Abstract
G-rich regions have the potential to form G4 DNA upon replication, which can lead to genomic instability. FANCJ, a G4 DNA helicase, has been shown to be critical for the stability of regions that match the G4 signature motif by experiments analyzing its nematode homolog.
DNA has considerable potential to form structures other than canonical Watson–Crick duplexes, and this can contribute to genomic instability. An example of such a structure is the triplet repeat, which forms a hairpin structure in vitro and which, in living cells, can undergo sequence expansion that manifests as neurodegenerative disease [1]. Guanine (G)-rich DNA also has structural potential and readily forms G4 DNA (also called G-quadruplex DNA), in which guanines joined in G-quartets stabilize interactions between four DNA strands [2,3]. The signature motif that predicts G4-DNA formation is four tracts of three guanines that may be separated by other bases: G3NxG3NxG3NxG3. Increased tract length and G-richness increases the number of Gs available to join G-quartets and the probability of G4-DNA formation, enabling a single sequence to form polymorphic structures of surprising variety [2,3].
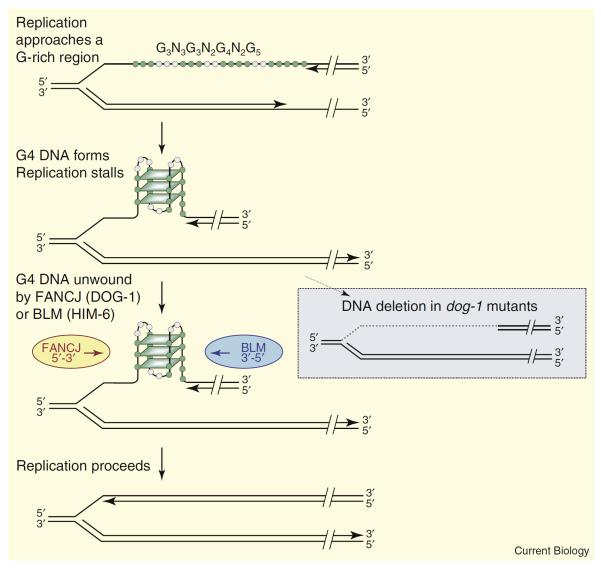
G4 DNA can form when the DNA duplex undergoes transient denaturation during the course of replication (Figure 1) or transcription. G4 DNA is predicted to block replication and, thus, cause genomic instability unless somehow repaired. An exciting report by Kruisselbrink et al. [4], in a recent issue of Current Biology, now demonstrates that the Caenorhabditis elegans dog-1 gene is critical for the stability of G-rich genomic regions. The dog-1 (deletion of guanine-rich regions) gene encodes a DEAH family helicase shown to be necessary for the stability of mononucleotide G tracts [5]. Using reporter constructs that allow direct visualization of cells that harbor destabilized tracts within whole animals, Kruisselbrink et al. [4] confirmed that result and showed that instability occurs both early and late in the worm's life cycle. Interrogating a custom array bearing the entire arm of one chromosome as well as a variety of sequences with structural potential (matches to the G4 signature, repeats, palindromes, telomeric DNA) revealed that only sequences that conform to the G4 signature are unstable in dog-1 mutants. Instability was proportional to tract length and G-richness. Thus, dog-1 directs a G4 repair pathway that is critical to genomic stability. Other repair pathways appeared not to contribute to the stability of G-rich regions, consistent with a previous genetic analysis [6].
Figure 1.
G4 DNA formation and repair at a replication fork.
The figure shows G4 DNA formed within the sequence G3N3G3N2G4N2G5. G, green; other bases, gray; parallelograms represent G-quartets. Structure formation is illustrated in the lagging strand, which is thought to be more prone to formation of alternative structures because it persists in a denatured state longer than the leading strand. Only a single structure is shown in the figure, but note that three of the guanines in the sequence that are shown outside of the quartets could be included by changing the register of individual G-runs, and thus contribute to structural polymorphism. FANCJ (C. elegans DOG-1) or BLM (C. elegans HIM-6) unwind G4 DNA to allow replication to proceed (left). In C. elegans dog-1 mutants, deletion occurs, with the 3′ boundary defined by the 3′ end of the G4 signature motif, and the 5′ boundary possibly determined by the location of the replication origin (right, dotted gray line indicates the deleted region).
Interest in DOG-1 has been fueled by the recent report from Rose, Boulton and collaborators [7] that C. elegans dog-1 is the homolog of human FANCJ. FANCJ is a member of one of 13 complementation groups associated with Fanconia anemia, a human genetic disease characterized by bone marrow failure, genomic instability, predisposition to cancer and developmental anomalies [8,9]. Even more recently, purified recombinant human FANCJ was shown to unwind G4 DNA in vitro, acting as a 5′→3′ helicase [10]. This activity provides the biochemical and human correlate for the genetic analysis in worms.
Figure 1 proposes a simplified model for replication-associated G4 DNA repair, building upon the relationship between two pairs of C. elegans and human homologs — DOG-1 and FANCJ, and HIM-6 and BLM. G4-DNA unwinding by FANCJ would relieve the block to replication, and allow a stalled fork to proceed. Deletions characterized in dog-1 mutants extend 5′ of the G-rich region, with a median length of about half that of an Okazaki fragment, suggesting that the 5′ deletion boundary falls at or near the replication origin [4], as shown (Figure 1, left). As an alternative to G4-DNA unwinding, specialized homologous recombination machinery could intervene [6,11], consistent with participation of the Fanconia anemia (FA) pathway in repair of interstrand crosslinks [8,9]; or FANCJ could recruit other factors to bypass the lesion. Nonetheless, unwinding is a straightforward mechanism for the elimination of G4 DNA, and one that been clearly defined in biochemical terms.
Human BLM helicase, a RecQ helicase, also unwinds G4 DNA [12]. The C. elegans homolog of BLM is HIM-6. Genetic analysis has shown that, while G-rich regions are not unstable in him-6 mutant animals, they are several-fold more unstable in dog-1; him-6 double mutants than in dog-1 single mutants [6]. Thus, HIM-6 (BLM) provides a secondary repair pathway that is active when DOG-1 (FANCJ) is absent but unnecessary in wild-type animals. In humans, another RecQ family member, WRN helicase, unwinds G4 DNA [13,14] and is active at G-rich telomeres [15].
The model in Figure 1 suggests an explanation for why FANCJ (DOG-1) and not BLM (HIM-6) may dominate replicative repair of G4 DNA. FANCJ and BLM both require single-stranded DNA tails to initiate unwinding but unwind with opposite polarities (5′→3′ and 3′→5′, respectively). When replication stalls, the DNA 5′ of the G4 structure unreplicated and single-stranded and may associate with replication protein A (RPA), which stimulates FANCJ [10]. In contrast, the 3′ end of the G4 structure may be replicated and therefore duplex or occluded by stalled replication factors and thus unable to provide the single-stranded tail necessary for BLM-mediated unwinding.
G4 DNA can form not only during replication but also upon transcription [16] and may block the progression of both prokaryotic and eukaryotic RNA polymerases [17]. Transcription-induced G4 structures may occur in either DNA or RNA, but they exhibit strand bias and form only in regions with G-rich non-template strands. The factors implicated thus far in G4 DNA repair are closely associated with replication. While these factors may also repair transcription-induced structures, additional, specialized G4 repair pathways may be associated with transcription or the transcription apparatus.
The genomic instability exhibited by dog-1 mutant worms [4–6] identifies G-rich regions as sequences at risk. This potential instability may explain why tumor suppressor genes — including FANCJ and BLM — are far less G-rich than the genomic average [18]. Nonetheless, there are hundreds of thousands of regions in the human genome with potential to form G4 DNA [19]. Many of these are at conserved positions in critical genes, consistent with positive selection for this feature of genomic structure [18,20]. G4 helicases provide a repair mechanism essential to overcome the inherent instability of such G-rich regions.
References
- 1.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 2.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 3.Phan AT, Kuryavyi V, Patel DJ. DNA architecture: from G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, Tijsterman M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ defective C. elegans. Curr. Biol. 2008;18:900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- 6.Youds JL, O'Neil NJ, Rose AM. Homologous recombination is required for genome stability in the absence of DOG-1 in Caenorhabditis elegans. Genetics. 2006;173:697–708. doi: 10.1534/genetics.106.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Youds JL, Barber LJ, Ward JD, Collis SJ, O'Neil NJ, Boulton SJ, Rose AM. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol. Cell. Biol. 2008;28:1470–1479. doi: 10.1128/MCB.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniguchi T, D'Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- 9.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 10.Wu Y, Shin-Ya K, Brosh RM., Jr. FANCJ helicase defective in Fanconi anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward JD, Barber LJ, Petalcorin MI, Yanowitz J, Boulton SJ. Replication blocking lesions present a unique substrate for homologous recombination. EMBO J. 2007;26:3384–3396. doi: 10.1038/sj.emboj.7601766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 13.Fry M, Loeb LA. Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 14.Mohaghegh P, Karow JK, Brosh RM, Jr., Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 16.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tornaletti S, Park-Snyder S, Hanawalt PC. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J. Biol. Chem. 2008;283:12756–12762. doi: 10.1074/jbc.M705003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]