negative comprehensive immunological investigation including normal T, B and NK cell populations, response to immunisations, normal serum immunoglobulins and sequencing of the IL10RA/B genes. An outline of the experimental approach is shown in figure 1.
Figure 1.
Flow diagram of NADPH oxidase genetic experiments. VEO-IBD, very early onset inflammatory bowel disease.
All exons and flanking intron sequences in NCF2, NCF4 and RAC2 were successfully sequenced in the 10 patients (data not shown). We found a novel NCF2 (encoding p67phox) heterozygote variant c.113 G→A (p67phox R38Q) in one patient. This male patient was diagnosed with IBD at 1 year of age with pancolitis (colonoscopy at age 1 and 3 years) and had developed perianal abscesses and small bowel disease by 2 years of age. He had low to normal reactive oxygen species (ROS) production as assessed by the nitroblue tetrazolium (NBT) test on two separate occasions (40 and 53; normal range 32–300). The patient had no history of chronic or significant infections. The identified NCF2 missense variant (c.113 G→A) has not previously been reported and is not known to cause CGD (NCF2base; Mutation Registry for Autosomal Recessive Chronic Granulomatous Disease, http://bioinf.uta.fi/NCF2base/). The c.113 G-A variant in NCF2 has been subsequently assigned the rs#-rs147415774 (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?rs=147415774).
To determine if this novel variant plays a role in VEO-IBD, we examined an additional 139 patients with VEO-IBD, 341 patients with IBD and 480 healthy controls. Four of the 139 patients with VEO-IBD were found to be heterozygous for the c.113 G→A variant compared with one of the 341 patients with IBD and one of the 480 controls. We next carried out genotyping of this novel variant in a second independent VEO-IBD cohort consisting of 119 patients. Four of these patients were found to be heterozygous and two homozygous for the c.113 G→A variant. Overall, 4% (11/268) of patients with VEO-IBD carried the c.113 G→A NCF2 variant compared with 0.2% (1/480) of healthy controls studied (p=1.3×10−5, OR 23.8 (95% CI 3.9 to 142.5); Fisher exact test). All patients with VEO-IBD carrying the NCF2 c.113 G→A variant had extensive colonic disease, five had perianal disease and one had significant arthritis (table 2).
Table 2. Phenotype of patients with inflammatory bowel disease with R38Q p67phox.
| VEO-IBD R38Q p67phox | Genotype of risk allele | Gender | Age at diagnosis (years) | Disease type at diagnosis | Disease location | Perianal disease | Other |
|---|---|---|---|---|---|---|---|
| 1 (index patient) | Heterozygote | M | <1 | CD | Ileocolonic (L3) | Yes | |
| 2 | Heterozygote | M | 2 | IBDU | Colonic (L2) | Yes | |
| 3 | Heterozygote | F | 4.2 | CD | Colonic (L2) | Yes | |
| 4 | Heterozygote | F | 4.6 | CD | Ileocolonic (L3) | No | Severe arthritis |
| 5 | Heterozygote | F | <1 | CD | Ileocolonic (L3) | Yes | |
| 6 | Heterozygote | M | 9 | CD | Ileocolonic (L3) | No | Extensive small bowel disease |
| 7 | Heterozygote | F | 9.7 | IBDU | Colonic (L2) | No | |
| 8 | Heterozygote | M | 8.8 | IBDU | Ileocolonic (L3) | No | |
| 9 | Heterozygote | M | 7.2 | IBDU | Colonic (L2) | No | |
| 10 | Homozygote | F | 5.5 | IBDU | Colonic (L2) | No | |
| 11 | Homozygote | F | 7.7 | CD | Ileocolonic (L3) | Yes |
L3, ileocolonic CD; L2, colonic only CD.
CD, Crohn's disease; IBDU, inflammatory bowel disease unclassified; VEO-IBD, very early onset inflammatory bowel disease.
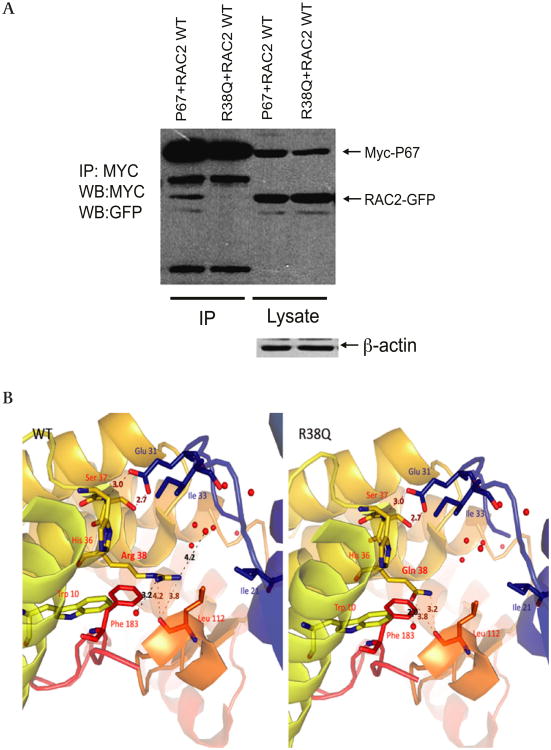
To determine the functional significance of the c.113 G→A NCF2 variant, we examined the protein product R38Q p67phox. The arginine at position 38 of p67phox has been shown to be important in RAC binding in a yeast two-hybrid assay25 and binding of these proteins is known to be an essential step in the assembly and activation of the NOX2 NADPH oxidase.3 Co-immunoprecipitation studies (figure 2A) showed that the p67phox R38Q variant has decreased binding to RAC2 in comparison with wild-type p67phox. Computational analysis showed that the reduced binding of the p67phox R38Q variant to RAC occurs as a result of a shorter side chain and loss of charge at amino acid 38 in p67phox; both are required to stabilise the interaction between p67phox and RAC (figure 2B).
Figure 2.
Functional studies of the p67phox R38Q polymorphism. (A) p67phox R38Q variant reduces binding to RAC. Interaction between RAC and wild-type p67phox and p67phox R38Q. GFP-tagged RAC2 and MYC-tagged p67phoxand p67phox R38Q were co-transfected into T293 cells. After 20 h p67phox was immunoprecipitated using anti-MYC antibody and blotted for MYC and GFP. RAC2 showed a 51% reduction of binding to p67phox R38Q in comparison with RAC2 binding to wild-type p67phox. Representative blot of three independent experiments. IP, immunoprecipitation; WB, western blot. (B) p67phox R38Q variant results in a conformational change. The model shows a close-up view of the RAC binding cavity of the wild-type p67phox and p67phox R38Q as adapted from the RAC/p67phox complex crystal structure (PDB 1E96). The proteins are shown in ribbon representation, coloured according to secondary structure, with RAC residues labelled in blue and p67phox residues labelled in red. Residues within 7×10-8 cm of p67phox R38 are represented by stick models with nitrogen and oxygen atoms indicated in blue and red, respectively. Water molecules found within the crystal structure are denoted by red spheres. Dashed lines denote putative hydrogen bond interactions between p67phox wild-type and p67phox L112 as well as water molecule 2013. In the p67phox R38Q variant, bond distances between p67phox R38Q and p67phox L112 are reduced, the interaction with water molecule 2013 is lost, and a new putative interaction between p67phox R38Q and water molecule 2025 forms. In addition, owing to the reduced carbon chain length between the arginine and glutamine side chains, a hydrophilic cavity is present perpendicular to the RAC/p67phox R38Q interface that is visible through electron density mapping (not shown). All distances labelled are ×10-8 cm. Images were generated with PyMOL (PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC.)
NADPH oxidase complex SNP analysis
We also examined the association of 62 genotyped SNPs in the NADPH oxidase complex, RAC2, CYBA, CYBB, NCF2 and NCF4 with IBD, CD and UC in the 2049 subjects in the discovery cohort (656 with CD, 544 with UC and 849 controls, table 1; see also table 3A–C in online supplement for full detailed analysis). The experimental approach is shown in figure 1. We found a novel association with RAC2 and CD after Bonferroni correction threshold for 62 SNPs examined in IBD/UC/CD (α=2.0×10−4; table 1). RAC2 is located on chromosome 22 (35 951 258 to 35 970 251; figure 1 in the online supplement shows the LD plot of RAC2 tag SNPs). To further examine this association, we genotyped four RAC2 SNPs with modest association in the discovery cohort (p<0.005; rs5756564, rs6572, rs9607431 and rs2239774) in two independent cohorts (figure 1). The replication cohort comprised 1836 Caucasian subjects including 443 patients with CD, 477 with UC and 916 controls recruited from North America (table 4A in online supplement) and a second validation cohort from Scotland comprised 2449 Caucasian subjects including 691 patients with CD, 615 with UC and 1143 controls (table 4B in online supplement). No RAC2 SNP association was replicated in both independent cohort (table 3). However, in a combined analysis of the discovery, replication and validation cohorts (1790 patients with CD, 1636 with UC and 2908 controls), the RAC2 association signal for rs6572 was significantly associated with CD and not UC (table 3; see also table 5 in online supplement). It is important to note that no RAC2 SNP achieved genome-wide association significance in this study nor was significantly associated in all three populations studied. One SNP (rs6572; pcombined=4.8×10−5, OR 1.2 (95% CI 1.09 to 1.29)) remained significantly associated with CD after the Bonferroni correction threshold for the number of SNPs genotyped in this study and the phenotypes examined (62 SNPs examined in IBD/UC/CD, α=2.0×10−4). Functional analysis showed that rs6572 did not affect RAC2 splicing or gene expression based on genotype, and imputation analysis did not provide further information regarding the possible casual variants (data not shown).
Table 3. Combined analysis for discovery, NIDDK replication and Scottish validation cohorts showing association between RAC2 SNPs and Crohn's disease.
| Discovery Toronto cohort | NIDDK cohort | Scottish cohort | Combined analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||
| RAC2 gene SNP | Chr 22 Position | p Value | OR (95% CI) | p Value | OR (95% CI) | p Value | OR (95% CI) | p Value | OR (95% CI) |
| rs5756564 | 35 942 049 | 6.5×10−4 | 0.75 (0.64 to 0.88) | 5.0×10−2 | 0.84 (0.74 to 0.1.0) | 5.4×10−2 | 0.95 (0.83 to 1.10) | 4.0×10-4 | 0.85 (0.78 to 0.93) |
| rs6572 | 35 951 391 | 3.8×10−4 | 1.33 (1.13 to 1.57) | 5.0×10−1 | 1.04 (0.96 to 1.22) | 1.0×10−2 | 1.18 (1.03 to 1.36) | 5.0×10−5 | 1.19 (1.09 to 1.29) |
| rs9607431 | 35 959 884 | 8.5×10−5 | 1.57 (1.25 to 1.98) | 2.7×10−2 | 0.75 (0.58 to 0.96) | 5.4×10−1 | 1.06 (0.87 to 1.30) | 4.2×10−2 | 1.36 (1.01 to 1.25) |
| rs2239774 | 35 967 599 | 1.5×10−4 | 1.49 (1.21 to 1.84) | 4.4×10−2 | 0.78 (0.62 to 0.99) | 6.9×10−2 | 1.03 (0.86 to 1.25) | 5.0×10−2 | 1.12 (0.96 to 1.21) |
p Values are presented as uncorrected.
Combined analysis of the discovery, replication and validation cohorts (1790 patients with Crohn's disease and 2908 controls).
In a secondary analysis we further examined the association of RAC2 with CD by disease phenotype using the IBD Montreal Classification criteria.16 In the combined analysis of the discovery replication and validation cohorts (1790 patients with CD, 1636 with UC and 2908 controls) we found further associations with the RAC2 SNPs, with the strongest signal in ileocolonic CD (Montreal Classification L3) with two SNPs: rs6572 (pcombined=6.0×10−6, OR 1.3 (95% CI 1.16 to 1.46)) and rs5756564 (pcombined=3.0×10−5, OR 0.77 (95% CI 0.68 to 0.87); see tables 5 and 6 in online supplement for summary). The association with RAC2 SNPs and phenotypes was not replicated in all three cohorts examined.
Replication of NCF4
We also replicated the association between the NCF4 rs4821544 and CD (p=5.1×10−3, OR 1.25 (95% CI 1.07 to 1.1.47); table 1). Figure 1 in the online supplement shows the LD plot of NCF4 tag SNPs used in this study. As the association with NCF4 rs4821544 was originally reported in an ileal CD GWAS and replication study,11,12 we examined ileal CD and also showed an association (Montreal Classification L1, p=3.5×10−3, OR 1.46 (95% CI 1.13 to 1.89); see tables 5 and 7 in online supplement for subphenotype analysis). The patients used in this study did not contribute to the original ileal CD NIDDK GWAS cohort that initially described the NCF4 association signal.11 The rs4821544 SNP did not alter gene expression or splicing of NCF4 based on genotype, and imputation analysis did not provide further information regarding the possible casual variants (data not shown).
Discussion
Phagocytic leucocytes (especially neutrophils) are critical for innate immunity and clear debris and prevent local damage in the bowel. The importance of phagocyte function in the development of non-infectious colitis is seen in a number of congenital disorders,26 particularly the impaired respiratory burst demonstrated in glycogen storage disease Ib and CGD.27–29 Patients with CGD often develop severe perianal disease, stricturing small bowel disease and discontinuous colitis that is endoscopically and histologically very similar to CD5 and even misdiagnosed as CD.30 A defective neutrophil respiratory burst has long been recognised as playing an important role in the pathogenesis of CD,8–10 but only recently have NADPH oxidase complex genes been implicated. We observed an association between both NCF2 and RAC2 SNPs with clinical phenotypes that resemble that seen in CGD. We chose to examine the VEO-IBD age group closely as patients diagnosed before the age of 10 years have a distinct phenotype with a high familial aggregation and greater tendency to present with severe colonic disease.31–33 This age group is most likely influenced by genetic alterations such as those seen in IL10RA/B and in other phenocopies of IBD such as CGD.34 We hypothesised that polymorphisms in NADPH oxidase complex genes that reduce NADPH oxidase function but do not cause CGD are associated with an increased risk of developing IBD. We identified a novel variant in NCF2 (encoding p67phox). Rare homozygous mutations in NCF2 are known to cause autosomal recessive CGD mostly through complete loss of p67phox expression and an inability to produce ROS, and these patients with CGD often present with a milder phenotype.29,35,36 The identified variant arginine at position 38 of p67phox has been shown to be important in RAC2 binding25; variation at this position reduces RAC binding. As p67phox binding to RAC2 is an essential step in the assembly and activation of NOX2 NADPH oxidase,3 we would expect that ROS production would be reduced but not absent as observed in our index patient. The patients with VEO-IBD with the p67phox R38Q variant all had extensive colonic involvement and five patients developed perianal disease. This phenotype resembles the colitis seen in CGD and in many patients with VEO-IBD. However, these patients had no evidence of immunodeficiency (as assessed by a comprehensive immunological investigation), indicating that the p67phox R38Q variant plays a role in the development of colitis only. Certainly, patients with CGD can present with CD-like colitis as the only manifestation of disease,37,38 and patients with CGD are at much greater risk of developing CD-like colitis.5–7 Interestingly, two patients were homozygotes for the p67phox R38Q variant. Neither patient had chronic infections or evidence to suggest CGD, and neither had worse disease at presentation. Long-term follow-up will be required to determine if there is a dosage effect, with more severe outcomes for individuals carrying two copies of this variant.
In this study we also identified the p67phox binding partner RAC2 as a CD susceptibility gene through a candidate gene approach. Although RAC2 mutations are not known to cause CGD, a rare dominant negative mutation in RAC2 has been shown to cause an immunodeficiency similar to CGD through inhibiting RAC2 from binding GTP39,40 The binding of RAC2 to p67phox is a critical step required for the activation of the NADPH oxidase complex and the production of ROS.41 We also replicated the association signal for NCF4 (encoding p40phox) and CD. The p40phox protein serves to enhance delivery of p47phox and p67phox to the membrane.3 Recently, a paediatric patient was found to have novel compound heterozygote NCF4 mutations which reduced the gene product p40phox binding to PtdIns(3)P, essential for NADPH oxidase phagocytosis-induced oxidant production in human neutrophils.38 Interestingly, this patient presented with granulomatous colitis and perianal disease without evidence of immune deficiency38 These studies demonstrate the importance of p40phox in the development of colitis.
Current estimates suggest that CD GWAS have explained 23% of the inherited contribution to the risk of CD.13 Thus, there remains substantial ‘missing heritability’ and it is broadly anticipated that this will not be fully explained by simply expanding the GWAS approach with larger numbers of patients.13 Our candidate gene approach suggests an association between RAC2 and CD, although these results need to be interpreted with caution as no RAC2 SNP replicated in all three cohorts examined and the association signal did not meet genome-wide significance. We observed both risk and protective signals for RAC2 suggesting that, similar to the IL23R signal, these SNPs are involved in alternate splicing or altered expression of RAC2 that may play opposing roles.42 However, this association was not reported in the International IBD Genetics Consortium (IIBDGC) meta-analysis as the gene is poorly tagged in that genome-wide array13 The SNPs used in that study did not provide adequate coverage for our lead SNP (GWAS SNPs rs132515, rs2413552 and rs5757362 r2<0.25 to rs6572); further large-scale analysis will therefore be required to validate these results. Our lead NCF4 SNP (rs4821544) was associated with CD in the NIDDK GWAS (pdiscovery=2.89×10−5; preplication=0.009; OR 1.19) which only included patients with ileal disease and was independently replicated in ileal CD.11,12 This association was not observed in a large European study43 In the first IIBDGC meta-analysis (including all disease locations of CD) the pmeta-analysis was 0.0078 for rs48215442; hence, this SNP was not taken forward for independent replication. In the expanded second IIBDGC meta-analysis, pmeta-analysis for rs4821544 was 1.80×10−5.13 It is therefore possible that NCF4 SNP is associated with CD and that disease location may be important for further large-scale replication, although previous European studies suggest that geographical factors may also be important.
Recent studies have provided insight into the role of the NOX2 NADPH oxidase complex in the development of colitis. Animal models have shown that defects in NADPH oxidase complex genes can impair bacterial killing44 and cause susceptibility to autoimmunity through defects in ROS production.45 Furthermore, studies have shown that bacterial pathogens can cause excessive ROS production leading to disease through tissue destruction and inflammation and, on the other hand, other bacteria can modulate ROS-dependent neutrophil apoptosis thereby surviving and causing disease.46 In human studies the non-infectious inflammation (especially colitis) that occurs in patients with CGD has been linked to the NADPH oxidase regulation of the inflammosome47 and autophagy48 pathways involved in the pathogenesis of CD. It is therefore plausible that polymorphisms that increase or decrease ROS production may be associated with the development of human colitis.
The NADPH oxidase complex has been implicated in the pathogenesis of human disease from arthritis to cancer.46 Overall, our studies of the NADPH oxidase complex provide further evidence supporting defective neutrophil ROS production in the pathogenesis of CD. The p67phox R38Q variant suggests that reduced ROS is important in the development of early onset disease, while the candidate gene approach shows the importance of the NADPH oxidase complex in the development of CD. Our study demonstrates that SNPs in the core NADPH oxidase complex genes are not associated with IBD. This is an interesting finding as variations in genes that are responsible for localisation of the NADPH oxidase complex (including p47phox and p67phox and RAC2) appear to be associated with IBD, and this mislocalisation or delay in localisation of the NADPH oxidase complex to the membrane may be important in the pathogenesis of IBD.
We propose that partial impairment of NADPH oxidase function coupled with other genetic variations, especially those in innate immunity genes, contribute to the pathogenesis of CD without development of the severe innate immune deficiency observed in CGD.
Supplementary Material
Significance of this study.
What is already known about this subject?
Defects in the NADPH oxidase complex genes cause X-linked and autosomal recessive chronic granulomatous disease (CGD).
Patients with CGD are more susceptible to developing Crohn's-like colitis and perianal disease.
Polymorphisms in the NADPH oxidase gene NCF4 were found to be associated with ileal Crohn's disease (CD).
What are the new findings?
Identification of a novel variant in NCF2 that is associated with very early onset inflammatory bowel disease (IBD) and results in reduced protein binding.
Genetic studies suggest the NADPH oxidase complex gene RAC2 as a CD susceptibility gene.
Replication of the NCF4 gene association with ileal CD.
How might it impact on clinical practice in the foreseeable future?
These results implicate the NADPH oxidase complex in the pathogenesis of IBD.
Identification of novel variants in these genes may lead to alternative therapies for a subgroup of patients with defective reactive oxygen species production.
Acknowledgments
The authors thank ET and his family, and all IBD patients and families for participating in this research. The authors also thank the NIDDK IBD Genetics Consortium for providing control samples and the International IBD Genetics Consortium (IIBDGC) for providing genotyping data for RAC2 and NCF4. They also acknowledge the work of Karoline Fiedler at the Hospital for Sick Children, Joanne Stempak at Mount Sinai and Dr Elaine Nimmo, Dr Johan Van Limbergen and Hazel Drummond in Edinburgh.
Funding AMM is supported by a transition award from the Crohn's and Colitis Foundation of Canada (CCFC)/Canadian Association of Gastroenterology (CAG)/Canadian Institute for Health Research (CIHR), a Canadian Child Health Clinician Scientist Program (Strategic Training Initiatives in Health Research Program—CIHR) Award, an Early Researcher Award from the Ontario Ministry of Research and Innovation and a CDHNF/NASPGHAN George Ferry Young Investigator Development Award. DCW is the holder of a Medical Research Council Patient Cohorts Research Initiative award (G0800675). Financial assistance was also provided by the Wellcome Trust Programme Grant (072789/Z/03/Z), Action Medical Research, the Chief Scientist Office of the Scottish Government Health Department and the GI/Nutrition Research Fund, Child Life and Health, University of Edinburgh. RHD is supported by NIH/NIDDK grant (DK062420). ADP holds a Canada Research Chair in Genetics of Complex Diseases. MSS is supported by the Gale and Graham Wright Research Chair in Digestive Diseases at Mount Sinai Hospital and funding from CCFC and NIH/NIDDK (DK-06-2423). JHB holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. Funding was provided by a CIHR operating grant (MOP97756) to AMM and JHB.
Footnotes
Competing interests None.
Ethics approval Hospital for Sick Children REB.
Contributors AMM, JHB and MSS conceived and designed all experiments. MSS, AMM, AMG, RHD, RR, JC, DCW and JS provided study samples. WX, TW and ADP analysed the data. VMW, GL, JCG, MS, RF, GL, JB, RM and CH performed functional analysis under supervision of AMM, JHB, MG. AMM wrote the manuscript with JHB and MSS and contributions from all authors.
Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–78. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;8:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nauseef WM. Biological roles for the NOX family NADPH oxidases. J Biol Chem. 2008;283:16961–5. doi: 10.1074/jbc.R700045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heyworth PG, Cross AR, Curnutte JT. Chronic granulomatous disease. Curr Opin Immunol. 2003;15:578–84. doi: 10.1016/s0952-7915(03)00109-2. [DOI] [PubMed] [Google Scholar]
- 5.Marks DJ, Miyagi K, Rahman FZ, et al. Inflammatory bowel disease in CGD reproduces the clinicopathological features of Crohn's disease. Am J Gastroenterol. 2009;104:117–24. doi: 10.1038/ajg.2008.72. [DOI] [PubMed] [Google Scholar]
- 6.Schappi MG, Smith VV, Goldblatt D, et al. Colitis in chronic granulomatous disease. Arch Dis Child. 2001;84:147–51. doi: 10.1136/adc.84.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werlin SL, Chusid MJ, Caya J, et al. Colitis in chronic granulomatous disease. Gastroenterology. 1982;82:328–31. [PubMed] [Google Scholar]
- 8.Ward M. The pathogenesis of Crohn's disease. Lancet. 1977;2:903–5. doi: 10.1016/s0140-6736(77)90835-2. [DOI] [PubMed] [Google Scholar]
- 9.Verspaget HW, Mieremet-Ooms MA, Weterman IT, et al. Partial defect of neutrophil oxidative metabolism in Crohn's disease. Gut. 1984;25:849–53. doi: 10.1136/gut.25.8.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segal AW, Loewi G. Neutrophil dysfunction in Crohn's disease. Lancet. 1976;2:219–21. doi: 10.1016/s0140-6736(76)91024-2. [DOI] [PubMed] [Google Scholar]
- 11.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts RL, Hollis-Moffatt JE, Gearry RB, et al. Confirmation of association of IRGM and NCF4 with ileal Crohn's disease in a population-based cohort. Genes Immun. 2008;9:561–5. doi: 10.1038/gene.2008.49. [DOI] [PubMed] [Google Scholar]
- 13.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anon The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 15.de Bakker PI, Yelensky R, Pe'er I, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 16.Silverberg MS, Satsangi J, Ahmad T, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a working party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19(Suppl A):5–36. doi: 10.1155/2005/269076. [DOI] [PubMed] [Google Scholar]
- 17.Silverberg MS, Cho JH, Rioux JD, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–20. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imielinski M, Baldassano RN, Griffiths A, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335–40. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freidlin B, Zheng G, Li Z, et al. Trend tests for case-control studies of genetic markers: power, sample size and robustness. Hum Hered. 2002;53:146–52. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- 22.Clayton D. Testing for association on the X chromosome. Biostatistics. 2008;9:593–600. doi: 10.1093/biostatistics/kxn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis. 2011;17:1314–21. doi: 10.1002/ibd.21493. [DOI] [PubMed] [Google Scholar]
- 25.Koga H, Terasawa H, Nunoi H, et al. Tetratricopeptide repeat (TPR) motifs of p67 (phox) participate in interaction with the small GTPase Rac and activation of the phagocyte NADPH oxidase. J Biol Chem. 1999;274:25051–60. doi: 10.1074/jbc.274.35.25051. [DOI] [PubMed] [Google Scholar]
- 26.Rahman FZ, Marks DJ, Hayee BH, et al. Phagocyte dysfunction and inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:1443–52. doi: 10.1002/ibd.20449. [DOI] [PubMed] [Google Scholar]
- 27.Couper R, Kapelushnik J, Griffiths AM. Neutrophil dysfunction in glycogen storage disease Ib: association with Crohn's-like colitis. Gastroenterology. 1991;100:549–54. doi: 10.1016/0016-5085(91)90229-e. [DOI] [PubMed] [Google Scholar]
- 28.Ament ME, Ochs HD. Gastrointestinal manifestations of chronic granulomatous disease. N Engl J Med. 1973;288:382–7. doi: 10.1056/NEJM197302222880802. [DOI] [PubMed] [Google Scholar]
- 29.Stasia MJ, Li XJ. Genetics and immunopathology of chronic granulomatous disease. Semin Immunopathol. 2008;30:209–35. doi: 10.1007/s00281-008-0121-8. [DOI] [PubMed] [Google Scholar]
- 30.Freudenberg F, Wintergerst U, Roesen-Wolff A, et al. Therapeutic strategy in p47-phox deficient chronic granulomatous disease presenting as inflammatory bowel disease. J Allergy Clin Immunol. 2011;125:943–6.e1. doi: 10.1016/j.jaci.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 31.Paul T, Birnbaum A, Pal DK, et al. Distinct phenotype of early childhood inflammatory bowel disease. J Clin Gastroenterol. 2006;40:583–6. doi: 10.1097/00004836-200608000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Griffiths AM. Specificities of inflammatory bowel disease in childhood. Best Pract Res Clin Gastroenterol. 2004;18:509–23. doi: 10.1016/j.bpg.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Heyman MB, Kirschner BS, Gold BD, et al. Children with early-onset inflammatory bowel disease (IBD): analysis of a pediatric IBD consortium registry. J Pediatr. 2005;146:35–40. doi: 10.1016/j.jpeds.2004.08.043. [DOI] [PubMed] [Google Scholar]
- 34.Glocker EO, Kotlarz D, Boztug K, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–45. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gentsch M, Kaczmarczyk A, van Leeuwen K, et al. Alu-repeat-induced deletions within the NCF2 gene causing p67-phox-deficient chronic granulomatous disease (CGD) Hum Mutat. 2011;31:151–8. doi: 10.1002/humu.21156. [DOI] [PubMed] [Google Scholar]
- 36.Francke U, Hsieh CL, Foellmer BE, et al. Genes for two autosomal recessive forms of chronic granulomatous disease assigned to 1q25 (NCF2) and 7q11.23 (NCF1) Am J Hum Genet. 1990;47:483–92. [PMC free article] [PubMed] [Google Scholar]
- 37.Marciano BE, Rosenzweig SD, Kleiner DE, et al. Gastrointestinal involvement in chronic granulomatous disease. Pediatrics. 2004;114:462–8. doi: 10.1542/peds.114.2.462. [DOI] [PubMed] [Google Scholar]
- 38.Matute JD, Arias AA, Wright NA, et al. A new genetic subgroup of chronic granulomatous disease with autosomal recessive mutations in p40 phox and selective defects in neutrophil NADPH oxidase activity. Blood. 2009;114:3309–15. doi: 10.1182/blood-2009-07-231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams DA, Tao W, Yang F, et al. Dominant negative mutation of the hematopoietic-specific Rho GTPase, Rac2, is associated with a human phagocyte immunodeficiency. Blood. 2000;96:1646–54. [PubMed] [Google Scholar]
- 40.Ambruso DR, Knall C, Abell AN, et al. Human neutrophil immunodeficiency syndrome is associated with an inhibitory Rac2 mutation. Proc Natl Acad Sci U S A. 2000;97:4654–9. doi: 10.1073/pnas.080074897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nauseef WM. Assembly of the phagocyte NADPH oxidase. Histochem Cell Biol. 2004;122:277–91. doi: 10.1007/s00418-004-0679-8. [DOI] [PubMed] [Google Scholar]
- 42.Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glas J, Seiderer J, Pasciuto G, et al. rs224136 on chromosome 10q21.1 and variants in PHOX2B, NCF4, and FAM92B are not major genetic risk factors for susceptibility to Crohn's disease in the German population. Am J Gastroenterol. 2009;104:665–72. doi: 10.1038/ajg.2008.65. [DOI] [PubMed] [Google Scholar]
- 44.Ellson CD, Davidson K, Ferguson GJ, et al. Neutrophils from p40phox-/- mice exhibit severe defects in NADPH oxidase regulation and oxidant-dependent bacterial killing. J Exp Med. 2006;203:1927–37. doi: 10.1084/jem.20052069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hultqvist M, Olofsson P, Holmberg J, et al. Enhanced autoimmunity, arthritis, and encephalomyelitis in mice with a reduced oxidative burst due to a mutation in the Ncf1 gene. Proc Natl Acad Sci U S A. 2004;101:12646–51. doi: 10.1073/pnas.0403831101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinn MT, Ammons MC, Deleo FR. The expanding role of NADPH oxidases in health and disease: no longer just agents of death and destruction. Clin Sci (Lond) 2006;111:1–20. doi: 10.1042/CS20060059. [DOI] [PubMed] [Google Scholar]
- 47.Meissner F, Seger RA, Moshous D, et al. Inflammasome activation in NADPH oxidase defective mononuclear phagocytes from patients with chronic granulomatous disease. Blood. 2010;116:1570–3. doi: 10.1182/blood-2010-01-264218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J, Canadien V, Lam GY, et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A. 2009;106:6226–31. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.