Abstract
Background and aims
Despite a recent new classification, a stable phylogeny for the cycads has been elusive, particularly regarding resolution of Bowenia, Stangeria and Dioon. In this study, five single-copy nuclear genes (SCNGs) are applied to the phylogeny of the order Cycadales. The specific aim is to evaluate several gene tree–species tree reconciliation approaches for developing an accurate phylogeny of the order, to contrast them with concatenated parsimony analysis and to resolve the erstwhile problematic phylogenetic position of these three genera.
Methods
DNA sequences of five SCNGs were obtained for 20 cycad species representing all ten genera of Cycadales. These were analysed with parsimony, maximum likelihood (ML) and three Bayesian methods of gene tree–species tree reconciliation, using Cycas as the outgroup. A calibrated date estimation was developed with Bayesian methods, and biogeographic analysis was also conducted.
Key Results
Concatenated parsimony, ML and three species tree inference methods resolve exactly the same tree topology with high support at most nodes. Dioon and Bowenia are the first and second branches of Cycadales after Cycas, respectively, followed by an encephalartoid clade (Macrozamia–Lepidozamia–Encephalartos), which is sister to a zamioid clade, of which Ceratozamia is the first branch, and in which Stangeria is sister to Microcycas and Zamia.
Conclusions
A single, well-supported phylogenetic hypothesis of the generic relationships of the Cycadales is presented. However, massive extinction events inferred from the fossil record that eliminated broader ancestral distributions within Zamiaceae compromise accurate optimization of ancestral biogeographical areas for that hypothesis. While major lineages of Cycadales are ancient, crown ages of all modern genera are no older than 12 million years, supporting a recent hypothesis of mostly Miocene radiations. This phylogeny can contribute to an accurate infrafamilial classification of Zamiaceae.
Keywords: Biogeography, Cycadales, Bowenia, Stangeria, Dioon, gymnosperms, molecular systematics
INTRODUCTION
With ten genera and 331 currently accepted species (Osborne et al., 2012), cycads (order Cycadales) can contribute to understanding the origin and evolution of seeds, cones and plant vegetative structures (Frohlich and Parker, 2000; Brenner et al., 2003a, b). Cycads also hold important clues that can be used to infer early molecular evolution trends of seed plants (Zhang et al., 2004; Wang et al., 2007), as well as provide evolutionary insights concerning ancestral plant–animal interactions such as ancient pollination and herbivory mechanisms (Schneider et al., 2002; Brenner et al., 2003a; Kono and Hiroshi, 2007; Terry et al., 2007; Hummel et al., 2008; Butler et al., 2009; Peñalver et al., 2012).
Cycads have a fossil record dating back to the Lower Permian of China, reaching their peak in abundance and diversity in the Mesozoic (Martínez et al., 2012). Due to their long evolutionary history and retention of ancestral characters such as flagellated sperm, cycads are considered the most primitive extant seed plant lineage (Brenner et al., 2003b) and are often characterized as ‘living fossils’. However, recent studies suggest that extant species are the result of Neogene speciation events (Crisp and Cook, 2011; Nagalingum et al., 2011).
Despite their extraordinary scientific importance and widespread horticultural popularity, phylogenetic relationships within Cycadales still are not fully resolved. At the infrageneric level, phylogenies incorporating molecular data have been published for Ceratozamia (González and Vovides, 2002; De Castro et al., 2006), Cycas (Chiang et al., 2009; Sangin et al., 2010; Xiao et al., 2010), Dioon (Moretti et al., 1993; González et al., 2008; Moynihan et al., 2012), Encephalartos (Treutlein et al., 2005; Rousseau, 2012) and Zamia (Caputo et al., 2004). There has also been some interest in using DNA sequences for bar-coding of cycad species (Sass et al., 2007; Nicolalde-Morejon et al., 2011). These phylogenetic reconstructions have been mostly based on restriction site data from the plastid genome, amplified fragment length polymorphisms (AFLPs) and sequences of several regions of plastid DNA and nuclear ribosomal DNA internal transcribed spacer (nrDNA ITS). Plastid sequences to date have not adequately resolved infrageneric relationships, while ITS data yield either phylogenies with few resolved nodes (Caputo et al., 2004; Treutlein et al., 2005; Sangin et al., 2010) or results highly incongruent with other molecular or morphological data (De Castro et al., 2006; González et al., 2008; Xiao et al., 2010; Moynihan et al., 2012).
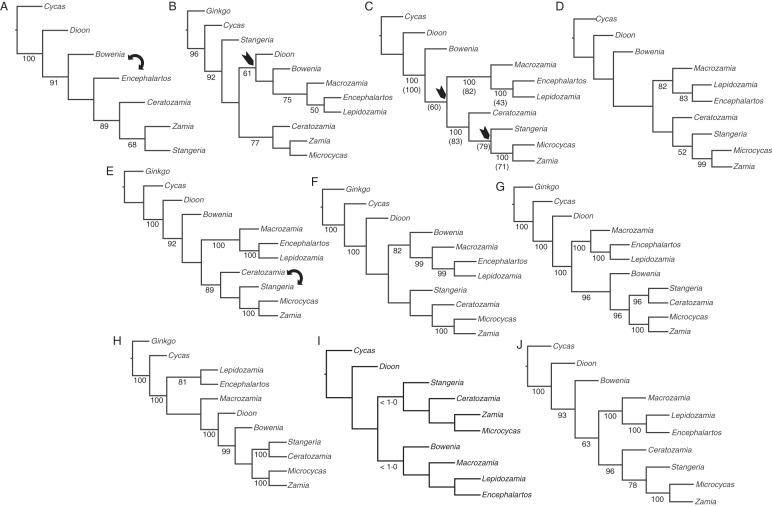
At the supra-generic level, questions still remain unresolved, particularly concerning the phylogenetic placement of three genera: Bowenia, Dioon and Stangeria. Phylogenies based on morphology, ITS sequences or several plastid regions yielded incongruent results for these three genera (e.g. Stevenson, 1990, 1992; Hill et al., 2003; Rai et al., 2003; Bogler and Francisco-Ortega, 2004; Chaw et al., 2005; Zgurski et al., 2008; Nagalingum et al., 2011; Griffith et al., 2012). However, all of these phylogenetic studies provided strong support for: (1) Cycas as sister to the rest of the Cycadales; (2) Encephalartos, Lepidozamia and Macrozamia forming a monophyletic group; and (3) Microcycas and Zamia (including Chigua) as a distinct clade (Figs 1 and 2).
Fig. 1.
Phylogeny of Cycacdales based on cladistic analysis of 30 morphological characters (Stevenson, 1990) and formal classification (Stevenson, 1992).
Fig. 2.
Molecular phylogenetic trees of Cycadales from various authors. Numbers below branches are bootstrap percentages ≥50 unless otherwise stated. (A) One of the two most-parsimonious trees from analysis of 17 plastid genes and non-coding regions (Rai et al., 2003). The second tree exchanged positions for Bowenia and Encephalartos (double-headed arrow). (B) One of the 81 most-parsimonious trees from analysis of concatenated plastid rbcL and trnL-F, and nrDNA ITS and 26S (Hill et al., 2003). (C) One of the 45 most-parsimonious trees from analysis of plastid matK (Chaw et al., 2005). Arrows indicate nodes that collapse in the strict consensus. With nrDNA ITS, the authors found the four most-parsimonious trees, the strict consensus of which was fully resolved at the generic level. Bootstrap values in parentheses are from the ITS analysis. (D) Single most-parsimonious tree found by Bogler and Francisco-Ortega (2004) using combined plastid trnL-F and nrDNA ITS2. (E) One of the two most-parsimonious trees found in the analysis of Zgurski et al. (2008) of the same 17 plastid genes and non-coding regions used by Rai et al. (2003). (F–H) Maximum likelihood trees from Nagalingum et al. (2011). (F) Plastid rbcL and matK. (G) Nuclear PHYP. (H) Combined plastid and nuclear genes. (I) Cladogram of Cycadales inferred from the maximum clade credability chronogram of the gymnosperms using plastid matK and 25S nrDNA (Crisp and Cook, 2011). Numbers below branches indicate those branches with posterior probabilities <1·0. (J) Strict consensus of the 96 most-parsimonious trees based on analysis of combined plastid matK, rpoB and rps4, mitochondrial mtITS and nad1, nrITS, nuclear NEEDLY sequences and 130 morphological characters (Griffith et al., 2012).
Since Doyle's (1992) admonishment of single gene phylogenies as ‘one character taxonomy’, systematists have striven to develop concatenated super-matrices of numerous gene sequences, thereby emulating the ‘total evidence’ approach of Kluge (1989, 2004). Over the last 15 years, the growing concern that gene histories are not necessarily congruent with species histories became more precisely understood (Avise and Wollenberg, 1997; Doyle, 1997; Maddison, 1997; Avise, 2000), and problems associated with concatenation, even with mixed models applied to the partitions (Nylander et al., 2004), have been a focus of discussion (Degnan and Rosenberg, 2006, 2009; Edwards et al., 2007; Kubatko and Degnan, 2007; Edwards, 2008). The heterogeneity of gene trees, and their conflict with species trees, can be caused by several phenomena, including: gene duplication/extinction, horizontal gene transfer and hybridization, and incomplete lineage sorting (Maddison, 1997; Degnan and Rosenberg, 2009). Further, one or several partitions of a concatenated super-matrix may bias the tree reconstruction results by the sheer number of phylogenetically informative characters (Edwards et al., 2007). Only recently has there been development of phylogenetic software tools optimized for estimating species trees from multiple gene trees without concatenation (e.g., Ané et al., 2007; Baum, 2007; Liu and Pearl, 2007; Liu, 2008; Liu et al., 2008, 2009; Wehe et al., 2008; Heled and Drummond, 2010; Maddison and Maddison, 2011). Such methods allow gene tree heterogeneity and estimate topologies which truly represent lineages of populations and species, rather than genes.
Single-copy nuclear genes (SCNGs) have been shown to provide a valid alternative to nrDNA and chlorolast DNA (cpDNA) regions for plant phylogenetic reconstructions (e.g. Popp and Oxelman, 2007; Roncal et al., 2008; Meerow et al., 2009; Rousseau-Gueutin et al., 2009; Albach and Meudt, 2010; Zhang et al., 2012; Zimmer and Wen, 2013). The utilization of multiple SCNGs can provide a whole selection of unlinked and independent characters which are extremely valuable in phylogenetics (Small et al., 2004). However, SCNGs have not been widely used mostly because of problems with isolation and amplification, and difficulty in distinguishing between paralogous and orthologous copies (Mort and Crawford, 2004). These molecular markers have rarely been used to resolve phylogenetic relationships within the Cycadales (e.g. Nagalingum et al., 2011; Moynihan et al., 2012), although at least six of them have been isolated and identified for Cycas or Zamia [i.e. vicilin-like (Braun et al., 1996), the largest subunit of RNA polymerase II (Nickerson and Drouin, 2004), the second largest subunit of RNA polymerase II (Oxelman et al., 2004), APETALA/EREBP (Shigyo et al., 2006), Floricaula/LEAFY (Frohlich and Parker, 2000) and Cycas-AGAMOUS (hereafter CyAG; Zhang et al., 2004)].
In this study, we apply five SCNGs to the phylogeny of the order Cycadales. We specifically aim to evaluate several gene tree–species tree reconciliation approaches for developing an accurate phylogeny of the order, contrasting them with concatenated parsimony and maximum likelihood (ML) analyses, and hopefully resolve the erstwhile problematic phylogenetic position of Bowenia, Dioon and Stangeria. We discuss our results in the contexts of clade age estimation, the cycad fossil record and ancestral area reconstruction.
MATERIALS AND METHODS
Taxonomic sampling
Twenty representative taxa of Cycadales were used in this study (Table 1). Up to three species were sampled in each genus to allow branch and bound (B&B) parsimony searches and to keep the duration of other analyses within a reasonable time frame (genera with one or two species were completely sampled). Several studies have indicated that sequence length and the number of loci are much greater factors in phylogenetic accuracy than the number of taxa (Rosenberg and Kumar, 2001; Rokas and Carroll, 2005), and that a denser taxon sample does not always reduce a pre-existent topological conflict (Zhao et al., 2013). There is agreement among cycad and gymnosperm specialists that the cycad genera with multiple species (i.e. Bowenia, Ceratozamia, Cycas, Dioon, Encephalartos, Lepidozamia, Macrozamia and Zamia) are monophyletic (Stevenson, 1990, 1992; Hill et al., 2003; Rai et al., 2003; Bogler and Francisco-Ortega, 2004; Chaw et al., 2005; Zgurski et al., 2008; Crisp and Cook, 2011; Nagalingum et al., 2011). All broad geographic areas in the range of the genera are represented by the samples in our study, with the exception of the outgroup Cycas, which has a single species in Africa and Madagascar (C. thouarsii R. Br. ex Gaudich), but extends throughout Southeast Asia, India, China, Australia, and the western Pacific.
Table 1.
List of taxa, origin, voucher information and GenBank accession numbers
| Genus | Species | Provenance* | Voucher (Herbarium) | GenBank accession number |
||||
|---|---|---|---|---|---|---|---|---|
| CyAG | COS26 | GroES | GTP | HTS | ||||
| Cycas | C. angulata. | Australia | M. Calonje MBC12-012 (FTG) | KF309296 | KF309276 | KF309316 | KF309336 | KF309356 |
| C. bifida | China | M. Calonje MBC12-005 (FTG) | KF309297 | KF309277 | KF309317 | KF309337 | KF309357 | |
| Bowenia | B. serrulata | Australia | D. P. Little & D.W. Stevenson 1004 (FTG) | KF309291 | KF309271 | KF309311 | KF309331 | KF309351 |
| B. spectabilis | Australia | M. Calonje MBC12-007(FTG) | KF309292 | KF309272 | KF309312 | KF309332 | KF309352 | |
| Stangeria | S. eriopus | South Africa | M. Calonje MBC12-001(FTG) | KF309307 | KF309287 | KF309327 | KF309347 | KF309361 |
| Zamia | Z. imperialis | Panama | N. Espinosa 2011–005 (FTG) | KF309308 | KF309288 | KF309328 | KF309348 | KF309368 |
| Z. chigua | Colombia | Wilderetal. s.n. (F) | KF309309 | KF309289 | KF309329 | KF309349 | KF309369 | |
| Z. erosa | Puerto Rico | Turnbull 21 (NY) | KF309310 | KF309290 | KF309330 | KF309350 | KF309370 | |
| Microcycas | M. calocoma | Cuba | M. Calonje MBC12-006 (FTG) | KF309306 | KF309286 | KF309326 | KF309346 | KF309366 |
| Ceratozamia | C. becerrae | Mexico | D. P. Little & D. W. Stevenson No.1103 (FTG) | KF309293 | KF309273 | KF309313 | KF309333 | KF309353 |
| C. hildae | Mexico | M. Calonje MBC12-008 (FTG) | KF309294 | KF309274 | KF309314 | KF309334 | KF309354 | |
| C. robusta | Belize | M. Calonje MBC12-009 (FTG) | KF309295 | KF309275 | KF309315 | KF309335 | KF309355 | |
| Dioon | D. mejiae | Honduras | M. Calonje MBC12-003 (FTG) | KF309298 | KF309278 | KF309318 | KF309338 | KF309358 |
| D. sonorense | Mexico | M. Calonje MBC12-002 (FTG) | KF309299 | KF309279 | KF309319 | KF309339 | KF309359 | |
| Encephalartos | E. altensteinii | South Africa | M. Calonje MBC12-013 (FTG) | KF309300 | KF309280 | KF309320 | KF309340 | KF309360 |
| E. macrostrobilus | Uganda | M. Calonje MBC12-014 (FTG) | KF309301 | KF309281 | KF309321 | KF309341 | KF309361 | |
| Lepidozamia | L. hopei | Australia | M. Calonje MBC12-004 (FTG) | KF309302 | KF309282 | KF309322 | KF309342 | KF309362 |
| L. peroffskyana | Australia | D. P. Little & D. W. Stevenson1050 (FTG) | KF309303 | KF309283 | KF309323 | KF309343 | KF309363 | |
| Macrozamia | M. diplomera | Australia | M. Calonje MBC12-010 (FTG) | KF309304 | KF309284 | KF309324 | KF309344 | KF309364 |
| M. macdonnellii | Australia | D. P. Little & D. W. Stevenson 1057 (FTG) | KF309305 | KF309285 | KF309325 | KF309345 | KF309365 | |
* All tissue used in this study was collected and processed from documented living collections at Montgomery Botanical Center, except for Zamia chigua, which was obtained from Lyon Arboretum, Manoa, Hawaii.
DNA extraction, gene amplification and sequencing
DNA extraction was performed using a FastDNA kit (MP Biomedicals, Santa Ana, CA, USA) or a DNeasy Plant Mini kit (Qiagen, Valencia, CA, USA) and 20–100 mg of dry leaf tissue. Five SCNGs (CyAG, COS26, GroES, GTP and HTS) were amplified using primers developed in our laboratories (see Supplementary Data Table S1). PCRs consisted of: 30 ng of total DNA, 1× PCR buffer, 0·2 mM of dNTPs, 0·2 mg mL−1 of bovine serum albumin (BSA) or 1× TBT-PAR (Samarakoon et al., 2013), 0·2 µM of each primer and 0·05 U μL−1of Taq polymerase (New England Biolabs, Ipswich, MA, USA). All the PCRs (except HTS) were carried out according to the following temperature profile: 95 °C for 2 min, 35 cycles of 95 °C for 30 s, annealing temperature (50–60 °C) for 1 min and 72 °C for 1 min, and final extension at 72 °C for 7 min. HTS was amplified using touchdown PCR (95 °C for 2 min; three cycles of 95 °C for 30 s, 58 °C for 1 min, 72 °C for 1 min; three cycles of 95 °C for 30 s, 57 °C for 1 min, 72 °C for 1 min; three cycles of 95 °C for 30 s, 56 °C for 1 min, 72 °C for 1 min; 30 cycles of 95 °C for 30 s, 54 °C for 1 min, 72 °C for 1 min; and final extension of 72 °C for 10 min). PCR products were checked using 1·2 % agarose gels with gel red (Biotium, Inc., Hayward, CA, USA) and a size standard ladder (100 bp New England Biolabs). Amplified products were cleaned up using exonuclease I (New England Biolabs) and shrimp alkaline phosphatase (USB Products- Affymetrix, Santa Clara, CA, USA), incubating at 37 °C for 1 h, followed by 80 °C for 20 min. The sequencing was done using ABI Big Dye Terminator v3·1 chemistry (Applied Biosystems, Carlsbad, CA, USA) followed by ethanol clean up. Labelled fragments were visualized on an ABI 3730 Automatic DNA Sequencer (Applied Biosystems). The nucleotide sequences were manually edited with Sequencher 4·9 (Gene Codes Corporation, Ann Arbor, MI, USA).
Phylogenetic methods
Cycas was used as the outgroup for all analyses, as we were only able to amplify DNA of Gingko with our CyAG primers. All previous phylogenetic analyses of Cycadales (e.g. Chaw et al., 2005; Zgurski et al., 2008; Nagalingum et al., 2011) have resolved Cycas as sister to all other genera in the order, thus its designation as the outgroup is appropriate. Sequences were aligned with MAFFT v.6 (Miyata et al., 2002) and/or manually in Sequencher 4·9. Nucleotide substitution models for each partition were evaluated with KAKUSAN 4·0 (Tanabe, 2007), and the corrected Akaike information criterion (AICc) (Sugiura, 1978) was used for model selection. Parsimony analysis of the concatenated matrix was conducted in PAUP v. 4·10b (Swofford, 2004) using a B&B search (Hendy and Penny, 1982) with simple addition, followed by generation of jackknife (JK) support values (1000 B&B iterations), and both total and partitioned Bremer indices, the latter with TreeRot v3.0 (Sorenson and Franzosa, 2007) and PAUP, using heuristic searches with 1000 rounds of random addition sequence and TBR branch swapping. B&B analyses were also conducted on each gene alone. The ML analyses were completed using Treefinder (Jobb, 2011). As the most likely model of nucleotide substitution was the same for all loci, a replicated (500 iterations) non-partitioned analysis was performed with bootstrap (1000 rounds). Species tree reconciliation analyses were performed with *BEAST (Heled and Drummond, 2010) as implemented in BEAST v.1.7.4 (Drummond et al., 2012); BEST (Liu, 2008), a modification of Mr. Bayes 3·1.2; and Bayesian Concordance Analysis (BCA; Ané et al., 2007) with BUCKy v.1.4.2 (Larget et al., 2010). A concordance factor can be defined as the proportion of the genome sampled that supports a given node in a species tree (Ané et al., 2007; Baum, 2007).
A partitioned analysis was run in *BEAST with 100 million Markov chain Monte Carlo (MCMC) iterations, under an uncorrelated relaxed clock (Drummond et al., 2006), with random starting trees for each partition generated under a constant population size coalescent model. A Yule model was applied as prior for the species tree likelihood. The MCMC statistics and trees were sampled every 1000 iterations. A maximum clade credibility (MCC) consensus species tree was created from 80 000 trees (100 000 trees saved minus 20 000 burn-in).
MrBayes v. 3.2.1 (Ronquist et al., 2012) was used to generate tree files for BCA in BUCKy. Two replicate analyses were run on each partition, with 10 million iterations and four chains, sampling every 500 iterations, and a burn-in of 1000 for summarizing posterior samples of both parameter values and trees. The resulting tree files from the two runs for each partition contained a total of 40 000 trees which were transformed to BUCKy infiles with a further burn-in of 10 000. BUCKy was run four times for 10 million MCMC updates each with four chains.
BEST was run twice concurrently for 100 million MCMC rounds with 32 chains on a 32-core parallel processing server. The log was written to and a tree sampled each 2500 iterations. The total number of trees sampled across both runs was 80 000, 50 % of which were discarded as further burn-in before generating an MCC consensus species tree.
Divergence age estimation
BEAST was used to perform an age estimation of divergence. Three fossils used for stratigraphic calibration points were from Hermsen et al. (2006) as used similarly by Nagalingum et al. (2011) with the following priors: stem node of Bowenia [lower = 33·9, upper = 265·7 million years (My)], stem node of Lepidozamia (lower = 33·9, upper = 265·7 My) and stem node of Dioon (lower = 55·8, upper = 265·7 My). The only monophyly constraint was placed on the outgroup, Cycas. A non-partitioned analysis was run since KAKUSAN found the same highest likelihood model for all five loci (see the Results). A random starting tree was used, and a random local clock model was applied for determination of tree likelihood, as all other models resulted in zero tree likelihood for the starting trees. A total of 100 million iterations were run in BEAST, with log and tree samples every 2500th round. The total number of trees sampled was 40 000, 10 % of which were discarded as further burn-in before generating an MCC consensus species tree. Log output was evaluated in Tracer 1·5. TreeAnnotator in BEAST was used to generate MCC consensus trees from both BEAST and BEST output. All species trees were visualized with FigTree v.1·4 (Rambout, 2012).
Biogeographic analyses
Biogeographic analyses were conducted using RASP v.2·1a (Yu et al., 2010, 2011). Both the S-DIVA (Yu et al. 2010) and the dispersal–extinction–cladogenesis (DEC; Ree and Smith, 2008) methods were applied, using the MCC tree obtained from the BEAST age estimation analysis, and limiting ancestral area reconstructions to three to avoid all possible areas being assigned to deep nodes in the tree. Areas and their coding included in the analyses were A = Australia, B = China, C = Africa, D = Caribbean, E = Mexico, F = Northern Central America, G = South America and H = Southern Central America. Cycas angulata was used as the functional outgroup as only a single outgroup taxon can be designated in RASP.
RESULTS
Parsimony
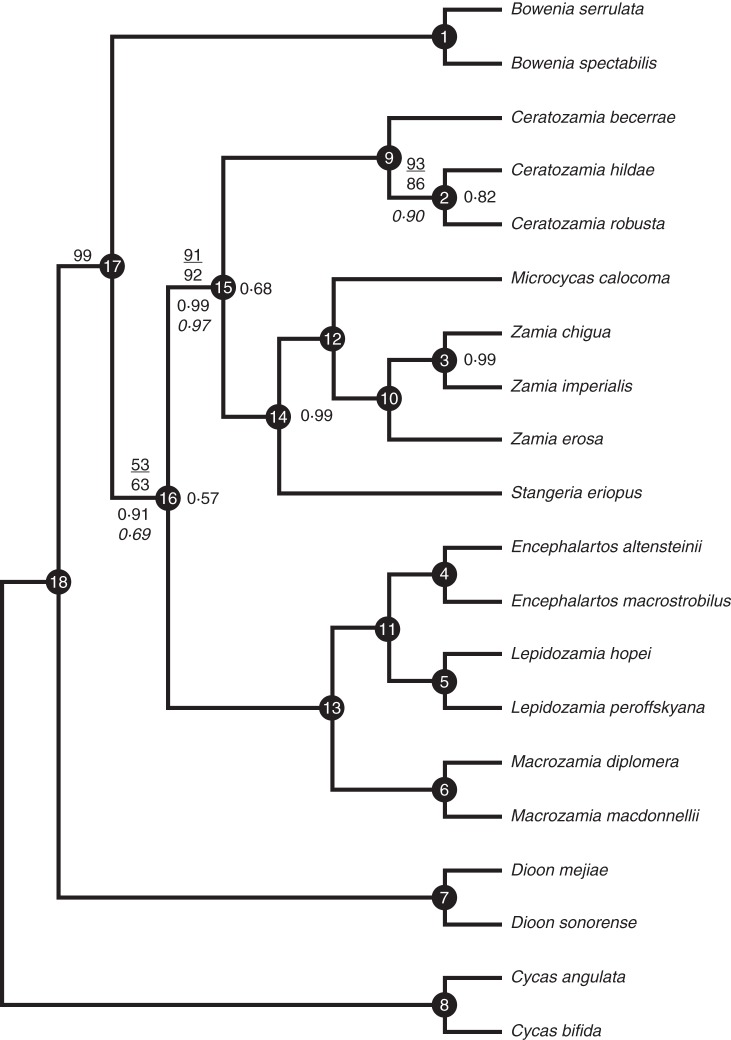
CyAG was the only locus that alone produced a single fully resolved tree (Table 2; Supplementary Data Fig. S1). A B&B search of the five locus concatenated matrix found a single fully resolved tree (Fig. 3) identical to the CyAG topology. Each non-monotypic genus is monophyletic with 100 % JK support at their crown nodes. Across 17 nodes of the ingroup, only two JK values were <92 % (nodes 2 and 16). Dioon is the first branch of the ingroup, with 100 % JK value at the stem node. Bowenia is the next branch of the tree with 99 % JK support at the stem. At the next node, two main clades are resolved with 63 % JK support at the stem, the lowest support in the topology. One unites the Old World Zamiaceae (with the exception of Stangeria) with 100 % JK support at the stem. Macrozamia is the first branch. Encephalartos and Lepidozamia are sister genera with 100 % JK support. The second clade (92 % JK support) unites the American Zamiaceae and Stangeria, with Ceratozamia as the first branch, and Stangeria as sister to Microcycas and Zamia with 100 % JK support. Non-partitioned Bremer indices ranged from 2 to 364 across the 17 nodes of the ingroup, with all but three >10 (Table 3). Both total and partitioned Bremer indices were weakest at nodes 2 and 16 (Table 3), which also received the lowest JK support.
Table 2.
Locus ID, locus description, alignment length, polymorphic and parsimony informative sites per locus, number of trees found, tree length, homoplasy index, consistency index and retention index
| Locus ID | Description | Alignment length | Polymorphic sites | Parsimony informative sites | Number of trees found | Tree length | Consistency index (CI)* | Homoplasy index (HI)* | Retention index (RI) |
|---|---|---|---|---|---|---|---|---|---|
| CyAG | MADS-box transcription factor family AGAMOUS | 1871 | 739 | 465 | 1 | 1048 | 0·7955 | 0·2045 | 0·8565 |
| COS26 | Uncharacterized protein | 752 | 204 | 138 | 32 | 266 | 0·8367 | 0·1633 | 0·8828 |
| GroES | GroES-like zinc-binding alcohol dehydrogenase family protein | 955 | 389 | 301 | 15 | 519 | 0·8442 | 0·1558 | 0·8870 |
| GTP | GTP-binding protein Era mRNA | 644 | 222 | 167 | 28 | 289 | 0·8534 | 0·1466 | 0·9045 |
| HTS | Histidyl-tRNA synthetase | 777 | 312 | 240 | 2 | 439 | 0·8623 | 0·1377 | 0·9025 |
| All loci combined | 4999 | 1866 | 1311 | 1 | 2569 | 0·8259 | 0·1741 | 0·8771 | |
* Excluding uninformative sites
Fig. 3.
Congruent tree topology found by concatenated branch and bound parsimony, likelihood, and three gene tree–species tree reconciliation methods for the five locus SCNG matrix across Cycadales with Cycas as outgroup. Only support values <1·0 or 100 % are shown. Numbers above branches are ML bootstrap (underlined) and parsimony jackknife support values for the single most-parsimonious tree; those at nodes are posterior probabilities (PP) from the *BEAST (Heled and Drummond, 2010) maximum clade credibility (MCC) consensus tree; those below branches are concordance factor scores from BUCKy (Larget et al., 2010) and PP (italic) from the BEST (Liu, 2008) MCC consensus species tree. Bremer (decay) indices are listed in Table 3 and reference the node numbers in black circles.
Table 3.
Total and partitioned Bremer indices for concatenated parsimony tree of Cycadales
| Node | Non-partitioned | Locus |
||||
|---|---|---|---|---|---|---|
| CyAG | COS26 | GroES | GTP | HTS | ||
| 1 | 115 | 47·5 | 13·5 | 32 | 8·5 | 21·5 |
| 2 | 3 | 4 | 0 | 0 | –1 | 0 |
| 3 | 18 | 6 | 0·5 | 10 | 0 | 1·5 |
| 4 | 53 | 28 | 4 | 4 | 5 | 12 |
| 5 | 48 | 19 | 1 | 11 | 7 | 10 |
| 6 | 67 | 32 | 7 | 11 | 5 | 12 |
| 7 | 115 | 47 | 12 | 28 | 8 | 20 |
| 8 | Outgroup | |||||
| 9 | 135 | 23 | 15 | 19 | 14 | 17 |
| 10 | 88 | 6 | 2 | 3 | 1 | –1 |
| 11 | 11 | 55 | 17 | 25 | 7 | 31 |
| 12 | 68 | 13 | 8 | 15 | 13 | 19 |
| 13 | 39 | 11 | 3 | 9 | 6 | 10 |
| 14 | 22 | 22 | –1 | 0 | 0 | 1 |
| 15 | 6 | 4 | 2 | 2 | 0 | –2 |
| 16 | 2 | –1 | 2 | 0 | 0 | 1 |
| 17 | 17 | 5 | 1 | 6 | 3 | 3 |
| 18 | 364 | 100 | 40 | 70 | 70 | 84 |
See Fig. 1 for the key to node numbers.
Negative numbers signify incongruous support at that node for that partition. A value of zero indicated neutral support.
Nucleotide substitution model
By the AICc, a general time-reversible (GTR) model with gamma correction was the best fit for all five loci. This model was applied to all MrBayes and BEAST analyses.
Maximum likelihood analyses
Maxinum likelihood resolved the same topology as parsimony, with similar support scores (Fig. 3). The mean tree score (log likelihood) for the analysis = –19 645·89. As with all other analyses, the most weakly supported node in the tree was number 16, which resolves a sister relationship between the encephalartoid and zamioid sub-clades.
Species tree estimation
All three gene tree–species tree reconciliation approaches converged on exactly the same tree topology as the concatenated parsimony analysis (Fig. 3). The effective sample size (ESS) scores from the *BEAST analysis were all >100, with most >1000. Posterior values in the MMC consensus species tree were <0·99 in only two nodes: the crown node for Old World and New World Zamiaceae (plus Stangeria) after the branching of Bowenia (node 16 in Fig. 3), and the stem node of Ceratozamia (node 15).
The MrBayes runs to develop gene tree sets for BUCKy achieved stationarity based on the internal diagnostics of MrBayes and examination in Tracer. The nodes in the MCC tree from the BCA had concordance factor (CF) scores of 1 in all but two nodes (Fig. 3): node 16 (CF = 0·91) and node 15 (0·99).
The ESS scores in Tracer from the BEST run were >1000 for all statistics except overall likelihood (LnL), all five gene tree likelihoods (TL) and gene mutation rate (GeneMu). These latter were <100, despite a 3 week run on 32 cores. The sub-optimal scores decreased if the burn-in of the log files in Tracer was increased, which suggested that the run duration should have been longer. We then increased the number of MCMC iterations to 150 million for a second run with little improvement of these same ESS scores. Despite this fact, the topology of MCC consensus species tree from BEST was exactly the same as for all the other analyses, thus we ceased running BEST at this point. The posterior probability (PP) was <1·0 at only three nodes of the BEST MCC consensus species tree: node 16 (PP = 0·56), and nodes 2 and 15 (0·9 and 0·97, respectively).
Age estimation
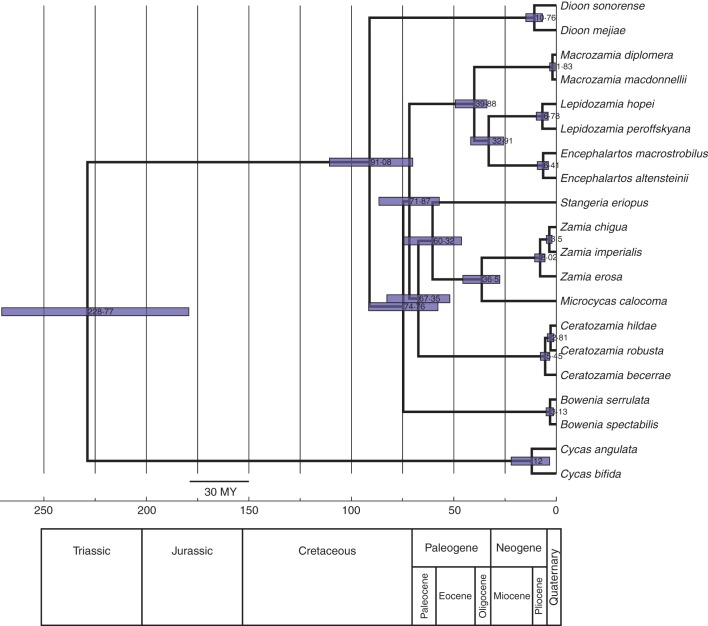
The ESS scores in Tracer for the age estimation analysis in BEAST were all >200, with many >1000. The mean estimated age of the crown node of Cycadales is 228·8 My (Fig. 4), with a 95 % highest posterior density (HPD) of 179·2–270·6. The crown node of Zamiaceae has a mean age of 91·1 My, with 95 % HPD of 70·1–110·8. The stem node of Bowenia is dated to a mean of 74·8 My (95 % HPD = 56·4–91·0), and the split between the Old World Zamiaceae (less Stangeria) and the New World (plus Stangeria) is estimated to have occurred approx. 3 My later (71·9 My; 95 % HPD = 55·7–87·2), followed 4–5 My later by the branching of Ceratozamia and Stangeria–Microcycas–Zamia (67·4 My; 95 % HPD = 52·0–82·7). The most recent common ancestor (MRCA) of Stangeria and Microcycas–Zamia is dated to 60·3 My (95 % HPD = 46·3–74·2) and that of Microcycas and Zamia at 36·5 My (95 % HPD = 27·6–45·6). Within the Encephalartaeae, the branching of Macrozamia is estimated at 39·9 My (95 % HPD = 33·9–49·2). The MRCA of Encephalartos and Lepidozamia has a mean age of 32·9 My (95 % HPD = 25·8–41·7). Crown nodes of all genera, with the exception of Cycas and Dioon (mid-Miocene), are dated to the late Miocene or to the Pliocene (Fig. 4), except Macrozamia (Pleistocene), with fairly narrow HPD ranges. However, the sampling of the larger cycad genera is limited. We tested removing all but one of each of the calibration points, respectively, and found a <10 % change in node ages in each run, but a corresponding increase in the length of the 9 5 % HPD intervals (not shown).
Fig. 4.
Chronogram from age estimation analysis in BEAST (Drummond et al., 2012). Blue bars at nodes are 95 % HPD.
Biogeographic analysis
The most likely area for the root node of the phylogeny is Australia–Mexico–Northern Central America (AEF) by the S-DIVA method, and Australia–China–Northern Central America (AEF) by DEC (Fig. 5). While both methods hypothesize two dispersal events and one extinction event subsequently (Table 4), probabilities are low, especially with DEC (P = 0·0082), though an ancestral presence in China (or somewhere in greater Asia) is more likely than in Mexico. In either case, there is little confidence for any area optimization for the root node.
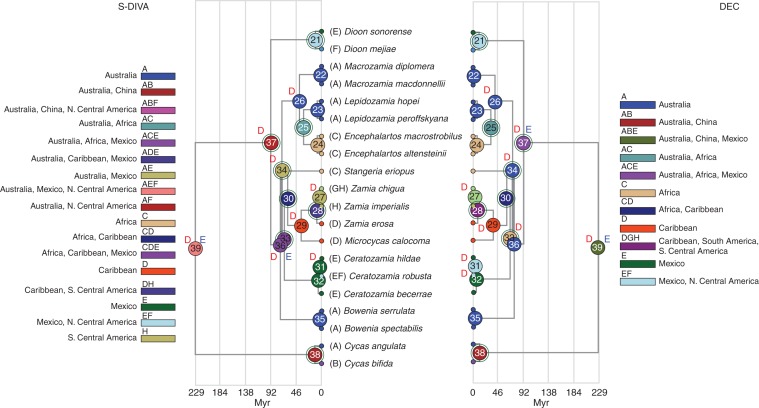
Fig. 5.
Chronogram from age estimation analysis in BEAST (Drummond et al., 2012) optimized with the most likely area on internal nodes as determined by S-DIVA (Yu et al., 2010) and DEC (Ree and Smith, 2008) biogeographic analyses. Green circles around nodes indicate vicariance, D = dispersal, E = extinction. See Table 4 for details.
Table 4.
Comparison of S-DIVA (Yu et al., 2010) and DEC (Ree and Smith, 2008) biogeographic analyses on the five gene sequence phylogeny of Cycadales
| Node | S-DIVA | DEC |
|---|---|---|
| 21 | Events: D: 0, V: 1, E: 0 | Events: D, 0; V, 1; E, 0 |
| Event route: EF → E|F | Event route: EF → E|F | |
| P = 1 | P = 1 | |
| 22 | Events: D, 0; V, 0; E, 0 | Events: D, 0; V, 0; E, 0 |
| Event route: A → A A → A|A | Event route: A → A A → A|A | |
| P = 1 | P = 1 | |
| 23 | Events: D, 0; V, 0; E, 0 | Events: D, 0; V, 0; E, 0 |
| Event route: A → A A → A|A | Event route: A → A A → A|A | |
| P = 1 | P = 1 | |
| 24 | Events: D, 0; V, 0; E, 0 | Events: D, 0; V, 0; E, 0 |
| Event route: C → C C → C|C | Event route: C → C C → C|C | |
| P = 1 | P = 1 | |
| 25 | Events: D, 0; V, 1; E, 0 | Events: D, 0; V, 1; E, 0 |
| Event route: AC → C|A | Event route: AC → C|A | |
| P = 1 | P = 0·7144 | |
| 26 | Events: D, 1; V, 0; E, 0 | Events: D, 1; V, 0; E, 0 |
| Event route: A → A A → AC A → AC|A | Event route: A → A A → AC A → AC|A | |
| P = 1 | P = 0·3881 | |
| 27 | Events: D, 1; V, 0; E, 0 | Events: D, 1; V, 0; E, 0 |
| Event route: H → H H → GH H → GH|H | Event route: GH → GH H → GH|H | |
| P = 1 | P = 0·6781 | |
| 28 | Events: D, 0; V, 1; E, 0 | Events: D, 0; V, 1; E, 0 |
| Event route: DH → D|H | Event route: DGH → D|GH | |
| P = 1 | P = 0·4069 | |
| 29 | Events: D, 1; V, 0; E, 0 | Events: D, 2; V, 0; E, 0 |
| Event route: D → D D → DH D → D|DH | Event route: D → D D → DGH D → D|DGH | |
| P = 1 | P = 0·4225 | |
| 30 | Events: D, 0; V, 1; E, 0 | Events: D, 0; V, 1; E, 0 |
| Event route: CD → C|D | Event route: CD → C|D | |
| P = 1 | P = 0·2214 | |
| 31 | Events: D, 1; V, 0; E, 0 | Events: D, 1; V, 0; E, 0 |
| Event route: E → E E → EF E → E|EF | Event route: EF → EF E → E|EF | |
| P = 1 | P = 0·5399 | |
| 32 | Events: D, 0; V, 0; E, 0 | Events: D, 1; V, 0; E, 0 |
| Event route: E → E E → E|E | Event route: E → E E → EF E → E|EF | |
| P = 1 | P = 0·2876 | |
| 33 | Events: D, 0; V, 1; E, 0 | Events: D, 3; V, 1; E, 0 |
| Event route: CDE → E|CD | Event route: C → ECD → E|CD | |
| P = 0·3333 | P = 0·0362 | |
| 34 | Events: D, 2; V, 1; E, 0 | Events: D, 2; V, 1; E, 0 |
| Event route: AE → CDEA → CDE|A | Event route: A → CA → C|A | |
| P = 0·0556 | P = 0·021 | |
| 35 | Events: D, 0; V, 0; E, 0 | Events: D, 0; V, 0; E, 0 |
| Event route: A → A A → A|A | Event route: A → A A → A|A | |
| P = 1 | P = 1 | |
| 36 | Events: D, 1; V, 0; E, 1 | Events: D, 0; V, 0; E, 0 |
| Event route: ACE → AE → AE A → A|AE | Event route: A → A A → A|A | |
| P = 0·0417 | P = 0·0427 | |
| 37 | Events: D, 3; V, 1; E, 0 | Events: D, 1; V, 1; E, 1 |
| Event route: AF → AF E → ACEF E → ACE|EF | Event route: ACE → AE → AEF → A|EF | |
| P = 0·0625 | P = 0·0393 | |
| 38 | Events: D, 0; V, 1; E, 0 | Events: D, 0; V, 1; E, 0 |
| Event route: AB → A|B | Event route: AB → A|B | |
| P = 1 | P = 1 | |
| 39 | Events: D, 2; V, 0; E, 1 | Events: D, 2; V, 0; E, 1 |
| Event route: | Event route: | |
| AEF → AF → AF A → ABF A → AB|AF | ABE → ABE A → ABCE A → AB|ACE | |
| P = 0·0625 | P = 0·0082 | |
| Global events | ||
| Global dispersal: 12 | Global dispersal: 14 | |
| Global vicariance: 8 | Global vicariance: 8 | |
| Global extinction: 2 | Global extinction: 2 | |
See Fig. 3 for node identity.
D = dispersal, V = vicariance, E = extinction, P = probability of events.
The crown node of Cycas (38) is situated in Australia and China with subsequent vicariance between the two areas by both methods with P = 1, but the minimal sampling of this genus weakens the optimization.
The crown node of Zamiaceae (37) is optimized quite differently between the two methods (Table 4), neither with high probability. S-DIVA calls Australia and Northern Central America, with three subsequent dispersal events [Africa, twice to Mexico, and one vicariance (Australia–Africa–Mexico|Mexico–Northern Central America)], while DEC considers Australia–Africa–Mexico as most likely, with subsequent single dispersal, vicariance and extinction events, the latter eliminating an early presence in Africa, dispersal to Northern Central America, and ultimately vicariance between Australia and Mexico–Northern Central America.
The stem node of Bowenia (36) is optimized differently by the two methods, but with similar probabilities (Table 4). S-DIVA hypothesizes an ancestral area comprising Australia, Africa and Mexico, with extinction eliminating Africa, a re-entry into Australia and vicariance between Australia and Australia–Mexico. DEC hypothesizes a simpler model: stasis in Australia. For the crown node of Bowenia (35), stasis in Australia is the optimal scenario with both methods (P = 1).
Node 34 (Fig. 5) is the ancestral node of the tribes Encephalarteae and Zamieae–Ceratozamieae [Stevenson's (1992) Zamioideae with the inclusion of Stangeria]. Both methods hypothesize two dispersal events and one vicariance event, but with different routes. From an ancestral Australia–Mexico, S-DIVA resolves two dispersals to Africa and the Caribbean, respectively, and vicariance between Australia and Africa–Mexico–Caribbean P = 0·0556). DEC optimizes two dispersals into Africa from ancestral Australia and later vicariance between the two (P = 0·0362).
The stem node of Ceratozamia (33) is resolved as a vicariance between Mexico and Africa–Mexico (P = 0·333). DEC envisages three dispersals, from Africa to Mexico, the Caribbean and again to Africa, with vicariance between Mexico and Africa–Caribbean (P = 0·0362). The crown node for Ceratozamia (32) is optimized as stasis in Mexico by S-DIVA (P = 1) or dispersal from Mexico to Central America by DEC (P = 0·2876).
Both area optimization methodologies converged on the same scenario for the crown node of Stangeria–Zamia–Microcycas (30), which involves vicariance between Africa and the Caribbean. The two regions together comprise the ancestral area. P = 1 with S-DIVA, and 0·2214 with DEC. The crown nodes of Zamia and Microcycas (29) were similar with both methods but differed in a second dispersal event with DEC (P = 0·4425), from the Caribbean to South America, while that of S-DIVA only included Southern Central America (P = 1). The crown node scenarios for Zamia (28) reflected this, in that DEC retained South America in the ancestral area with DEC (P = 0·4069), with the single vicariance between the Caribbean and Southern Central America–South America, vs. only Southern Central America and the Caribbean with S-DIVA (P = 1).
Optimization of the crown node of Encephalarteae (26) was exactly the same with both methods (S-DIVA P = 1, DEC P = 0·3881) with dispersal to Africa from ancestry in Australia. The split between Lepidozamia and Encephalartos (25) was also the same in both S-DIVA (P = 1) and DEC (P = 0·7144), with vicariance between Australia and Africa.
The remaining nodes (21–24) had exactly the same scenarios and probabilities (P = 1) with both methods (Fig. 5, Table 3), involving geographic stasis in the ancestral area except for a vicariance event between the two ancestral areas of Dioon (21).
DISCUSSION
Supra-generic arrangements in the Cycadales and the placement of Bowenia, Dioon and Stangeria
Complete congruence of species tree topology from three different gene tree–species tree reconciliation approaches with the single tree resolved by parsimony analysis of the concatenated matrix (Fig. 3) supports the following conclusions concerning phylogenetic placement for three formerly sedis incertae genera of the Cycadales: (1) Dioon is sister to all other members of sub-order Zamiineae sensu Stevenson (1992); (2) Bowenia is the next branch in Zamiineae; and (3) Stangeria is sister to Microcycas/Zamia. Our tree topology (Fig. 3) is identical to that of Bogler and Francisco-Ortega (2004) based on combined nrDNA ITS2 and plastid trnL-F (Fig. 2C), the ITS tree of Chaw et al. (2005) and the combined analysis of Griffith et al. (2012), but with improved support values.
Several morphological synapomorphies support the clades recovered in our study. For instance, the large sister clade to Dioon includes the vast majority of cycad genera, all of which have stomata on their sporangia (Dehgan et al., 1993). The presence of lateral lobes in the megasporophylls and vascular bundles in the pith defines the clade comprised of Encephalartos, Lepidozamia and Macrozamia (Stevenson, 1990). Unequally branched trichomes are shared only by Microcycas and Zamia (Stevenson, 1990). The sister relationship of African Encephalartos and Australian Lepidozamia is supported by common mucilage chemistry that is unique to these two genera (De Luca et al., 1982).
We are not aware of morphological synapomorphies supporting two of the main clades resolved in the species tree (Fig. 3): (1) Ceratozamia–Stangeria–Microcycas–Zamia; and (2) Stangeria–Microcycas–Zamia. The molecular topologies of Bogler and Francisco-Ortega (2004), Chaw et al. (2005), Zgurski et al. (2008), Nagalingum et al. (2011) and Crisp and Cook (2011) supported the Ceratozamia–Stangeria–Microcycas–Zamia clade. Not all of the gene topologies recovered in our study support this clade; indeed, based on the partitioned Bremer index analysis (Table 3, node 15), support for this group came mostly from three of the loci (i.e. CyAG, COS26 and GroES). Only the molecular studies by Bogler and Francisco-Ortega (2004), Chaw et al. (2005), Zgurski et al. (2008) in part, and the combined molecular and morphology analysis of Griffith et al. (2012) recovered the clade composed of Stangeria, Microcycas and Zamia. The partitioned Bremer index analysis (Table 3, node 14) showed that this group is mostly supported by the CyAG data set.
The last comprehensive systematic arrangement of the Cycadales is that of Stevenson (1992), based on cladistic analyses of morphological traits (Stevenson, 1990), with clades formally labelled from the rank of sub-order to sub-tribe (Fig. 1). More recently, Christenhusz et al.,(2011), following the plastid-based sequence phylogeny of Zgurski et al. (2008), treated the Cycadales as consisting of two families: Cycadaceae including only the genus Cycas, and Zamiaceae including the rest of the genera with no recognized subfamilial ranks.
No available molecular-based phylogenies (Fig. 2) support the placement of Stangeria and Bowenia within the same clade [i.e. Stangeriaceae in Stevenson's (1992) classification]. In contrast, all available molecular phylogenies support the Cycadineae, Encephalarteae and Zamieae [excluding Chigua, which has been sunk back into Zamia (Chaw et al., 2005; Lindstrom, 2009)] as three monophyletic groups, but are highly discordant concerning the monophyly of the other suprageneric taxa proposed by Stevenson (1992). Interestingly, all molecular studies placed the tribes Ceratozamieae and Zamieae in the same clade (Fig. 2); however, several of the recovered topologies showed Stangeria as an anomalous member of this clade (Hill et al., 2003; Rai et al., 2003; Bogler and Francisco-Ortega, 2004; Chaw et al., 2005; Zgurski et al., 2008; Nagalingum et al., 2011, in part).
The chronogram of Nagalingum et al. (2011) was based on a single nuclear gene (PHYP), which resolved Macrozamia as sister to a clade of Stangeria and the American Zamioideae sensu Stevenson (1992), but including Dioon. Only when two chloroplast regions were included in the matrix, despite their much lower taxonomic coverage, did Macrozamia resolve as it does in all of our gene trees and species trees (Fig. 3, Supplementary Data Fig. S1), as sister to the remainder of Encephalarteae. However, none of the trees shown in Nagalingum et al. (2011) is identical to ours (Fig. 3).
Age estimations and biogeographic associations
The earliest known cycad fossils date to the Early Permian of north China, Crossozamia (Norstog and Nichols, 1997) and Pseudotenis (Pott et al., 2010), approx. 300–280 million years ago (Mya). Our mean crown age for modern Cycadales of approx. 230 My (95 % HPD = 271–179; Fig. 4), may thus be a reasonable estimate. This would also support a putative ancestral distribution in China for the order (Tang, 2004), which appears at the crown node of our optimization with DEC, at least in part (Fig. 5, Table 4). North American fossils from this era, previously assigned to Cycadales, are now considered to represent different but related orders (Anderson et al., 2007).
The Mesozoic was characterized by an increased occurrence and diversity of fossil cycads (Hermsen et al., 2009), broadly distributed throughout the relatively uniform climate of the super-continent Pangaea. No relationships to living cycad genera have been proposed for these fossil genera (Anderson and Anderson, 1989). Fossils identified as Cycas, sister to the rest of the extant Cycadales, have been described from the late Cretaceous of Greenland (Osborne, 2002), and from the early Cenozoic of Japan and China (Liu et al., 1991). The lack of Cycas fossils from the Southern hemisphere suggests that the genus was absent from southern Pangaea. A crown age of 12 My for Cycas (Fig. 4) supports the hypothesis of Hill (1999) that the modern presence of the genus in Africa, Australia and the Pacific islands probably represents a relatively recent dispersal from the ancestral area.
The dating of the stem node of Dioon in the Cretaceous (Fig. 2), with area optimizations (Fig. 5) of Australia–Northern Central America (S-DIVA) or Australia–Africa–Mexico (DEC), probably reflects a broad distribution of an ancestral cycad flora across Laurasia, the northern half of Pangaea, which began to split apart in the Jurassic, of which Dioon remains a surviving relict. The lineage (tribe Diooeae) has fossils from the Triassic of western Laurasia (Lyssoxylon) and southwestern Gondwana (Micheliilloa) (Artabe et al., 2005). South American (Bororoa) elements of this tribe disappeared after the Cenozoic (Artabe et al., 2005).
Gondwana, the southern portion of Pangaea, also hosted a great diversity of cycads and cycad-like plants in the area that would become Australia (Delevoryas, 1975). Bowenia, the stem node of which is estimated at 75 My, may represent a surviving remnant of this diversity.
During the late Cretaceous period through the early Tertiary, the remaining major clades of extant cycads [Encephalartaeae and Zamioideae sensu Stevenson (1992) + Stangeria] were established. The dispersal/vicariance is dated at approx. 72 Mya (Fig. 4), with an ancestral distribution in Australia with DEC or Australia–Mexico with S-DIVA (Fig. 5, Table 4). This split may represent the termination of direct exchange between Laurasia and Gondwana.
Evaluated on the basis of extant distribution of genera, the Encephalarteae would appear to represent an eastern Gondwana lineage the stem age of which (approx. 72 My) suggests was initially isolated by the break up of that continent. While the sister relationships of Australian endemic Lepidozamia and the African endemic Encephalartos is optimized by both S-DIVA and DEC as vicariance between Africa and Australia (Fig. 5, Table 4), by the late Paleogene, the stem age of this clade, the two continents were already situated well apart from each other (Raven and Axelrod, 1974). This relationship does not fit any of the Southern hemisphere biogeographic scenarios recognized by Sanmartin and Ronquist (2004), and for now remains an enigma. However, fossil remains for the Encephalarteae (Cantrill, 2000; Artabe et al., 2004, 2005; Martínez et al., 2012) have been found from the Jurassic of India (Fascivarioxylon), the Cretaceous of Antarctica (Centricycas) and Argentina (Neochamberlainia, Worsdellia and Wintucycas), and the Tertiary of Argentina (Menucoa). It has been suggested that the tribe originated in the Triassic of western Laurasia (Charmorgia; Artabe et al., 2005), but this latter interpretation implies Dioon as a member of the group (Zamiaceae subfamily Encephalartoideae), which it most certainly is not (Fig. 3).
The asteroid impact at the Cretaecous/Tertiary (K–T) boundary, approx. 65·5 Mya (Schulte et al., 2010), resulted in extinction of nearly a third of terrestrial vegetation and great declines in species abundance (Nichols and Johnson, 2008). The extinction rate in tropical North America may have been as high as 60–70 % (Nichols and Johnson, 2008). The effects of the K–T boundary impact on cycads are not as clear, but the time period represented a major diversification time for the Cycadales. The branch lengths of nodes dated from approx. 75–60 Mya are very short (Fig. 4), suggesting rapid diversification (Kubatko and Degnan, 2007), perhaps in this case associated with drastic environmental changes. Tang (2012) hypothesizes that modern cycad genera in the Americas represent lineages that evolved from the survivors of the K–T extinction. The stem nodes of Ceratozamia (67 Mya) and the Zamia–Microcyas–Stangeria clade (60 Mya) may reflect the influence of this catastrophic event. North American leaf remains identified as cycads have been dated to the first half of the Paleogene, apparently representing taxa that were not known from before the K–T boundary (Mustoe, 2008).
Fossils from the early Cenozoic have been variously related to modern taxa, e.g. Dioonopsis, which has been classified as Dioon, Ceratozamia or Zamia, but has unique cuticular characteristics (Erdei et al., 2012). An undescribed fossil from an Eocene deposit in Oregon resembles Dioon (Manchester, 1981; Tang, 2012), at least superficially. Younger fossils from Oligocene and Miocene strata in Europe bear the diagnostic cuticle morphology of Ceratozamia (Kvaĉek, 2002, 2004). Pseudodioon, a Miocene fossil from Turkey, bears macromorphological characteristics of Dioon and anatomical features of Cycas (Erdei et al., 2010). Eostangeria, described from Paleocene deposits in Wyoming, and Eocene fossils from Europe and Oregon, greatly resembles Stangeria but has divergent cuticle morphology (Kvaĉek and Manchester, 1999; Uzunova et al., 2001). A recent fossil from Patagonia, Argentina pushes a leaflet mid-ribbed zamioid lineage to the early Cretaceous (Passalia et al., 2010).
In the late Eocene, there were major extinction events linked to global cooling and a decrease in atmospheric carbon dioxide (Jaramillo et al., 2006; Zachos et al., 2008; Kunzig, 2011). These climatic changes led to latitudinal shifts of the main vegetation belts of the planet and probably the elimination of cycads from higher latitudes of North America and Eurasia (Tang, 2012).
Our results indicate that extant diversification of cycad species occurred in the relatively recent past (Fig. 4), in agreement with Nagalingum et al. (2011). No crown node of any modern genus of cycad in our chronogram has a mean date estimate >12 Mya. Crisp and Cook (2011) also showed that most of the current species diversification within the Cycadales occurred relatively recently, although many of their age estimates placed these speciation events in the Eocene and Oligocene rather than in the Miocene.
The sister relationship between the South African endemic monotypic Stangeria eriopus and Zamia–Microcycas, dated to the early Paleocene but with a 95 % HPD that extends back into the late Cretaceous (Fig. 4), is also enigmatic, implying an African–Caribbean vicariance at the ancestral node (Fig. 5, Table 4). Mesodescolea, a Cretaceous fossil genus described from Argentina, has some features in common with Stangeria (Artabe et al., 2004), as does the Laurasian Eostangeria (Kvaĉek and Manchester, 1999; Uzunova et al., 2001). Eostangeria also has elements in common with Zamia (Usunova et al., 2001). We conclude that S. eriopus represents the only surviving branch of a lineage that was once more widely distributed. The available fossil evidence suggests that while Gondwana once enjoyed a cosmopolitan tropical cycad flora from east to west (Sabato, 1990; Artabe and Stevenson, 1999; Tang, 2006), the South American elements of this flora were extirpated as Africa and South America separated, while in Africa and Australia, related taxa evolved and persisted.
The split between the Cuban endemic Microcycas and the more broadly distributed Zamia is estimated in our chronogram at approx. 36 Mya when Cuba was already a well-established and isolated island (Graham, 2003a, b). This is more or less the same time in the late Eocene/early Oligocene to which the stem nodes of the modern Encephalarteae are dated (Fig. 4), a period of global climate change (Jaramillo et al., 2006) that seems to have resulted in cycad cladogenesis in both hemispheres. Tang (2002) hypothesized that modern cycads colonized the Greater Antilles from Mesoamerica during an interval during the late Cretaceous/early Cenozoic when they may have formed a land bridge with the south of Mesoamerica (Pindell and Kennan, 2009), ground zero for the K–T asteroid impact at about the same time.
Massive extinctions of the Cycadales meant that many areas where several of the genera existed in the past do not currently have any of their living representatives. This might explain why the inferences in the historical biogeographic reconstructions for many deeper nodes of our five gene phylogeny were poorly supported (Fig. 5, Table 4).
Concluding remarks
Our five SCNG phylogeny of Cycadales provides the most congruent estimate of the phylogeny of the order yet presented. An emerging picture for the biogeography and evolutionary history of Cycadales is supported by both the recent molecular phylogenies and the fossil record. Our results suggest that the current supra-generic classification of the order needs to be revisited. Further research is underway to determine if there are morphological and anatomical features to support some of the clades detected in our study and others with congruent results (Bogler and Francisco-Ortega, 2004; Chaw et al., 2005; Griffith et al., 2012). The extant diversity of cycad species is recent and post-dates the Cretaceous–Paleogene boundary. Our topology, resolved by both concatenation of five loci and three methods of gene tree–species tree reconciliation, suggests massive extinctions prior to the most recent diversification events, and the elimination of certain lineages from entire geographic areas, including close relatives of modern cycads. This hypothesis is supported by the fossil record. Biogeographic reconstructions of cycad ancestral areas are compromised by these extinction episodes and the fact that current distributions do not mirror those from the past.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank S. Wang, M. Alford and T. Samarakoon for providing a sample and recipe for the PCR adjuvant TBT-PAR 5X. This is contribution number 261 from the Tropical Biology Program of Florida International University. This work was supported by National Science Foundation Grant DEB 1050340 and by a Christiane Tyson Research Fellowship to D.S-L. through Montgomery Botanical Center.
LITERATURE CITED
- Albach DC, Meudt HM. Phylogeny of Veronica in the Southern and Northern Hemispheres based on plastid, nuclear ribosomal and nuclear low-copy DNA. Molecular Phylogenetics and Evolution. 2010;54:457–471. doi: 10.1016/j.ympev.2009.09.030. [DOI] [PubMed] [Google Scholar]
- Álvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Anderson JM, Anderson HM. Rotterdam: A.A. Balkema; 1989. Palaeoflora of southern Africa, Molteno Formation (Triassic), volume 2: Gymnosperms (excluding Dicroidium). [Google Scholar]
- Anderson J, Anderson H, Cleal C. Brief history of the gymnosperms: classification, biodiversity, phytogeography and ecology. Strelitzia. 2007;20:1–280. [Google Scholar]
- Ané C, Larget B, Baum DA, Smith SD, Rokas A. Bayesian estimation of concordance among gene trees. Molecular Biology and Evolution. 2007;24:412–426. doi: 10.1093/molbev/msl170. [DOI] [PubMed] [Google Scholar]
- Artabe AE, Stevenson DW. Fossil Cycadales of Argentina. Botanical Review. 1999;65:219–238. [Google Scholar]
- Artabe AE, Zamuner AB, Stevenson DW. Two new petrified cycad stems, Brunoa gen. nov. and Worsdellia gen. nov., from the Cretaceous of Patagonia (Bajo de Santa Rosa, Río Negro Province), Argentina. Botanical Review. 2004;70:121–133. [Google Scholar]
- Artabe AE, Zamuner AB, Stevenson DW. A new genus of Late Cretaceous cycad stem from Argentina, with reappraisal of known forms. Alcheringa. 2005;29:87–100. [Google Scholar]
- Avise JC. Phylogeography: the history and formation of species. Cambridge, MA: Harvard University Press; 2000. [Google Scholar]
- Avise JC, Wollenberg K. Phylogenetics and the origin of species. Proceedings of the National Academy of Sciences, USA. 1997;94:7748–7755. doi: 10.1073/pnas.94.15.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum DA. Concordance trees, concordance factors, and the exploration of reticulate genealogy. Taxon. 2007;56:417–426. [Google Scholar]
- Bogler DJ, Francisco-Ortega J. Molecular systematic studies in cycads: evidence from trnL intron and ITS2 rDNA sequences. Botanical Review. 2004;70:260–273. [Google Scholar]
- Braun H, Czihal A, Shutov AD, Baumlein H. A vicilin-like seed protein of cycads: similarity to sucrose-binding proteins. Plant Molecular Biology. 1996;31:35–44. doi: 10.1007/BF00020604. [DOI] [PubMed] [Google Scholar]
- Brenner ED, Stevenson DW, Twigg RW. Cycads: evolutionary innovations and the role of plant-derived neurotoxins. Trends in Plant Science. 2003a;8:446–452. doi: 10.1016/S1360-1385(03)00190-0. [DOI] [PubMed] [Google Scholar]
- Brenner ED, Stevenson DW, McCombie RW, et al. Expressed sequence tag analysis in Cycas, the most primitive living seed plant. Genome Biology. 2003b;4:R78. doi: 10.1186/gb-2003-4-12-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler RJ, Barrett PM, Kenrick P, Penn MG. Testing co-evolutionary hypothesis over geological timescales: interactions between Mesozoic non-avian dinosaurs and cycads. Biological Reviews. 2009;84:73–89. doi: 10.1111/j.1469-185X.2008.00065.x. [DOI] [PubMed] [Google Scholar]
- Cantrill DJ. A petrified cycad trunk from the Late Cretaceous of the Larsen Basin, Antarctica. Alcheringa. 2000;24:307–318. [Google Scholar]
- Caputo P, Cozzolino S, De Luca P, Moretti A. Cycad classification. Concepts and recommendations. Wallingford, UK: CABI Publishing; 2004. Molecular phylogeny of Zamia (Zamiaceae). In: Walters T, Osborene R; pp. 149–157. [Google Scholar]
- Chaw S-M, Walters TW, Chang C-C, Hu S-H, Chen S-H. A phylogeny of cycads (Cycadales) inferred from chloroplast matK gene, trnK intron, and nuclear rDNA ITS region. Molecular Phylogenetics and Evolution. 2005;37:214–234. doi: 10.1016/j.ympev.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Chiang Y-C, Hung K-H, Moore S-J, et al. Paraphyly of organelle DNAs in Cycas sect. Asiorientalis due to ancient ancestral polymorphisms. BMC Evolutionary Biology. 2009;9:161. doi: 10.1186/1471-2148-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenhusz MJM, Reveal JL, Farjon A, Gardner MF, Mill RR, Chase MW. A new classification and linear sequence of extant gymnosperms. Phytotaxa. 2011;19:5570. [Google Scholar]
- Crisp MD, Cook LG. Cenozoic extinctions account for the low diversity of extant gymnosperms compared with angiosperms. New Phytologist. 2011;192:997–1009. doi: 10.1111/j.1469-8137.2011.03862.x. [DOI] [PubMed] [Google Scholar]
- De Castro O, Vazquez-Torres M, De Luca P. Utility of AFLP markers for the assessment of molecular relationships in Ceratozamia Brongn. (Zamiaceae) Plant Biosystems. 2006;140:221–228. [Google Scholar]
- Degnan JH, Rosenberg NA. Discordance of species trees with their most likely gene trees. PLoS Genetics. 2006;2 doi: 10.1371/journal.pgen.0020068. e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends in Ecology and Evolution. 2009;24:332–340. doi: 10.1016/j.tree.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Dehgan B, Schutzman BA, Almira F. Utilization of scanning electron microscopy on the study of surface features in Cycadales. In: Stevenson DW, Norstog KJ, editors. The biology, structure and systematics of the Cycadales. Milton, Queensland: Palm & Cycad Societies of Australia; 1993. pp. 228–235. Proceedings of the Second International Conference on Cycad Biology. [Google Scholar]
- De Luca P, Moretti A, Sabato S, Gigliano GS. A comparative study of cycad mucilages. Phytochemistry. 1982;21:1609–1611. [Google Scholar]
- Delevoryas T. Mesozoic cycadophytes. In: Campbell KSW, editor. Papers presented at the 3rd Gondwana symposium. Canberra: Australian National University Press; 1975. [Google Scholar]
- Doyle JJ. Gene trees and species trees – molecular systematics as one-character taxonomy. Systematic Botany. 1992;17:144–163. [Google Scholar]
- Doyle JJ. Trees within trees: genes and species, molecules and morphology. Systematic Biology. 1997;46:537–553. doi: 10.1093/sysbio/46.3.537. [DOI] [PubMed] [Google Scholar]
- Drummond A, Ho S, Phillips M, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biology. 2006;4 doi: 10.1371/journal.pbio.0040088. e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Suchard M, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1·7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV. Is a new and general theory of molecular systematics emerging? Evolution. 2008;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Liu L, Pearl DK. High-resolution species trees without concatenation. Proceedings of the National Academy of Sciences, USA. 2007;104:5936–5941. doi: 10.1073/pnas.0607004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdei B, Argün F, Barone Lumaga MR. Pseudodioon akyoli gen. et sp. nov., an extinct member of Cycadales from Turkish Miocene. Plant Systematics and Evolution. 2010;285:33–49. [Google Scholar]
- Erdei B, Manchester SR, Kvaček Z. Dioonopsis Horiuchi et Kimura leaves from the Eocene of Western North America: a cycad shared with the Paleogene of Japan. International Journal of Plant Sciences. 2012;173:81–95. [Google Scholar]
- Frohlich MW, Parker DS. The mostly male theory of flower evolutionary origins: from genes to fossils. Systematic Botany. 2000;25:155–170. [Google Scholar]
- González D, Vovides AP. Low intralineage divergence in Ceratozamia (Zamiaceae) detected with nuclear ribosomal DNA ITS and chloroplast DNA trnL-F non-coding region. Systematic Botany. 2002;27:654–661. [Google Scholar]
- González D, Vovides AP, Bárcenas C. Phylogenetic relationships of the neotropical genus Dioon (Cycadales, Zamiaceae) based on nuclear and chloroplast DNA sequence data. Systematic Botany. 2008;33:229–236. [Google Scholar]
- Graham A. Historical phytogeography of the Greater Antilles. Brittonia. 2003a;55:357–383. [Google Scholar]
- Graham A. Geohistorical models and Cenozoic paleoenvironments of the Caribbean region. Systematic Botany. 2003b;28:378–386. [Google Scholar]
- Griffith MP, Calonje MA, Stevenson DW, Husby CE, Little DP. Time, place, and relationships: cycad phenology in a phylogenetic and biogeographic context. Memoirs of the New York Botanical Garden. 2012;106:59–81. [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Molecular Biology and Evolution. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendy MD, Penny D. Branch and bound algorithms to determine minimal evolutionary trees. Mathematical Biosciences. 1982;59:277–290. [Google Scholar]
- Hermsen EJ, Taylor TN, Taylor EL, Stevenson DW. Cataphylls of the Middle Triassic cycad Antarcticycas schopfii and new insights into cycad evolution. American Journal of Botany. 2006;93:724–738. doi: 10.3732/ajb.93.5.724. [DOI] [PubMed] [Google Scholar]
- Hermsen EJ, Taylor EL, Taylor TN. Morphology and ecology of the Antarcticycas plant. Review of Palaeobotany and Palynology. 2009;153:108–123. [Google Scholar]
- Hill KD. Cycas – an evolutionary perspective. In: Chen CJ, editor. Biology and conservation of cycads. Beijing: International Academic Publishers; 1999. pp. 98–115. Proceedings of the Fourth International Conference on Cycad Biology. [Google Scholar]
- Hill KD, Chase MW, Stevenson DW, Hills HG, Schutzman B. The families and genera of cycads: a molecular phylogenetic analysis of Cycadophyta based on nuclear and plastid DNA sequences. International Journal of Plant Sciences. 2003;164:933–948. [Google Scholar]
- Hummel J, Gee CT, Südekum K-H, Sander PM, Nogge G, Clauss M. In vitro digestibility of fern and gymnosperm foliage: implications for sauropod feeding ecology and diet selection. Proceedings of the Royal Society B: Biological Sciences. 2008;275:1015–1021. doi: 10.1098/rspb.2007.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo C, Rueda MJ, Mora G. Cenozoic plant diversity in the Neotropics. Science. 2006;311:1893–1896. doi: 10.1126/science.1121380. [DOI] [PubMed] [Google Scholar]
- Jobb G. TREEFINDER version of March 2011. Munich, Germany: 2011. Distributed by the author at www.treefinder.de . [Google Scholar]
- Kluge AG. A concern for evidence and a phylogenetic hypothesis of relationships among Epicrates (Boidae, Serpentes) Systematic Zoology. 1989;38:7–25. [Google Scholar]
- Kluge AG. On total evidence: for the record. Cladistics. 2004;20:205–207. doi: 10.1111/j.1096-0031.2004.00020.x. [DOI] [PubMed] [Google Scholar]
- Kono M, Hiroshi T. Is Cycas revoluta (Cycadaceae) wind- or insect-pollinated? American Journal of Botany. 2007;94:847–855. doi: 10.3732/ajb.94.5.847. [DOI] [PubMed] [Google Scholar]
- Kubatko LS, Degnan JH. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Systematic Biology. 2007;56:17–24. doi: 10.1080/10635150601146041. [DOI] [PubMed] [Google Scholar]
- Kunzig R. World without ice. National Geographic. 2011;220:90–109. [Google Scholar]
- Kvaček Z. A new Tertiary Ceratozamia (Zamiaceae, Cycadopsida) from the European Oligocene. Flora. 2002;197:303–316. [Google Scholar]
- Kvaček Z. A noteworthy cycad, Ceratozamia hofmannii Ettinghausen 1881, from the Lower Miocene of Austria re-examined. Neues Jahrbuch für Geologie und Paläontologie, Monatshefte. 2004;240:111–118. [Google Scholar]
- Kvaček Z, Manchester S. Eostangeria barthel (extinct Cycadales) from the Paleogene of Western North America and Europe. International Journal of Plant Sciences. 1999;16:621–629. [Google Scholar]
- Larget BR, Kotha SK, Dewey CN, Ane C. BUCKy: gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics. 2010;26:2910–2911. doi: 10.1093/bioinformatics/btq539. [DOI] [PubMed] [Google Scholar]
- Lindstrom AJ. Typification of some species names in Zamia L. (Zamiaceae), with an assessment of the status of Chigua D. Stev. Taxon. 2009;58:265–270. [Google Scholar]
- Liu L. BEST: Bayesian estimation of species trees under the coalescent model. Bioinformatics. 2008;24:2542–2543. doi: 10.1093/bioinformatics/btn484. [DOI] [PubMed] [Google Scholar]
- Liu L, Pearl DK. Species trees from gene trees: reconstructing Bayesian posterior distributions of a species phylogeny using estimated gene tree distributions. Systematic Biology. 2007;56:504–514. doi: 10.1080/10635150701429982. [DOI] [PubMed] [Google Scholar]
- Liu L, Pearl DK, Brumfield RT, Edwards SV. Estimating species trees using multiple-allele DNA sequence data. Evolution. 2008;62:2080–2091. doi: 10.1111/j.1558-5646.2008.00414.x. [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L, Kubatko L, Pearl DK, Edwards SV. Coalescent methods for estimating phylogenetic trees. Molecular Phylogenetics and Evolution. 2009;53:320–328. doi: 10.1016/j.ympev.2009.05.033. [DOI] [PubMed] [Google Scholar]
- Liu YS, Zhou ZY, Li HM. First discovery of Cycas fossil leaf in northeast China. Science Bulletin. 1991;22:1758–1759. [Google Scholar]
- Maddison WP. Gene trees in species trees. Systematic Biology. 1997;46:523–536. [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. 2011 Version 2.75. http://mesquiteproject.org . [Google Scholar]
- Manchester SR. Fossil plants of the Eocene Clarno nut beds. Oregon Geology. 1981;43:75–81. [Google Scholar]
- Martínez LCA, Artabe AEE, Bodnar J. A new cycad stem from the Cretaceous in Argentina and its phylogenetic relationships with other Cycadales. Botanical Journal of the Linnean Society. 2012;170:436–458. [Google Scholar]
- Meerow AW, Noblick L, Borrone JW, et al. Phylogenetic analysis of seven WRKY genes across the palm subtribe Attaleinae (Arecaceae) identifies Syagrus as sister group of the coconut. PLoS One. 2009;4:e7353. doi: 10.1371/journal.pone.0007353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata KK, Kazuharu M, Kei-ichi K, Takashi M. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A, Caputo P, Cozzolino S, et al. A phylogenetic analysis of Dioon (Zamiaceae) American Journal of Botany. 1993;80:204–214. [Google Scholar]
- Mort ME, Crawford DJ. The continuing search: low-copy nuclear sequences for lower-level plant molecular phylogenetic studies. Taxon. 2004;53:257–261. [Google Scholar]
- Moynihan J, Stevenson DW, Lewis CE, Vovides AP, Caputo P, Francisco-Ortega J. A phylogenetic study of Dioon Lind. (Zamiaceae, Cycadales), based on morphology, nuclear ribosomal DNA, a low copy nuclear gene and plastid RFLPs. Memoirs of the New York Botanical Garden. 2012;106:448–479. [Google Scholar]
- Mustoe GE. Cycadophyte fossils from the Pacific Northwest. Cycad Newsletter. 2008;31(2/3):28–32. [Google Scholar]
- Nagalingum NS, Marshall CR, Quental TB, Rai HS, Little DP, Mathews S. Recent synchronous radiation of a living fossil. Science. 2011;334:796–799. doi: 10.1126/science.1209926. [DOI] [PubMed] [Google Scholar]
- Nichols D, Johnson L. Plants and the K–T boundary. Cambridge: Cambridge University Press; 2008. [Google Scholar]
- Nickerson J, Drouin G. The sequence of the largest subunit of RNA polymerase II is a useful marker for inferring seed plant phylogeny. Molecular Phylogenetics and Evolution. 2004;31:403–415. doi: 10.1016/j.ympev.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Nicolalde-Morejón F, Vergara-Silva F, González-Astorga J, Stevenson DW, Vovides AP, Sosa V. A character-based approach in the Mexican cycads supports diverse multigene combinations for DNA barcoding. Cladistics. 2011;27:150–164. doi: 10.1111/j.1096-0031.2010.00321.x. [DOI] [PubMed] [Google Scholar]
- Norstog KJ, Nicholls TJ. The biology of the cycads. Ithaca, NY: Cornell University Press; 1997. [Google Scholar]
- Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. Bayesian phylogenetic analysis of combined data. Systematic Biology. 2004;53:47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
- Osborne R. Cycad fossils. Encephalartos. 2002;69:4–13. [Google Scholar]
- Osborne R, Calonje M, Hill KD, Stanberg L, Stevenson DW. The world list of cycads. Memoirs of the New York Botanical Garden. 2012;106:480–510. [Google Scholar]
- Oxelman B, Yoshikawa N, McConaughy BL, Luo J, Denton AL, Hall DB. RPB2 gene phylogeny in flowering plants, with particular emphasis on asterids. Molecular Phylogenetics and Evolution. 2004;32:462–479. doi: 10.1016/j.ympev.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Passalia MG, Del Fueyo G, Archangelsky S. An Early Cretaceous zamiaceous cycad of South West Gondwana: Restrepophyllum nov. gen. from Patagonia, Argentina. Review of Palaeobotany and Palynology. 2010;161:137–150. [Google Scholar]
- Peñalver E, Labandeira CC, Barrón E, et al. Thrips pollination of Mesozoic gymnosperms. Proceedings of National Academy of Sciences, USA. 2012;22:8623–8628. doi: 10.1073/pnas.1120499109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pindell J, Keenan K. Tectonic evolution of the Gulf of Mexico, Caribbean and northern South America in the mantle reference frame: an update. In: James K, Lorente M, Pindell J, editors. The origin and evolution of the Caribbean plate. London: Geological Society of London; 2009. pp. 1–55. [Google Scholar]
- Poczai P, Hyvönen J. Nuclear ribosomal spacer regions in plant phylogenetics: problems and prospects. Molecular Biology Reports. 2010;37:1897–1912. doi: 10.1007/s11033-009-9630-3. [DOI] [PubMed] [Google Scholar]
- Popp M, Oxelman B. Origin and evolution of North American polyploid Silene (Caryophyllaceae) American Journal of Botany. 2007;94:330–349. doi: 10.3732/ajb.94.3.330. [DOI] [PubMed] [Google Scholar]
- Pott C, McLoughlin S, Lindström A. Late Palaeozoic foliage from China displays affinities to Cycadales rather than to Bennettitales necessitating re-evaluation of the Paleozoic Pterophyllum species. Acta Palaeontologica Polonica. 2010;55:157–168. [Google Scholar]
- Rai HS, Obrien HE, Reeves PA, Olmstead RG, Graham SW. Inference of higher-order relationships in the cycads from a large chloroplast data set. Molecular Phylogenetics and Evolution. 2003;29:350–359. doi: 10.1016/s1055-7903(03)00131-3. [DOI] [PubMed] [Google Scholar]
- Rambout A. FigTree v. 1.4. 2012 Available from http://tree.bio.ed.ac.uk/software/figtree/ [Google Scholar]
- Raven PH, Axelrod DI. Angiosperm biogeography and past continental movements. Annals of the Missouri Botanical Garden. 1974;61:539–673. [Google Scholar]
- Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- Rokas A, Carroll AB. More genes or more taxa? The relative contribution of gene number and taxon number to phylogenetic accuracy. Molecular Biology and Evolution. 2005;22:1337–1344. doi: 10.1093/molbev/msi121. [DOI] [PubMed] [Google Scholar]
- Roncal J, Zona S, Lewis CE. Molecular phylogenetic studies of Caribbean palms (Arecaceae) and their relationships to biogeography and conservation. Botanical Review. 2008;74:78–102. [Google Scholar]
- Ronquist F, Teslenko M, van der Bank MP, et al. MrBayes 3·2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–42. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MS, Kumar S. Incomplete taxon sampling is not a problem for phylogenetic inference. Proceedings of the National Academy of Sciences, USA. 2001;98:10751–10756. doi: 10.1073/pnas.191248498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau P. A molecular systematic study of the African endemic cycads. Unpublished Masters thesis, Department of Botany and Microbiology, University of Johannesburg. 2012 [Google Scholar]
- Rousseau-Gueutin M, Gaston A, Ainouche A, et al. Tracking the evolutionary history of polyploidy in Fragaria L. (strawberry): new insights from phylogenetic analyses of low-copy nuclear genes. Molecular Phylogenetics and Evolution. 2009;51:515–530. doi: 10.1016/j.ympev.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Sabato S. West Indian and South American cycads. Memoirs of the New York Botanical Garden. 1990;57:173–185. [Google Scholar]
- Samarakoon T, Wang SY, Alford MH. Enhancing PCR amplification of DNA from recalcitrant plant specimens using a trehalose-based additive. Applications in Plant Sciences. 2013;1:1200236. doi: 10.3732/apps.1200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangin P, Lindstrom A, Kokubugata G, Chaiprasongsuk M, Mingmuang M. Phylogenetic relationships within Cycadaceae inferred from non-coding regions of chloroplast DNA. Kasetsart Journal. 2010;44:544–557. [Google Scholar]
- Sanmartin I, Ronquist F. Southern Hemisphere biogeography inferred by event-based models: plant versus animal patterns. Systematic Biology. 2004;53:216–243. doi: 10.1080/10635150490423430. [DOI] [PubMed] [Google Scholar]
- Sass C, Little DP, Stevenson DW, Specht CD. DNA barcoding in the Cycadales: testing the potential of proposed barcoding markers for species identification of cycads. PLoS One. 2007;2 doi: 10.1371/journal.pone.0001154. pe1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Wink M, Sporer F, Lounibos P. Cycads: their evolution, toxins, herbivores and insect pollinators. Naturwissenschaften. 2002;89:281–294. doi: 10.1007/s00114-002-0330-2. [DOI] [PubMed] [Google Scholar]
- Schulte P, Alegret L, Arenillas I, et al. The Chicxulub asteroid impact and mass extinction at the Cretaceous–Paleogene boundary. Science. 2010;327:1214–1218. doi: 10.1126/science.1177265. [DOI] [PubMed] [Google Scholar]
- Shigyo M, Hasebe M, Ito M. Molecular evolution of the AP2 subfamily. Gene. 2006;366:256–265. doi: 10.1016/j.gene.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Small RL, Cronn RC, Wendel JF. LAS Johnson Review No. 2. Use of nuclear genes for phylogeny reconstruction in plants. Australian Systematic Botany. 2004;17:145–170. [Google Scholar]
- Sorenson MD, Franzosa EA. TreeRot, version 3. Boston, MA: Boston University; 2007. [Google Scholar]
- Stevenson DW. Morphology and systematics of the Cycadales. Memoirs of the New York Botanical Garden. 1990;57:8–55. [Google Scholar]
- Stevenson DW. A formal classification of the extant cycads. Brittonia. 1992;44:220–223. [Google Scholar]
- Sugiura N. Further analysis of data by Akaikes information criterion and finite corrections. Communications in Statistics Part a – Theory and Method, 1978;7:13–26. [Google Scholar]
- Swofford DL. PAUP* phylogenetic analysis using parsimony (*and other methods). v. 4.0 beta 10. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- Tanabe AS. KAKUSAN: a computer program to automate the selection of a nucleotide substitution model and the configuration of a mixed model on multilocus data. Molecular Ecology Notes. 2007;7:962–964. [Google Scholar]
- Tang W. The greater Antilles and the cycads of Puerto Rico. Encephalartos. 2002;71:16–22. [Google Scholar]
- Tang W. Continental drift and the evolution of Asian cycas. Encephalartos. 2004;80:23–28. [Google Scholar]
- Tang W. Continental drift and the evolution of South American cycads. Encephalartos. 2006;86:26–31. [Google Scholar]
- Tang W. The evolutionary history of North American cycads. Cycad Newsletter. 2012;35:7–13. [Google Scholar]
- Terry I, Gimme HW, Moore C, Roemer R, Hull C. Odor-mediated push–pull pollination in cycads. Science. 2007;318:70. doi: 10.1126/science.1145147. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Vorster P, Wink M. Molecular relationships in Encephalartos (Zamiaceae, Cycadales) based on nucleotide sequences of nuclear ITS 1 and 2, rbcl, and genomic ISSR fingerprinting. Plant Biology. 2005;7:79–90. doi: 10.1055/s-2004-830478. [DOI] [PubMed] [Google Scholar]
- Uzunova K, Palamarev E, Kvaĉek Z. Eostangeria ruzinciniana (Zamiaceae) from the Middle Miocene of Bulgaria and its relationship to similar taxa of fossil Eostangeria, and extant Chigua and Stangeria (Cycadales) Acta Palaeobotanica. 2001;41:177–193. [Google Scholar]
- Wang D, Wu YW, Shih ACC, Wu CS, Wang YN, Chaw SM. Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 mya. Molecular Biology and Evolution. 2007;24:2040–2048. doi: 10.1093/molbev/msm133. [DOI] [PubMed] [Google Scholar]
- Wehe A, Bansal MS, Burleigh JG, Eulenstein O. DupTree: a program for large-scale phylogenetic analyses using gene tree parsimony. Bioinformatics. 2008;24:1540–1541. doi: 10.1093/bioinformatics/btn230. [DOI] [PubMed] [Google Scholar]
- Xiao LQ, Möller M, Zhu H. High nrDNA ITS polymorphism in the ancient extant seed plant Cycas: incomplete concerted evolution and the origin of pseudogenes. Molecular Phylogenetics and Evolution. 2010;55:168–177. doi: 10.1016/j.ympev.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Yu Y, Harris AJ, He X. S-DIVA (Statistical Dispersal-Vicariance Analysis): a tool for inferring biogeographic histories. Molecular Phylogenetics and Evolution. 2010;56:848–50. doi: 10.1016/j.ympev.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Yu Y, Harris AJ, He X-J. RASP (Reconstruct Ancestral State in Phylogenies) 2·0 beta. 2011 doi: 10.1016/j.ympev.2015.03.008. Available at http://mnh.scu.edu.cn/soft/blog/RASP . Last accessed 24 December 2012. [DOI] [PubMed] [Google Scholar]
- Zachos JC, Dickens GR, Zeebe RE. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature. 2008;451:279–283. doi: 10.1038/nature06588. [DOI] [PubMed] [Google Scholar]
- Zgurski JM, Rai HS, Fai QM, Bogler DJ, Francisco-Ortega J, Graham SW. How well do we understand the overall backbone of cycad phylogeny? New insights from a large, multigene plastid data set. Molecular Phylogenetics and Evolution. 2008;47:1232–1237. doi: 10.1016/j.ympev.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Zhang N, Zeng L, Hongyan S, Ma H. Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. New Phytologist. 2012;195:923–937. doi: 10.1111/j.1469-8137.2012.04212.x. [DOI] [PubMed] [Google Scholar]
- Zhang PY, Tan HTW, Pwee KH, Kumar PP. Conservation of class C function of floral organ development during 300 million years of evolution from gymnosperms to angiosperms. The Plant Journal. 2004;37:566–577. doi: 10.1046/j.1365-313x.2003.01983.x. [DOI] [PubMed] [Google Scholar]
- Zhao L, Annie ASH, Amrita S, Yi SKF, Rudolf M. Does better taxon sampling help? A new phylogenetic hypothesis for Sepsidae (Diptera: Cyclorrhapha) based on 50 new taxa and the same old mitochondrial and nuclear markers. Molecular Phylogenetics and Evolution. 2013;69:153–164. doi: 10.1016/j.ympev.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Zimmer EA, Wen J. Using nuclear gene data for plant phylogenetics: progress and prospects. Molecular Phylogenetics and Evolution. 2013;66:539–550. doi: 10.1016/j.ympev.2013.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.