Abstract
Background and Aims
Understanding the factors that shape variation in genetic diversity across the geographic ranges of species is an important challenge in the effort to conserve evolutionary processes sustaining biodiversity. The historical influences leading to a central–marginal organization of genetic diversity have been explored for species whose range is known to have expanded from refugia after glacial events. However, this question has rarely been addressed for Mediterranean endemic plants of azonal habitats such as rocky slopes or screes. In this context, this comprehensive study examined molecular and field data from Arenaria provincialis (Caryophyllaceae), a narrow endemic plant of south-eastern France.
Methods
Across the whole geographic range, an investigation was made of whether high levels of abundance and genetic diversity (estimated from amplified fragment length polymorphism markers) are centrally distributed, to evaluate the relevance of the central–marginal hypothesis. Phylogeographic patterns inferred from chloroplast DNA (cpDNA) were used, applying Bayesian methods to test the influence of past biogeographic events. Multivariate analysis combining phylogeographic and ecological data was used to reveal the historical and ecological distinctiveness of populations.
Key Results
Despite the narrow distribution of A. provincialis, a high level of nucleotide variation is found within cpDNA loci, supporting its persistence throughout the Pleistocene period. The area characterized by the highest genetic diversity is centrally located. Structured phylogeography and Bayesian factor analysis supported the hypothesis that the central area of the distribution was the source of both westward and eastward migrations, probably during arid periods of the Pleistocene, and more recently was a crossroads of backward migrations. By contrast, the two areas located today at the range limits are younger, have reduced genetic diversity and are marginal in the ecological gradients.
Conclusions
This study highlights a case of strong population distinctiveness within a narrow range. Phylogeography sheds light on the historical role of the areas centrally situated in the distribution. The current range size and abundance patterns are not sufficient to predict the organization of genetic diversity.
Keywords: Arenaria provincialis, Caryophyllaceae, central–marginal hypothesis, genetic diversity, geographic range, ecological niche, Pleistocene, Provence, conservation, migration, population distinctiveness
INTRODUCTION
A fundamental issue in ecology and conservation biology is how evolutionary processes shape variation across the whole geographic range of a species (Brown, 1984; Lesica and Allendorf, 1995; Gaston, 2003; Guo, 2012). According to the central–marginal hypothesis (Eckert et al., 2008), genetic diversity decreases and genetic differentiation increases towards the periphery of the range. The ecological and evolutionary features of populations living at range limits of species (Sexton et al., 2009) are of particular importance in the context of global change (Sexton et al., 2011). Low habitat quality at the range margins may lead to higher rates of extinction, while increased isolation reduces the rate of recolonization as well as that of gene flow. However, the decline in genetic diversity at range limits is not a ubiquitous trend, either in multispecific comparisons (Eckstein et al., 2006; Eckert et al., 2008; Hardie and Hutchings, 2010; Guo, 2012) or within intraspecific case studies (Hoban et al., 2010). As emphasized by several authors (Garner et al., 2004; Eckert et al., 2008; Moeller et al., 2011), looking for the causes of a central–marginal structure of genetic diversity is particularly challenging because of the diverse alternative models and the effects of current demographic and ecological factors combined with persistent historical influences. Exploring historical influences, e.g. whether marginal populations are younger than core populations (Eckert et al., 2008), has been seen as relevant in the northern hemisphere, where Pleistocene climatic oscillations are still perceptible in the current patterns of widespread species distribution, through persistence in refugial areas and post-glacial migration roads (Taberlet et al., 1998; Hewitt, 2000, 2004; Petit et al., 2003; Hampe and Petit, 2005; Soltis et al., 2006). Thus, range shifts and distance from areas ensuring the persistence of populations – notably refugia (Stewart et al., 2010) – are often invoked to explain the coarse geographic changes in genetic variation (Garner et al., 2004; Hoban et al., 2010; references in Hardie and Hutchings, 2010).
Yet neither the historical influences shaping genetic variation nor the central–marginal hypothesis have been invoked in connection with Mediterranean endemic plants. Our aim here was to explore these questions, for three reasons. First, as underlined recently by Nieto Feliner (2011), the post-glacial colonization paradigm pattern is insufficient as a basis for exploring Mediterranean evolutionary patterns and processes during the Pleistocene. Second, in the highly structured landscapes of the Mediterranean, narrow endemic plants are so frequent (Médail and Verlaque, 1997; Casazza et al., 2005) that they are one of the main features of Mediterranean biodiversity (Thompson et al., 2005). Third, because their distribution is structured in small areas, narrow endemic plants could make it easier to test the central–marginal hypothesis at the scale of the whole range; moreover, they constitute key candidates for the study of persistence and speciation (Stebbins and Major, 1965; Chown, 1997). Recent studies of some narrow endemic Mediterranean plants (e.g. Medrano and Herrera, 2008; Molins et al., 2011; Mayol et al., 2012; Nicoletti et al., 2012) support the hypothesis of their long-term persistence during the Pleistocene and their fluctuating genetic diversity. Nevertheless, the link between historical influences, geographic range and genetic diversity in Mediterranean narrow endemic plants remains relatively unelucidated in the context of the central–marginal hypothesis. To explore this link, we focus on Arenaria provincialis (Caryophyllaceae) (Chater and Halliday, 1964), which inhabits the calcareous outcrops of Southern Provence in south-eastern France.
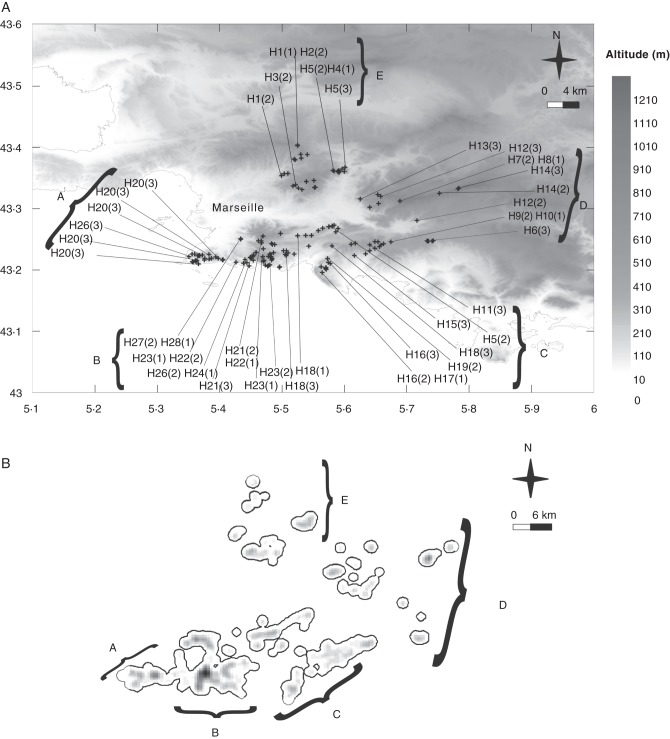
Recent studies on geographic distribution and ecological niches (Véla et al., 2008; Youssef et al., 2011) provide the basis for examining central and marginal populations of A. provincialis throughout its entire range, taking into account both abundance and ecological aspects. According to the central–marginal hypothesis, populations of A. provincialis living near the centre of the distribution are expected to exhibit more genetic diversity than populations living further afield (Fig. 1). Moreover, at range limits, populations of A. provincialis experience contrasting ecological conditions (Youssef et al., 2011) that could raise conservation issues if, concurrently, their genetic diversity decreased and their isolation increased with respect to more central populations.
Fig. 1.
Table 1.
| Group | Code | Latitude, longitude | Altitude (m) | Haplotypes |
|---|---|---|---|---|
| A (south-west) | M16 | 43·2091, 5·36656 | 39 | H20 (3) |
| M3A | 43·2232, 5·36799 | 365 | H20 (3) | |
| MCC | 43·2251, 5·36003 | 361 | H20 (3) | |
| MG4 | 43·2208, 5·35103 | 143 | H26 (3) | |
| MHM | 43·2184, 5·38723 | 395 | H20 (3) | |
| MS | 43·2231, 5·36841 | 408 | H20 (3) | |
| B (south) | Car8 | 43·2555, 5·52511 | 303 | H18 (1) |
| Carp | 43·2408, 5·48494 | 325 | H21 (2); H22 (1) | |
| Cer | 43·2467, 5·46236 | 239 | H21 (3) | |
| Gar10 | 43·2154, 5·48339 | 276 | – | |
| Gar14 | 43·2066, 5·47836 | 196 | H23 (1) | |
| Gar2 | 43·2278, 5·48654 | 398 | H21 (2) | |
| Gar4 | 43·2254, 5·48099 | 438 | – | |
| Gar8 | 43·2212, 5·47848 | 282 | – | |
| Gard9 | 43·2309, 5·50711 | 169 | H18 (3) | |
| LMO | 43·2113, 5·44408 | 35 | H26 (2); H24 (1) | |
| LSO | 43·2122, 5·42633 | 91 | H25 (1); H27 (2) | |
| Vau | 43·2503, 5·43408 | 200 | H27 (2); H28 (1) | |
| C (Centre) | Cap | 43·2044, 5·56306 | 316 | H16 (3) |
| Cap10 | 43·2101, 5·57739 | 194 | H19 (2) | |
| Cap13 | 43·2384, 5·58016 | 284 | H18 (3) | |
| Cap3 | 43·1954, 5·56473 | 282 | H16 (2); H17 (1) | |
| Car3 | 43·2637, 5·55819 | 299 | H5 (2) | |
| Cey5 | 43·2430, 5·61752 | 465 | – | |
| Cey12 | 43·2229, 5·61626 | 215 | H15 (3) | |
| FB6 | 43·2374, 5·65623 | 413 | H11 (3) | |
| D (east) | Cas3 | 43·2468, 5·73241 | 409 | H6 (3) |
| ColB | 43·3114, 5·68911 | 1017 | H7 (2); H8 (1) | |
| Cu1 | 43·2799, 5·71646 | 296 | H12 (2) | |
| FB4 | 43·2450, 5·67466 | 433 | H9 (2); H10 (1) | |
| Gem18 | 43·3193, 5·65844 | 735 | H12 (3) | |
| Gem2 | 43·3149, 5·62552 | 277 | H13 (3) | |
| Jou | 43·3326, 5·78356 | 1089 | H14 (3) | |
| Pcab | 43·3239, 5·75241 | 954 | H14 (2) | |
| E (north) | Cad11 | 43·3798, 5·51971 | 361 | H1 (2) |
| Cad15 | 43·3531, 5·49952 | 345 | – | |
| G10 | 43·3433, 5·53942 | 587 | H3 (2) | |
| Roq3 | 43·3667, 5·60136 | 294 | H5 (3) | |
| Roq5 | 43·3595, 5·60011 | 388 | H5 (2); H4 (1) | |
| Sav2 | 43·4028, 5·52514 | 582 | H1 (1); H2 (2) |
Here, occurrence data are used to show that abundance is not uniformly distributed across the range of A. provincialis, which confirms the importance of exploring the central–marginal hypothesis. First, we test the consistency of our hypotheses on a central–marginal structure of genetic diversity using multilocus markers, through analysis of amplified fragment length polymorphism (AFLP) markers. Second, we infer the historical influences throughout the distribution from the phylogeography, based on the nucleotide variation of four chloroplast loci analysed in a Bayesian framework. Finally, we use ecological data to link marginality in the ecological gradient with range position and phylogeography, thus revealing the conservation significance of our results.
MATERIALS AND METHODS
Study system and occurrence data
In south-eastern France, the winter annual plant Arenaria provincialis (Chater and Halliday, 1964) is distributed within an area of less than 145 km2 along an altitudinal gradient and in different rocky habitats (Véla et al., 2008; Baumel et al., 2009). Its southern limits occur in the driest context of steep scree slopes at low altitudes and short distances from the coastline, whereas its northern limits lie in colder conditions near the summits of the outcrops.
Variation in abundance of A. provincialis throughout its entire range was estimated from the variation in density of occurrence in samples collected during several field campaigns. Data were filtered to ensure that occurrences were at least 100 m apart, to avoid redundancy and overestimation of abundance due to oversampling. Five hundred and ninety-one occurrences were used to model abundance variation by applying an isotropic Gaussian kernel function (density.ppp function of the spatstat package; R Development Core Team, 2011).
Occurrence data were also used to set discrete, non-overlapping geographic units to organize measures of genetic diversity and for phylogeographic analysis (see below). Preliminary analyses distinguished five geographic units (A–E in Fig. 1A; Table 1). These geographic units were delineated by non-hierarchical clustering analysis (kmeans function in the stats package; R Development Core Team, 2011) from coordinates of A. provincialis occurrences. The barycentre of the distribution is situated in the northern part of area C.
DNA extraction and molecular methods
Forty populations of A. provincialis were sampled throughout its entire geographic range (Table 1) between April 2009 and May 2010. Leaves were dried with silica gel and then stored at −20 °C. Total DNA extraction followed the CTAB protocol of Doyle and Doyle (1987).
The polymorphism of the chloroplast genome was examined on two or three randomly sampled individuals per population (n = 94) for four plastid regions: the internal part of the matK gene; the trnK intron plus the 5′ part of matK; trnL-trnF; and the trnT-trnL intergenic spacer (Supplementary Data Table S1). Sequence characterization of the four chloroplast sequences and the combined matrix are reported in Table 2. According to the most recent phylogeny published on this genus (Youssef et al., 2011), A. provincialis and A. cinerea are sister species. The same chloroplast DNA (cpDNA) regions were sequenced for three samples of A. cinerea sampled at three different locations, as well as one sample of A. ciliata and one sample of A. hispida (GenBank accession numbers are given in Supplementary Data Table S2).
Table 2.
Characterization and diversity of the four chloroplast DNA sequences separately and for the combined matrix (Total) examined for A. provincialis
| Fragment length | Number of haplotypes | Number of polymorphic sites | Haplotype diversity (h) | Nucleotide diversity (π) | |
|---|---|---|---|---|---|
| matK | 711 | 14 | 19 | 0·90 | 0·00787 |
| trnK-matK | 830 | 16 | 12 | 0·88 | 0·00297 |
| trnL-trnF | 267 | 9 | 12 | 0·85 | 0·01099 |
| trnT-trnL | 359 | 13 | 8 | 0·77 | 0·00613 |
| Total | 2150 | 28 | 51 | 0·94 | 0·00626 |
PCRs were performed in a 50 μl volume reaction that contained 2 μl of each primer (5 mM), 1·25 units of Taq polymerase (Q-Biogen), 5 μl of incubation buffer, 6 μl of MgCl2 (10 mM), 4 μl of dNTP (25 mM) and 5 μl of DNA (50 ng μl−1). Amplification was performed using a PTC-200 thermocycler (MJ Research; cycling conditions are shown in Supplementary Data Table S3). Direct sequencing (ABI 3730 xl, Applied Biosystems) was carried out by LGC Genomics (Germany). After quality checking and alignment, the cpDNA data set contained 94 sequences.
Variation in genetic diversity throughout the range was investigated by multilocus genotyping based on the AFLP technique developed by Vos et al. (1995). Two primer combinations for the selective PCR (fluorescent dyes in brackets; Eurofins MWG Operon, Ebersberg, Germany) were used: EcoRI (6-FAM)-AAGG/MseI-CCAG; and EcoRI (6-FAM)-AACG/MseI-CAAG. Selective amplification was performed in 20 μL volumes with 5 pmol of each primer, 0·65 mm of MgCl2, 0·5 mm of dNTPs, 1 unit of Taq DNA polymerase (Q-Biogen) and 5 μL of 100× diluted preamplification. The selective amplification thermocycle profile was 94 °C for 2 min, 10 cycles of 94 °C for 30 s, 65 °C for 30 s (step –0·7 °C per cycle), 72 °C for 1 min, followed by 20 cycles at 94 °C for 30 s, 56 °C for 30 s, 72 °C for 1 min and 72 °C for 5 min. PCR products were separated and quantified on an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA). The reliability of AFLP markers was checked by repeating the complete analysis from DNA amplification to AFLP screening on ten samples for each pair of primers.
More than 40 polymorphic markers were generated, the minimum threshold for the resolution of a genetic structure with AFLPs (Gaudeul et al., 2004). Amplified labelled fragments were revealed on an ABI 3730 DNA Analyzer (PE Applied Biosystems) and fingerprint profiles were scored with Genemapper 3·7 (PE Applied Biosystems). The results of the scoring were exported as a presence/absence matrix and used for further analysis. Two individuals per population were studied for all populations. However, AFLP failed for some samples, and the final AFLP data set comprised 73 genotypes.
Variation in AFLP across the geographic range
Genetic diversity was estimated using the percentage of polymorphic loci, the number of private markers and the Shannon entropy index, which were computed from AFLP data for each of the five geographic units (A–E) described above (Table 3), using GenAlEx 6·41 (Genetic Analysis in Excel; Peakall and Smouse, 2006). Genetic differentiation was estimated as the mean of genotype divergence within each geographic unit. Genotype divergences were based on the Jaccard index (dist.binary function in the ade4 package; R Development Core Team, 2011).
Table 3.
Summary statistics of AFLP data for the geographic groups of populations of A. provincialis
| A | B | C | D | E | |
|---|---|---|---|---|---|
| N | 11 | 24 | 12 | 16 | 10 |
| Fragtot | 133 | 212 | 215 | 186 | 160 |
| Private | 1 | 9 | 13 | 2 | 4 |
| %poly | 50·2 | 81·67 | 80·48 | 72·11 | 61·35 |
| Shan | 0·20 ± 0·02 | 0·28 ± 0·01 | 0·33 ± 0·02 | 0·26 ± 0·01 | 0·25 ± 0·02 |
| Diff | 0·697 ± 0·1 | 0·703 ± 0·1 | 0·644 ± 0·08 | 0·716 ± 0·09 | 0·701 ± 0·1 |
N, number of investigated individuals; Fragtot, number of AFLP fragments per population; Private, number of fragments that only occur in one population; %poly, percentage of fragments exhibiting intrapopulation polymorphism; Shan, Shannon diversity index; Diff, genetic differentiation.
Phylogeography of A. provincialis
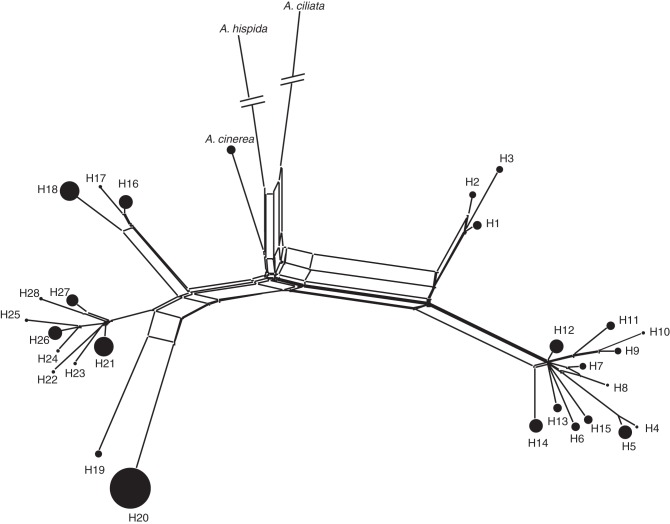
A network of relationships among cpDNA haplotypes was established from a NeighborNet analysis (Bryant and Moulton, 2004) using SplitsTree4 (Huson and Bryant, 2006). Arenaria cinerea, A. hispida and A. ciliata were included in the analysis so as to identify the branch representing the oldest divergence in A. provincialis. To compute pairwise distances between haplotypes, a distance using a GTR + gamma substitution model was chosen according to jModelTest (Posada, 2008) with empirical estimation of nucleotide frequencies (Fig. 2).
Fig. 2.
NeighborNet diagram generated using SplitsTree4 connecting the 28 haplotypes detected for the 94 individuals of A. provincialis by combining the polymorphisms of four cpDNA loci (internal part of matK gene, trnK intron and the 5′ part of matK, trnL-trnF and trnT-trnL intergenic spacer; total of 2150 bp). Each circle represents a haplotype, with size proportional to the number of individuals (see Table 1).
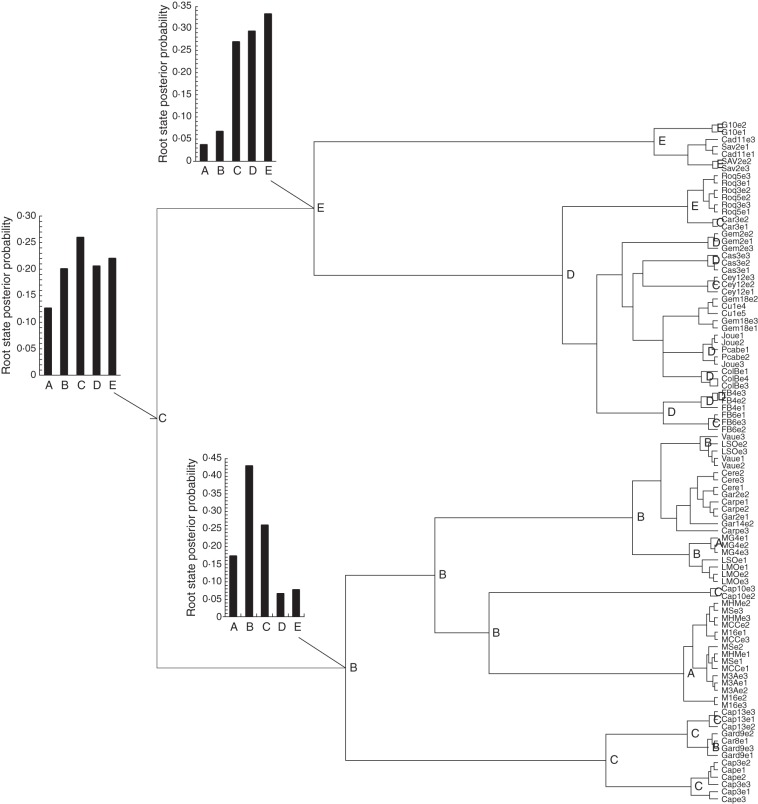
The phylogeography of A. provincialis was based on the Bayesian MCMC methods of BEAST 1·7 (Drummond et al., 2012), using all sequences obtained for A. provincialis (four cpDNA genes and n = 94 samples). All five geographic units (A–E) were used to set a geographic state for each sample. The ancestral geographic states were inferred at each node using the Bayesian stochastic search variable selection (BSSVS; Lemey et al., 2009) (Fig. 3). Without a priori knowledge about the history of A. provincialis, we decided not to specify a prior rate for the ancestral geographic states; thus, geographic state transitions (dispersal) are estimated solely from the genealogical processes. This method was recently used to build discrete phylogeographies in intra-specific studies (e.g. Winkler et al., 2012). We used Bayes factor (BF) analysis to identify well-supported geographic state transitions with strong posterior support (i.e. BF values >3; Lemey et al., 2009).
Fig. 3.
Maximum clade credibility tree based on sequence variation at four cpDNA loci of A. provincialis and reconstruction of ancestral areas by the BSSVS method. Nodes are named according to the highest posterior probability of the location state. Node histograms for the three principal nodes show posterior probability distributions of location state.
The maximum clade credibility (MCC) tree produced by BEAST 1·7 was used to map the ancestral distributions reconstructed for each node via the BSSVS method. We ran the MCMC analysis for 10 million steps, sampling states every 1000 generations. The coalescent model for molecular data was set with a GTR + gamma substitution model, an empirical estimation of nucleotide frequencies, a coalescent-constant size tree prior, a random starting tree, a strict molecular clock and default programme prior distribution on other model parameters. The coalescent model for the discrete geographic data was set with a symmetrical substitution model and the BSSVS was then enabled. Convergence and acceptable mixing of the sampled parameters were checked using TRACER (part of BEAST; Supplementary Data Fig. S1); all ESS values were higher than 200. TREEANNOTATOR (part of BEAST) was used to choose the MCC tree after a 10 % burn-in.
The most probable location of each node was labelled on the MCC tree with the corresponding geographic state. For the main phylogroups and the root nodes, the distribution of posterior probabilities of geographic states was also shown to account for uncertainties in ancestral reconstructions (Fig. 3). The history of migration events within the distribution of A. provincialis is summarized in Fig. 4 according to ancestral geographic state reconstruction.
Fig. 4.
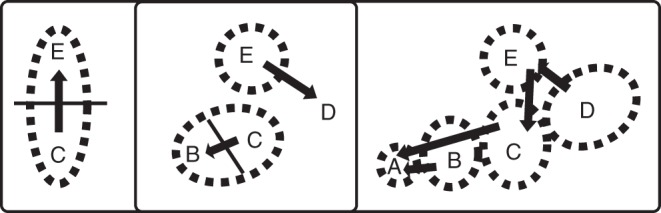
Migration patterns in three steps mapped using the ancestral distributions reconstructed at each node by the BSSVS method. Arrows indicate direction of migration.
Correlation between phylogeography and ecological gradients
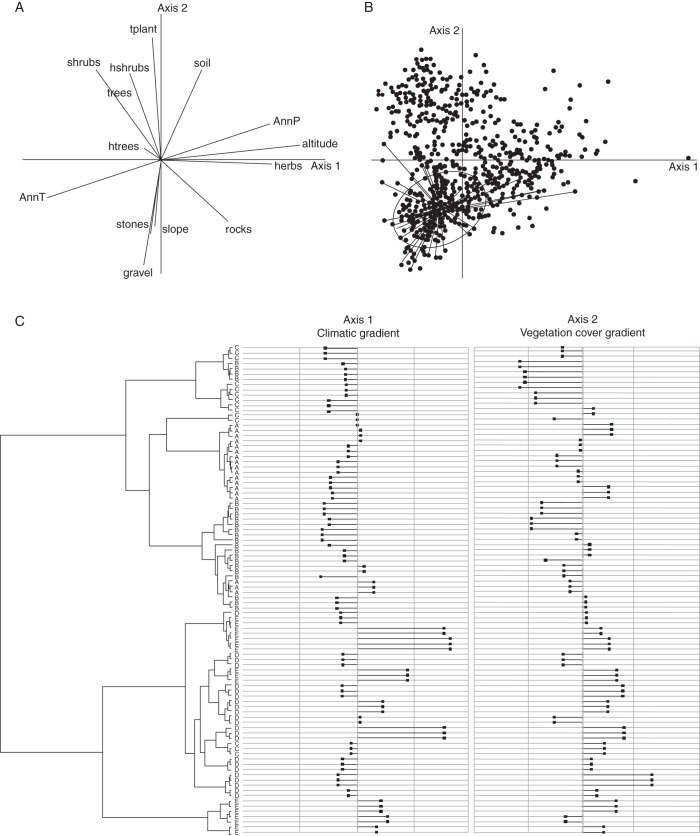
Youssef et al. (2011) found that the flora of open rocky habitats in southern Provence is organized along two ecological gradients, according to an outlying mean index analysis (OMI; Dolédec et al., 2000) based on 623 floristic relevés and 14 variables: altitude, slope, soil, mean annual temperature and precipitation (BIOCLIM variables; Hijmans et al., 2005), cover percentage of high and low shrubs, high and low trees, herbaceous plants and total plant cover, and the proportions of rocks, stones and gravel. The first gradient is mainly climatic, related to altitude and distance from the coastline, from thermophilous coastline habitats to colder and wetter habitats located at high altitude in the study area. The second gradient is in vegetation cover, from scrublands to scree slopes, measured within a 100 m2 circular area. This vegetation structure is mainly determined by substrate granulometry and slope steepness.
In our analysis, each location, corresponding to a leaf of the MCC tree of cpDNA sequences, was reported with respect to its contribution to the two ecological gradients described above using the dotchart.phylog function (adephylo and ade4 packages; R Development Core Team, 2011). The contributions were centred; therefore the mean value on each gradient corresponded to the barycentre of the ecological distribution of A. provincialis. Finally, the phylogenetic autocorrelation on the two gradients (n = 94) was tested using the Abouheif test with the abouheif.moran function (adephylo package, R Development Core Team, 2011), based on the Moran statistic and a randomization procedure (1000 permutations) (Pavoine et al., 2008).
RESULTS
Abundance variation across the geographic range
The density graph obtained by the Kernel method (Fig. 1B) shows, in different intensities of grey, the smoothing of occurrence density inside a 500 × 500 m plot, with a maximum of 17 occurrences. Density was higher and more continuous in the south-western part of the distribution and lower, as well as more fragmented, in the north-eastern part. The core of abundance was not in line with the geographic barycentre, but located in the south-western part (mainly area B). Moreover, abundance decreased on the northern and eastern margins but not on the southern and western margins.
Variation in AFLP across the geographic range
When AFLP data were collated according to the five geographic units (Table 3), area C was distinguished as having the highest number of private markers (13) and the highest Shannon entropy index (0·33). Area B was also characterized by high diversity, while the lowest diversity was observed in area A for all indexes (Table 3). Medium diversity was observed in both areas D and E. Genetic differentiation, estimated by mean pairwise AFLP genotype divergence within each geographic unit, was lowest in area C.
Phylogeography of A. provincialis
When the four cpDNA sequences for A. provincialis (Table 2) were combined, the alignment was 2151 bp in length. Twenty-eight haplotypes (H1–H28; Table 1) were identified for the 94 samples of A. provincialis from 59 variable sites (51 parsimony-informative positions), and five indels. The trnL-trnF DNA region presented the highest nucleotide diversity (0·011), in spite of having the smallest number of nucleotides (267 bp) and haplotypes (only nine). Conversely, the longest region, trnK-matK (830 bp), was characterized by the highest number of haplotypes (16) and the lowest nucleotide diversity (0·003).
NeighborNet analysis revealed two main phylogroups within A. provincialis. The outgroup taxa (A. cinerea, A. ciliata and A. hispida) were connected to the branch separating these two main phylogroups (Fig. 2). The geographic distribution of cpDNA haplotypes revealed a strong phylogeographic signal: the first phylogroup (H1–H15) was mainly situated in the northern part of the distribution range (groups D and E), while the second phylogroup (H15–H28) was restricted to the south (areas A, B and C). The two phylogroups were separated by the Huveaune river valley, but a few haplotypes of the first phylogroup were also found in the southern part of the distribution range (group C, haplotypes H5, H11 and H15).
The MCC tree provided by BEAST and based on the 94 samples (Fig. 3) is congruent with the result of the NeighborNet analysis. The root of the MCC tree is located on the branch separating the two main phylogroups. The BSSVS method used to reconstruct the ancestral areas of A. provincialis identified area C as ancestral (highest posterior probability for the root; Fig. 3). After the initial divergence of the two phylogroups, the history of A. provincialis was characterized by migrations to areas E and B. The first phylogroup originated from area E and migrated eastwards to area D. The second phylogroup originated from area B, migrated westwards to area A and then migrated back to area C. There were at least three dispersal events back towards former areas [e.g. from D to E (H4 and H5), from E to C (H5) or from B to C; Figs 3 and 4], but no migration event from the southern areas to the north occurred after the divergence of the phylogroups. BF analysis revealed that the geographic state transitions with strongest posterior support were between C and B (BF = 24) and between C and E (BF = 22).
Correlation between ecological and phylogenetic structures
The Abouheif tests exhibited significant phylogenetic autocorrelation on the two ecological gradients provided by OMI analysis for all haplotypes (P = 0·001) as well as for the two phylogroups separately (P = 0·001). Descriptive results of the Abouheif tests are given in Supplementary Data Table S4. Figure 5 shows that samples closely related in phylogeny tended to share the same position in the ecological gradients.
Fig. 5.
(A) Projection of the first two axes of the OMI analysis following Youssef et al. (2011) realized on 14 variables and 254 stations. The 14 variables are described in the text, with the following abbreviations used on the figure: AnnP = annual preciptitation; AnnT = annual temperature; htrees, hshrubs = percentage cover of high trees and shrubs, respectively; tplant = total plant cover. (B) Position of the 156 stations in the presence of A. provincialis inside the two axes. (C) Dot chart representing the MCC tree (Fig. 2) and the position of each haplotype within the OMI axes relative to the mean (values above the mean on the right and values below the mean on the left); individuals are named by geographic group.
Across the first gradient, samples from sites with above average climatic conditions (first OMI axis) were mainly in the first phylogroup, while those below average were mainly in the second phylogroup. In the former areas of both phylogroups, i.e. C and E (H16, H17, H18, H1, H2 and H3), climatic conditions differed but were not extreme. The most marginal values, corresponding to either the coldest and wettest or the warmest and driest climatic conditions, were for locations at the periphery, where the most recent haplotypes were found (D versus A areas) and for which migrations from former areas (B, C and E) were inferred (see above results).
Across the second gradient, that of vegetation cover, the distribution of samples was also phylogenetically structured. Samples belonging to the second phylogroup were mainly distributed in sites characterized by low vegetation cover, high slope values and high gravel cover values (corresponding to screes), whereas samples belonging to the first phylogroup were mainly distributed in sites with low vegetation and low gravel cover values and high rock cover values (representing lapiaz). Again, the most marginal values were observed in areas A, B and D, while values for areas C and E were at the centre of the gradient.
DISCUSSION
A hotspot of biodiversity, the Mediterranean Basin has been described as a mosaic of refugial areas harbouring numerous endemic taxa (Médail and Diadema, 2009). Few studies have investigated precisely the phylogeography of narrow endemic plants, especially outside the major regional Mediterranean biodiversity hotspots (Médail and Quézel, 1997). Furthermore, annual plants are generally not well represented in phylogeographic studies. Therefore, our study focusing on a winter annual plant of southern Provence, a region that remains relatively unexplored in molecular studies, represents a contribution towards more comprehensive knowledge of plant evolution in the Mediterranean region.
Unexpected chloroplast DNA variation
In a recent study of its ecological niche and phylogenetic position (Youssef et al., 2011), A. provincialis was shown to be the only one of the six Arenaria species of Provence able to colonize scree slopes. This suggests a probable relationship between its history and the emergence of lapiaz and scree slopes derived from the intense frost erosion processes occurring during the Pleistocene glacial periods.
Here, NeighborNet analysis based on the combined sequences of four cpDNA loci detected a well-resolved genealogy with two phylogroups in A. provincialis, themselves connected to the outgroups A. cinerea, A. hispida and A. ciliata (Fig. 2). After the divergence of the two main phylogroups of A. provincialis, cpDNA haplotype diversity accumulated, leading to the current high numbers of haplotypes (28) observed within a small area (less than 145 km2). In comparison, a study based on Arenaria humifusa, endemic to the Arctic Arc, showed only two substitutions and one indel for 1578 bp in two cpDNA loci, revealing three haplotypes distributed over more than 5,000 km2 (Westergaard et al., 2010).
Our study, like other Mediterranean case studies (e.g. in insular endemics: Molins et al., 2011; Mayol et al., 2012), confirms that strong genetic structuring is plausible in any part of the distribution range of Mediterranean endemics, and therefore that the size of a distribution range is not correlated with levels and structure of genetic diversity. The well-known climatic impact of the last glacial maximum (21 000 years before the present) on species distributions (Hewitt, 2000) has also been shown for vegetation in the western Mediterranean area (Cheddadi et al., 2009; Médail and Diadema, 2009; Fady and Connors, 2010). However, the high cpDNA diversity and polymorphism of A. provincialis we observed in such a restricted area support the idea that the preservation and accumulation of genetic diversity during the Ice Ages could be a common pattern in the Mediterranean, as underlined recently by Nieto Feliner (2011). Our study yields new evidence that extreme or azonal habitats, i.e. rocky terrain, cliffs and steep slopes, may have promoted the persistence of endemic plants and enhanced specific traits (Thompson, 2005). Arenaria provincialis, with its phylogeographic structure, therefore affords an excellent opportunity to relate long-term persistence and patterns of genetic diversity to the central–marginal hypothesis (Eckert et al., 2008; Sexton et al., 2009).
Spatial distribution and the central–marginal hypothesis
The kernel model of density (Fig. 1B) clearly shows that abundance of A. provincialis is not homogeneous across its geographic range. Area B has the highest abundance. In the northern part of its distribution, A. provincialis is markedly less abundant and more fragmented. A bias in field inventories is unlikely to explain this pattern, since inventories of A. provincialis have been made for several years and its northern and eastern range limits have been intensively investigated. The north-eastern parts of the distribution correspond to isolated patches of open rocky habitats most frequently embedded in mattoral or forest matrixes. A decrease in abundance is observed towards the northern and eastern range limits.
Genetic diversity showed a different pattern from that of contemporary abundance: measured as the polymorphism and Shannon entropy of AFLP markers (Table 3), it increased towards the centre (areas B and C), whereas AFLP genotype divergence, used here to describe genetic differentiation, was significantly lower in area C (Table 3). Molecular markers thus show that the populations of A. provincialis with the highest diversity and that are the least differentiated are near the central part of the distribution (mostly C and secondary B areas), whereas marginal populations with the lowest genetic diversity are in areas A, D and E, near the western, eastern and northern parts of the distribution respectively. The structure of genetic diversity seems roughly consistent with the hypotheses formulated here (see Introduction), and we cannot rule out the contemporary effects of isolation and small effective population size, notably at the northern and eastern limits. However, we make the point below that this pattern of contemporary genetic diversity is also well correlated with Pleistocene range dynamics and expansion.
Past migrations from the centre to the periphery
The structured phylogeography of A. provincialis and the reconstruction of ancestral areas, based on the BSSVS method of Lemey et al. (2009), enabled us to reconstruct a step-by-step history of the migrations responsible for the current range of A. provincialis. The ancestral area of A. provincialis was thus found to be situated near the centre of the distribution (area C), with evidence that, after migrations to the west (area B) and north (area E), migrations occurred towards the eastern and western peripheries (areas A and D; Figs 3 and 4). Moreover, phylogeography and the reconstruction of ancestral states revealed that backward migrations also occurred to areas C and E (Figs 3 and 4). BF analysis revealed strong posterior support for dispersal between areas C and B and between areas E and C, thus confirming the central position of C as both an ancestral and a crossroads area. In this phylogeographic context, it should be noted that the frequency of AFLP private markers (Table 3) supported a longer persistence of A. provincialis in area C, and secondarily in area B. Consequently, area C, which is central in the distribution, appears to have been crucial in the history of A. provincialis.
The history of A. provincialis therefore reveals that the central–marginal pattern is strongly associated with a history of expansion from former areas (Fig. 4). Such correlations between genetic diversity and distance from refugia have been found elsewhere (e.g. Garner et al., 2004; Hoban et al., 2010), but over a coarser scale and in the context of glaciations. To the best of our knowledge, this correlation has never before been observed for a Mediterranean narrow endemic plant. In the light of the extensive cpDNA nucleotide variation and marked phylogeographic structure revealed here (28 haplotypes in two phylogroups), it is likely that the rocky habitats situated in the central parts (C, and subsequently areas B and E) of the distribution range have sheltered A. provincialis for a long time (i.e. probably since the early Pleistocene). Originating from these central, ancestral areas, the populations situated today at the range limits (areas A and D) are younger and have more limited AFLP diversity (Table 3).
An extensive survey of the literature yielded no conclusive evidence from the recent geological history of the focus area that might have helped to calibrate precisely in time the haplotype genealogy of A. provincialis. Regarding past environmental change, paleoecological data are also very scarce for rocky outcrops of the focus area, and ancestral distribution modelling would be difficult to realize for a narrow endemic plant living in azonal habitats. Of the plants common to the rocky habitats of A. provincialis, the pioneer tree Juniperus phoenicea has been hypothesized to have undergone expansion during arid and cold periods of the Pleistocene, such as the Last Glacial Maximum (Boratynski et al., 2009). Similarly, it has been proposed that Cheirolophus intybaceus reached southern France from the Iberian Peninsula during the mid-late Pleistocene (Garnatje et al., 2013). The sole relevant hypothesis, therefore, is that a decrease in vegetation cover due to climate cooling and drying during the major glaciation phases beginning around 0·9 Mya (De Beaulieu et al., 2005) may have favoured the expansion of pioneer plants like A. provincialis.
Conservation significance of the phylogeographic pattern
The habitat of A. provincialis is structured around two main ecological gradients mainly determined by altitude and vegetation cover (Baumel et al., 2009; Youssef et al., 2011; Fig. 5A). As supported by the significant phylogenetic autocorrelation on these two ecological gradients (Abouheif test), the populations having the most divergent haplotypes are now living at the extremes of its two ecological gradients (Fig. 5C). During the history of its expansion, A. provincialis reached the top of its elevation gradient in southern Provence (i.e. a range of 1000 m), where its populations are currently experiencing contrasting climatic conditions (axis 1; Fig. 5A, C). For example, A. provincialis is the sole Arenaria species living both near the littoral and near the summit of the Sainte Baume mountain range (almost 1000 m above sea level), in contrast to its congeners A. grandiflora and A. aggregata, which are present only in the second location. Yet populations located near the littoral, and belonging mainly to the second phylogroup, are living in the most open and steep habitats of the species, such as huge south-facing seaside screes (axis 2; Fig. 5A, C). This means that today's populations of A. provincialis are organized across an ecological range explained both by the history of the species and by the geographic structure of its genetic diversity.
In conclusion, population distinctiveness based on molecular variation and ecological data is well supported here: populations from the two phylogroups are exchangeable neither genetically nor ecologically (Crandall et al., 2000). Further study will be needed in order to evaluate the probable adaptive polymorphism of A. provincialis suggested by the strong correlation between phylogeography and ecological gradients (Moritz, 2002). Our results point to key issues in terms of conservation. First, the two phylogroups of A. provincialis have different ecologies and exhibit different patterns of fragmentation and abundance (Figs 1 and 5). The populations situated near the south-western and north-eastern range limits contribute significantly to this fragmented structure. Second, the central populations have persisted the longest and been the source of expansions, but they are not currently characterized by the highest abundance.
Exploring the abundance, genetic diversity and phylogeography of a Mediterranean narrow endemic plant, this study highlights the fact that current range size and abundance patterns are not sufficient to predict the organization of genetic diversity. However, our study reveals the crucial role played in the history of A. provincialis by the areas situated centrally in its distribution.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank the Conservatoire Botanique National Méditerranéen de Porquerolles for providing unpublished data about Arenaria taxon locations and allowing us to collect Arenaria material. We are grateful to Errol Véla for his help in the field and in the botanical assessment. Our thanks to Marjorie Sweetko for English language editing. This research was funded by the CNRS, the French National Office of Forests (ONF) and the General Council of Bouches du Rhône district (CG13). Marine Pouget was supported by the Provence–Alpes–Côte d'Azur region (PACA).
LITERATURE CITED
- Baumel A, Affre L, Véla E, Auda P, et al. Ecological magnitude and fine scale dynamics of the Mediterranean narrow endemic therophyte, Arenaria provincialis (Caryophyllaceae) Acta Botanica Gallica. 2009;156:259–272. [Google Scholar]
- De Beaulieu J-L, Miras Y, Andrieu-Ponel V, Guiter F. Vegetation dynamics in north-western Mediterranean regions: instability of the Mediterranean bioclimate. Plant Biosystems. 2005;139:114–126. [Google Scholar]
- Boratynski A, Lewandowski A, Boratynska K, Montserrat JM, Romo A. High level of genetic differentiation of Juniperus phoenicea (Cupressaceae) in the Mediterranean region: geographic implications. Plant Systematics and Evolution. 2009;277:163–172. [Google Scholar]
- Brown JH. On the relationship between abundance and distribution of species. American Naturalist. 1984;124:255. [Google Scholar]
- Bryant D, Moulton V. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Molecular Biology and Evolution. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Casazza G, Barberis G, Minuto L. Ecological characteristics and rarity of endemic plants of the Italian Maritime Alps. Biological Conservation. 2005;123:361–371. [Google Scholar]
- Chater AO, Halliday G. Arenaria L. In: Tutin TG, Heywood VH, Burges NA, Valentine DH, Walters SM, Webb DA, editors. Flora Europaea 1. Cambridge: Cambridge University Press; 1964. pp. 140–148. [Google Scholar]
- Cheddadi R, Fady B, François L, et al. Putative glacial refugia of Cedrus atlantica deduced from Quaternary pollen records and modern genetic diversity. Journal of Biogeography. 2009;36:1361–1371. [Google Scholar]
- Chown SL. Speciation and rarity: separating cause from consequence. In: Kunin WE, Gaston KJ, editors. The biology of rarity. London: Chapman & Hall; 1997. pp. 91–109. [Google Scholar]
- Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK. Considering evolutionary processes in conservation biology. Trends in Ecology and Evolution. 2000;15:290–295. doi: 10.1016/s0169-5347(00)01876-0. [DOI] [PubMed] [Google Scholar]
- Dolédec S, Chessel D, Gimaret-Carpentier C. Niche separation in community analysis: a new method. Ecology. 2000;81:2914–2927. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation method for small quantities of fresh tissues. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1·7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert CG, Samis KE, Lougheed SC. Genetic variation across species' geographical ranges: the central-marginal hypothesis and beyond. Molecular Ecology. 2008;17:1170–1188. doi: 10.1111/j.1365-294X.2007.03659.x. [DOI] [PubMed] [Google Scholar]
- Eckstein RL, O'Neill RA, Danihelka J, Otte A, Köhler W. Genetic structure among and within peripheral and central populations of three endangered floodplain violets. Molecular Ecology. 2006;15:2367–2379. doi: 10.1111/j.1365-294X.2006.02944.x. [DOI] [PubMed] [Google Scholar]
- Fady B, Conord C. Macroecological patterns of species and genetic diversity in vascular plants of the Mediterranean basin. Diversity and Distributions. 2010;16:53–64. [Google Scholar]
- Garnatje T, Perez-Collazos E, Pellicer J, Catalan P. Balearic insular isolation and large continental spread framed the phylogeography of the western Mediterranean Cheirolophus intybaceus s.l. (Asteraceae) Plant Biology. 2013;15:166–175. doi: 10.1111/j.1438-8677.2012.00632.x. [DOI] [PubMed] [Google Scholar]
- Garner TWJ, Pearman PB, Angelone S. Genetic diversity across a vertebrate species' range: a test of the central-peripheral hypothesis. Molecular Ecology. 2004;13:1047–1053. doi: 10.1111/j.1365-294X.2004.02119.x. [DOI] [PubMed] [Google Scholar]
- Gaston KJ. The structure and dynamics of geographic range. Oxford, UK: Oxford University Press; 2003. [Google Scholar]
- Gaudeul M, Till-Bottraud I, Barjon F, Manel S. Genetic diversity and differentiation in Eryngium alpinum L. (Apiaceae): comparison of AFLP and microsatellite markers. Heredity. 2004;92:508–518. doi: 10.1038/sj.hdy.6800443. [DOI] [PubMed] [Google Scholar]
- Guo Q. Incorporating latitudinal and central-marginal trends in assessing genetic variation across species ranges. Molecular Ecology. 2012;21:5396–5403. doi: 10.1111/mec.12012. [DOI] [PubMed] [Google Scholar]
- Hampe A, Petit RJ. Conserving biodiversity under climate change: the rear edge matters. Ecology Letters. 2005;8:461–467. doi: 10.1111/j.1461-0248.2005.00739.x. [DOI] [PubMed] [Google Scholar]
- Hardie DC, Hutchings JA. Evolutionary ecology at the extremes of species' ranges. Environmental Reviews. 2010;18:1–20. [Google Scholar]
- Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society of London B, Biological Sciences. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005;25:1965–1978. [Google Scholar]
- Hoban SM, Borkowski DS, Brosi SL, et al. Range-wide distribution of genetic diversity in the North American tree Juglans cinerea: a product of range shifts, not ecological marginality or recent population decline. Molecular Ecology. 2010;19:4876–4891. doi: 10.1111/j.1365-294X.2010.04834.x. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Computational Biology. 2009;5 doi: 10.1371/journal.pcbi.1000520. pe1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesica P, Allendorf FW. When are peripheral populations valuable for conservation? Conservation Biology. 1995;9:753–760. [Google Scholar]
- Mayol M, Palau C, Rosselló JA, González-Martínez SC, Molins A, Riba M. Patterns of genetic variability and habitat occupancy in Crepis triasii (Asteraceae) at different spatial scales: insights on evolutionary processes leading to diversification in continental islands. Annals of Botany. 2012;109:429–441. doi: 10.1093/aob/mcr298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médail F, Diadema K. Glacial refugia influence plant diversity patterns in the Mediterranean Basin. Journal of Biogeography. 2009;36:1333–1345. [Google Scholar]
- Médail F, Quézel P. Hot-spots analysis for conservation of plant biodiversity in the Mediterranean basin. Annals of the Missouri Botanical Garden. 1997;84:112–127. [Google Scholar]
- Médail F, Verlaque R. Ecological characteristics and rarity of endemic plants from southeast France and Corsica: implications for biodiversity conservation. Biological Conservation. 1997;80:269–281. [Google Scholar]
- Medrano M, Herrera CM. Geographical structuring of genetic diversity across the whole distribution range of Narcissus longispathus, a habitat-specialist, Mediterranean narrow endemic. Annals of Botany. 2008;102:183–194. doi: 10.1093/aob/mcn086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller DA, Geber MA, Tiffin P. Population genetics and the evolution of geographic range limits in an annual plant. American Naturalist. 2011;178(Suppl):S44–S57. doi: 10.1086/661783. [DOI] [PubMed] [Google Scholar]
- Molins A, Bacchetta G, Rosato M, Rosselló JA, Mayol M. Molecular phylogeography of Thymus herba-barona (Lamiaceae ): insight into the evolutionary history of the flora of the western Mediterranean islands. Taxon. 2011;60:1295–1305. [Google Scholar]
- Moritz C. Strategies to protect biological diversity and the evolutionary processes that sustain it. Systematic Biology. 2002;51:238–254. doi: 10.1080/10635150252899752. [DOI] [PubMed] [Google Scholar]
- Nicoletti F, De Benedetti L, Airò M, et al. Spatial genetic structure of Campanula sabatia, a threatened narrow endemic species of the Mediterranean Basin. Folia Geobotanica. 2012;47:249–262. [Google Scholar]
- Nieto Feliner G. Southern European glacial refugia: a tale of tales. Taxon. 2011;60:365–372. [Google Scholar]
- Pavoine S, Ollier S, Pontier D, Chessel D. Testing for phylogenetic signal in phenotypic traits: new matrices of phylogenetic proximities. Theoretical Population Biology. 2008;73:79–91. doi: 10.1016/j.tpb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit J, Aguinagalde I, De Beaulieu J-L, et al. Glacial refugia: hotspots but not melting pot of genetic diversity. Science. 2003;300:1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. Vienna: R Foundation for Statistical Computing; 2011. R: a language and environment for statistical computing. http://www.r-project.org . [Google Scholar]
- Sagarin RD, Gaines SD. The ‘abundant centre’ distribution: to what extent is it a biogeographical rule? Ecology Letters. 2002;5:137–147. [Google Scholar]
- Sexton JP, McIntyre PJ, Angert AL, Rice KJ. Evolution and ecology of species range limits. Annual Review of Ecology, Evolution, and Systematics. 2009;40:415–436. [Google Scholar]
- Sexton JP, Strauss SY, Rice KJ. Gene flow increases fitness at the warm edge of a species' range. Proceedings of the National Academy of Sciences, USA. 2011;108:11704–11709. doi: 10.1073/pnas.1100404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS. Comparative phylogeography of unglaciated eastern North America. Molecular Ecology. 2006;15:4261–4293. doi: 10.1111/j.1365-294X.2006.03061.x. [DOI] [PubMed] [Google Scholar]
- Stebbins GL, Major J. Endemism and speciation in the California flora. Ecological Monographs. 1965;35:1–35. [Google Scholar]
- Stewart JR, Lister AM, Barnes I, Dalén L. Refugia revisited: individualistic responses of species in space and time. Proceedings of the Royal Society B: Biological Sciences. 2010;277:661–671. doi: 10.1098/rspb.2009.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy A-G, Cosson J-F. Comparative phylogeography and postglacial colonization. Molecular Ecology. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Lavergne S, Affre L, Gaudeul M, Debussche M. Ecological differentiation of Mediterranean endemic plants. Taxon. 2005;54:967–976. [Google Scholar]
- Véla E, Auda P, Léger JF, Gonçalves V, Baumel A. Exemple d'une nouvelle évaluation du statut de menace suivant les critères de l'UICN version 3·1.: le cas de l'endémique provençale Arenaria provincialis Chater & Halliday (= Gouffeia arenarioides DC., Caryophyllaceae) Acta Botanica Gallica. 2008;155:547–562. [Google Scholar]
- Vos P, Hogers R, Bleeker M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard KB, Alsos IG, Popp M, Engelskjøn T, Flatberg KI, Brochmann C. Glacial survival may matter after all: nunatak signatures in the rare European populations of two west-arctic species. Molecular Ecology. 2010;20:376–393. doi: 10.1111/j.1365-294X.2010.04928.x. [DOI] [PubMed] [Google Scholar]
- Winkler M, Tribsch A, Schneeweiss GM, et al. Tales of the unexpected: phylogeography of the arctic-alpine model plant Saxifraga oppositifolia (Saxifragaceae) revisited. Molecular Ecology. 2012;21:4618–4630. doi: 10.1111/j.1365-294X.2012.05705.x. [DOI] [PubMed] [Google Scholar]
- Youssef S, Baumel A, Véla E, et al. Factors underlying the narrow distribution of the Mediterranean annual plant Arenaria provincialis (Caryophyllaceae) Folia Geobotanica. 2011;46:327–350. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.