Abstract
Purpose
Mutations in the SNRNP200 gene have been reported to cause autosomal dominant retinitis pigmentosa (adRP). In this study, we evaluate the mutation profile of SNRNP200 in a cohort of southern Chinese RP patients.
Methods
Twenty adRP patients from 11 families and 165 index patients with non-syndromic RP with mixed inheritance patterns were screened for mutations in the mutation hotspots of SNRNP200. These included exons 12–16, 22–32, and 38–45, which covered the two helicase ATP-binding domains in DEAD-box and two sec-63 domains. The targeted regions were amplified by polymerase chain reaction and analyzed by direct DNA sequencing, followed by in silico analyses.
Results
Totally 26 variants were identified, 18 of which were novel. Three non-synonymous variants (p.C502R, p.R1779H and p.I698V) were found exclusively in patients. Two of them, p.C502R and p.R1779H, were each identified in one simplex RP patient, whereas p.I698V occurred in one patient with unknown inheritance pattern. All three residues are highly conserved in SNRNP200 orthologs. Nevertheless, only p.C502R and p.R1779H were predicted to affect protein function by in silico analyses, suggesting these two variants are likely to be disease-causing mutations. Notably, all mutations previously identified in other study populations were not detected in this study.
Conclusions
Our results reveal a distinct mutation profile of the SNRNP200 gene in a southern Chinese cohort of RP patients. The identification of two novel candidate mutations in two respective patients affirmed that SNRNP200 contributes to a proportion of overall RP.
Keywords: retinitis pigmentosa, SNRNP200, mutation
Introduction
Retinitis pigmentosa (RP; MIM268000; Mendelian Inheritance in Man; National Center for Biotechnology Information, Bethesda, MD, USA) refers to a clinically and genetically diverse group of diffuse retinal dystrophies. The prevalence of RP is about 1/4000 to 1/6000 in certain populations1, 2, 3 with no significant ethnic difference and affects more than 1 million individuals worldwide.4 Presenting symptoms of RP are various, but the typical ones include the following: (1) nyctalopia (night blindness) as a hallmark, most commonly the earliest symptom in RP; (2) constricted visual fields, usually peripheral, and even central visual loss in advanced cases; and (3) photopsia (seeing flashes of light).5 These symptoms usually occur during the third decade,6 although they can occur earlier depending on the inheritance pattern.7 The fundus hallmarks include arteriolar attenuation, mid-peripheral perivascular bone-spicule pigmentation, tessellated fundus appearance, and waxy disc pallor.8 RP can occur as a simplex disorder without known family history (35–50%) or inherit in autosomal dominant (ad) (10–30%), autosomal recessive (ar) (10–45%), and X-linked (xl) (0–15%) patterns.4, 9 Also it may present as part of systemic disorders, which are usually autosomal recessive.10
Genetically RP is a highly heterogeneous condition. More than 60 RP genes have been identified, while 23, 36, and 3 genes were responsible for adRP, arRP, and xlRP, respectively (RetNet, the Retinal Information Network, University of Texas Houston Health Science Center, Houston, TX, accessed on 22 November 2012). Despite this large number of genes, the vast majority was mapped in populations other than Chinese. Up to date no single gene or mutation contributes to a large proportion of RP in Chinese has been reported,11, 12, 13, 14, 15, 16, 17, 18, 19 as does p.P23H of rhodopsin (RHO, MIM 180380) in Caucasians. The RP33 locus, initially mapped to chromosomal region 2cen-q12.1, was identified in a large Chinese family with adRP.20 Subsequent studies had identified two mutations, p.S1087L21 and p.R1090L,22 in the small nuclear ribonucleoprotein 200 kDa (U5) (SNRNP200, MIM 601664) gene to be segregated with RP in two respective large Chinese adRP families. SNRNP200 is a member of the RNA intron-splicing factor protein family and is widely expressed in human tissues. It encodes a U5 small nuclear ribonucleoprotein 200 kDa helicase, which is a 2136-amino acid splice-complex enzyme containing two pairs of DEAD-box, each followed by a Secretory(sec)-63 domain. DEAD-box proteins are ubiquitous enzymes that use ATP to rearrange RNA and RNA–protein structures. They consist of a helicase core containing 12 conserved motifs involved in ATP binding, RNA binding, and ATP hydrolysis.23 They all possess an ATPase activity that is stimulated by RNA, giving rise to a broad range of biochemical effects in vitro. Thus, they can often unwind short RNA duplexes. This property, together with their structural similarity to the well-characterized DNA helicases, has led to their common designation as ‘RNA helicases.' However, they also can displace RNA-bound proteins, accelerate RNA strand annealing or RNA folding, and act as RNA clamps to form stable ribonucleoprotein (RNP) complexes.23 In yeast, the sec-63 domain is required to reconstitute post-translational translocation in protoliposomes.24 Yeast BiP/Kar2p binds to a DnaJ domain of sec-63 and mediates the import of polypeptides into the endoplasmic reticulum (ER) lumen25 and the export of misfolded proteins into the cytoplasm.26 The DnaJ domain of sec-63 is known to face the ER lumen.27 The large cytoplasmic portion of sec-63 has no known homolog in other proteins. Sequence analysis demonstrated significant similarity between this region and two regions of the helicases family typified by yeast Brr2p and human U5-200 kD.28, 29 These RNA helicases contain two sec63-like domains, each lying C-terminal to an ATPase domain. The Brr2p N-terminal ATPase domain is essential for in vitro RNA unwinding in U4/U6 small ribonucleoprotein particles (snRNPs).30, 31 Although the functions of the helicase sec-63-like domains remain unknown, the associations of the translocon and Brr2p with RNA–protein complexes (the ribosome and spliceosome, respectively) indicate that sec-63 may act as a ribosome receptor. The two initially reported SNRNP200 mutations are both located in the first sec-63 domain, suggesting this domain could be a mutation hotspot in individuals of Chinese descent. Further, the contribution of SNRNP200 to RP has also been investigated in a group of Americans with mixed descent.32 The p.S1087L was also identified in this study, along with another three disease-causing mutations (DCM), p.R681C, p.R681H, and p.Y689C. Besides, a new mutation p.Q885E was recently identified in another four generations of Chinese family.33 All four mutations (p.R681C, p.R681H, p.Y689C, and p.Q885E) are located in the first DEAD-box of SNRNP200.
In this study, we screened the regions containing all the DEAD-box and sec-63 domains in SNRNP200 that may cover the mutation hotspots in a southern Chinese cohort of 185 RP patients and 178 control subjects in order to further evaluate the contributions of SNRNP200 in Chinese RP.
Materials and methods
Study subjects
Twenty adRP patients from 11 families and 165 unrelated non-syndromic RP patients were recruited from the Hong Kong Eye Hospital and the Prince of Wales Hospital, Hong Kong. All were Han Chinese. The diagnosis was based upon typical RP presentations according to ocular examinations that included slit-lamp biomicroscopy, full-field electroretinography, visual fields, and fundus photography. Besides, best-corrected visual acuity (BCVA) and intraocular pressure as assessed by applanation tonometry were recorded. Patients with syndromic RP, such as Bardet-Biedl syndrome and Usher syndrome, or end-stage syphilitic neuroretinitis, and cancer-associated retinopathy were excluded. A group of 178 unrelated healthy control subjects, aged above 60 years, were also recruited. They were given complete ophthalmic examinations and confirmed free of RP or other major eye diseases except for mild senile cataract. This study was approved by the Ethics Committee on Human Research, the Chinese University of Hong Kong. Informed consents were obtained from all study subjects. All procedures were conducted in accordance with the tenets of the Declaration of Helsinki.
Mutational screening of the SNRNP200 gene
In order to evaluate the contribution of the potentially mutation hotspots in SNRNP200, we screened 24 exons of the gene, including exons 12–16, 22–32, and 38–45, in all the study subjects. Exons 12–15 and exons 30–32 cover the two helicase ATP-binding domains, whereas exon 16 covers the first connection of the helicase ATP-binding domain and the helicase CTER, which could be the hotspot in Americans.32 Exons 22–29 and exons 38–45 cover the two sec-63 domains.
Genomic DNA from whole blood was extracted using the QIAamp DNA Blood kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Polymerase chain reaction (PCR) primers were designed according to the sequence of human SNRNP200 (ENSG00000144028) from the Ensembl database (primer information provided upon request). A total of 14 amplicons covering the regions, including exons 12–16, 22–32, and 38–45, and the corresponding exon–intron boundaries were analyzed by PCR and direct DNA sequencing with the Big-Dye Terminator Cycle Sequencing Reaction Kit (version 3.1, Applied Biosystems, Inc. (ABI), Foster City, CA, USA) on an automated DNA sequencer (3130XL; ABI) according to the manufacturer's protocol.
Analyses of variants
The detected variants were defined as ‘novel' if it has neither been reported in the literature nor registered in the single nucleotide polymorphism database (dbSNP). Several criteria were observed to select and prioritize potentially disease-causing variants: (1) the variant was predicted to alter the amino-acid sequence of the protein; (2) it occurred exclusively in patients and was absent in controls; and (3) it was predicted to alter the protein structure or function by in silico analyses. In this study, we applied five in silico programs. Three of them, Polyphen, SIFT, and PMUT, had been described in our previous study.11 Here, two new programs were used to predict the effect of each missense mutation: PolyPhen-2 (Polymorphism Phenotyping version 2, http://genetics.bwh.harvard.edu/pph2/; accessed on 10 November 2012)34 and SIFT BLink (Sorts Intolerant From Tolerant Blink, http://sift.jcvi.org/www/SIFT_BLink_submit.html; accessed on 10 November, 2012).35 In PolyPhen-2, a position-specific independent counts (PSIC) score, based on multiple sequences alignment of observation and three-dimensional protein structural information, was given to each variant in the context of two data sets: HumDiv and HumVar. We chose HumVar because it is the preferred model for diagnosis of Mendelian diseases. A mutation with a PSIC score of >2.0 was classified as ‘probably damaging,' a score of 1.5 to 2.0 was classified as ‘possibly damaging,' and a score of <1.5 was classified as ‘benign'.34 In SIFT BLink, the protein sequence ID NP_054733.2 (GI: 40217847) was used with default parameter setting. The specific amino-acid location in the protein was compared among various organisms from pre-computed NCBI BLAST searches. The output score represented a normalized probability for how the amino-acid substitution was tolerated. A score of <0.05 gave a prediction of ‘affect protein function' and a score of >0.05 gave a prediction of ‘tolerated'.35
The influence of a substitution on any functional motifs was studied by the PROSITE database (ScanProsite, http://prosite.expasy.org Swiss Institute of Bioinformatics, Lausanne, Switzerland; accessed on 10 November 2012). The helicase ATP-binding domain and the helicase CTER domain were revealed by the program. Further, the SMART software (Simple Modular Architecture Research Tool, European Molecular Biology Laboratory, Heidelberg, Germany; accessed on 10 November 2012 http://smart.embl-heidelberg.de/) was used to locate the sec-63 domain in the SNRNP200 protein. To predict coding exons and splice sites in SNRNP200, we used NetGene2 Server (Technical University of Denmark, Lyngby, Copenhagen, Denmark; accessed on 10 November 2012 http://www.cbs.dtu.dk/services/NetGene2/).36 We also investigated evolutionary conservation of amino acids in SNRNP200 using the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway, accessed on 9 January 2012).37 An amino acid that is shown to be highly conserved among different species indicated its importance in maintaining protein function.38
Statistical analysis
Genotype and allele frequencies of any common variants were compared between patients and controls using the χ2-test in SPSS (ver.16.0, SPSS Inc., Chicago, IL, USA). The Bonferroni method was used to correct the P-values in multiple comparisons. A corrected P-value (Pcorr) of less than 0.05 was defined as statistically significant.
Results
Characteristics of the RP patients
Among the 11 recruited adRP families, there were 10 male and 10 female affected members. Whereas the 165 non-syndromic RP patients consisted of 87 males and 78 females, with age ranging from 6 to 84 years. On the basis of the family history, 11 (6.7%) of them were classified as adRP, 26 (15.8%) as arRP, 2 (1.2%) as xlRP, and 97 (58.8%) as simplex RP (sRP). Besides, 29 (17.6%) patients could not be classified because of the absence of family information and were denoted as ‘unknown' inheritance pattern. Therefore, these 165 patients represented a group of southern Chinese non-syndromic RP patients with mixed inheritance.
Sequence variants detected in the SNRNP200 gene
A total of 26 variants were identified in the sequenced regions that cover the entire two helicase ATP-binding domains and the two Sec-63 domains (Table 1). Eight of them (p.L1184L, p.S1218S, c.5134-6C>G, p.L1773L, c.5324-31G>C, p.A1819A, c.5488+82C>T, and c.5755-20A>G) were common SNPs registered in the dbSNP database. They were found in both patients and control subjects. Association analyses showed that all these eight SNPs were not associated with RP (Pcorr>0.05). Further, none of these variants leads to a missense change, implying that they are unlikely to be disease causing.
Table 1. Sequence variants detected in exons 12–16, 22–32, and 38–45 of the SNRNP200 gene among 165 Chinese RP patients and 20 affected members from 11 adRP families.
| Number | Location | Nucleotide change | Residual change | Description | MAF in HapMap (CHB) |
Variation frequencya |
||
|---|---|---|---|---|---|---|---|---|
| Mixed RP Patients | adRP family Affected members | Controls | ||||||
| 1 | Exon 12 | c.1431C>T | p.G477G | Novel | / | 0/1/165 | 0/0/20 | 0/0/178 |
| 2 | Exon 12 | c.1482G>A | p.E494E | Novel | / | 0/0/165 | 0/1/20 | 0/0/178 |
| 3 | Exon 12 | c.1504T>C | p.C502R | Novel | / | 0/1/165 | 0/0/20 | 0/0/178 |
| 4 | Intron 12 | c.1515+67T>C | / | Novel | / | 0/1/165 | 0/0/20 | 0/0/178 |
| 5 | Intron 15 | c.2036+74A>G | / | Novel | / | 0/0/165 | 0/0/20 | 0/2/178 |
| 6 | Intron 15 | c.2036+93G>T | / | Novel | / | 0/0/165 | 0/0/20 | 0/2/178 |
| 7 | Intron 15 | c.2036+107delT | / | Novel | / | 0/0/165 | 0/0/20 | 0/1/178 |
| 8 | Exon 16 | c.2092A>G | p.I698V | Novel | / | 0/1/165 | 0/0/20 | 0/0/178 |
| 9 | Intron 25 | c.3365+145 G>A | / | Novel | / | 0/1/165 | 0/0/20 | 0/0/178 |
| 10 | Intron 25 | c.3366-25 A>G | / | Novel | / | 0/1/165 | 0/0/20 | 0/0/178 |
| 11 | Exon 27 | c.3550T>C | p.L1184L | rs3171927 | 0.155 | 5/50/165 | 0/3/20 | 1/51/178 |
| 12 | Intron 27 | c.3639+53_c.3639+93del | / | Novel | / | 0/0/165 | 0/0/20 | 0/1/178 |
| 13 | Exon 28 | c.3654T>C | p.S1218S | rs3214060 | 0.088 | 0/25/165 | 0/5/20 | 1/14/178 |
| 14 | Exon 28 | c.3690C>T | p.S1230S | Novel | / | 0/1/165 | 0/0/20 | 0/0/178 |
| 15 | Intron 36 | c.5134-17C>T | / | Novel | / | 0/1/165 | 0/0/20 | 0/0/178 |
| 16 | Intron 36 | c.5134-6C>G | / | rs3214062 | 0.005 | 0/2/165 | 0/0/20 | 0/2/179 |
| 17 | Exon 37 | c.5317C>T | p.L1773L | rs772175 | 0.474 | 33/81/165 | 2/10/20 | 25/78/178 |
| 18 | Exon 37 | c.5336G>A | p.R1779H | Novel | / | 0/1/165 | 0/0/20 | 0/0/178 |
| 19 | Intron 37 | c.5324-41G>A | / | Novel | / | 0/1/165 | 0/0/20 | 0/0/178 |
| 20 | Intron 37 | c.5324-31G>C | / | rs78519182 | 0.026 | 0/4/165 | 0/0/20 | 0/2/178 |
| 21 | Exon 38 | c.5457C>T | p.A1819A | rs148995951 | N/A | 0/0/165 | 0/0/20 | 0/1/178 |
| 22 | Intron 38 | c.5488+82C>T | / | rs72937668 | 0 | 0/2/165 | 0/0/20 | 0/0/178 |
| 23 | Intron 40 | c.5755-20A>G | / | rs3214063 | 0.216 | 10/57/165 | 2/5/20 | 5/61/178 |
| 24 | Exon 42 | c.5983G>A | p.A1995T | Novel | / | 0/1/165 | 0/0/20 | 0/1/178 |
| 25 | Exon 42 | c.6057A>G | p.L2019L | Novel | / | 0/0/165 | 0/0/20 | 0/1/178 |
| 26 | Exon 42 | c.6087C>A | p.I2029I | Novel | / | 0/1/165 | 0/0/20 | 0/0/178 |
Abbreviations: CHB, Chinese Han Beijing in HapMap populations; MAF, minor allele frequency; N/A, not available.
The variation frequency was presented in ‘Homozygote variant genotypes/Heterozygote variant genotypes/Total genotypic counts' fashion.
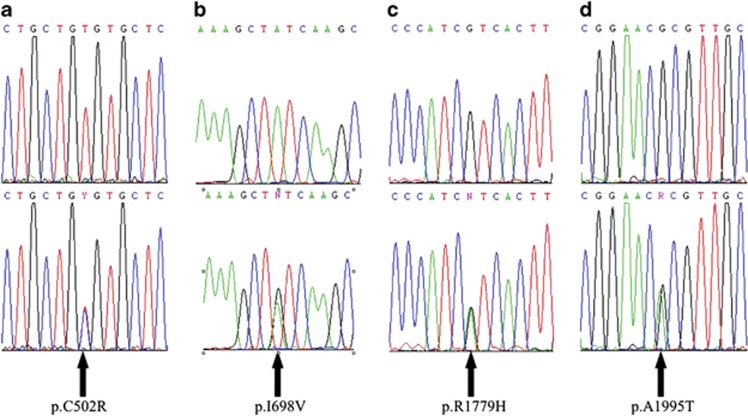
Apart from the eight SNPs, we identified 18 novel variants. Nine were intronic, of which five (c.1515+67T>C, c.3365+145G>A, c.3366-25A>G, c.5134-17C>T, and c.5324-41G>A) were each found in one patient but not in controls. Although we cannot rule out the pathogenicity of these intronic changes as family members were not available for segregation analysis, none of these variants is located in splicing sites according to the NetGene2 Server prediction (data not shown), making them less likely to be disease causing. Two intronic variants (c.2036+74A>G and c.2036+93G>T) were each found in two control subjects, whereas the remaining two (c.2036+107delT and c.3639+53_c.3639+93del) were found in one control each. They are less likely to be pathogenic. Of the nine coding variants, four synonymous ones, p.G477G, p.E494E, p.S1230S, and p.I2029I, were found in one patient each, whereas p.L2019L was found in one control subject. By using SIFT and SIFT BLink, the only in silico analysis available for synonymous variants showed these five variants were benign with low potential of disease causing (data not shown). Among the four non-synonymous variants, p.A1995T was found in both patients and controls, whereas p.C502R, p.I698V, and p.R1779H were each found in one patient but not in controls (Figure 1).
Figure 1.
The forward sequence chromatograms of (a) p,C502R; (b) p.I698V; (c) p.R1779H; and (d) p.A1995T. Chromatograms for the normal allele are shown above those for the mutant allele. Arrows indicate positions of mutations.
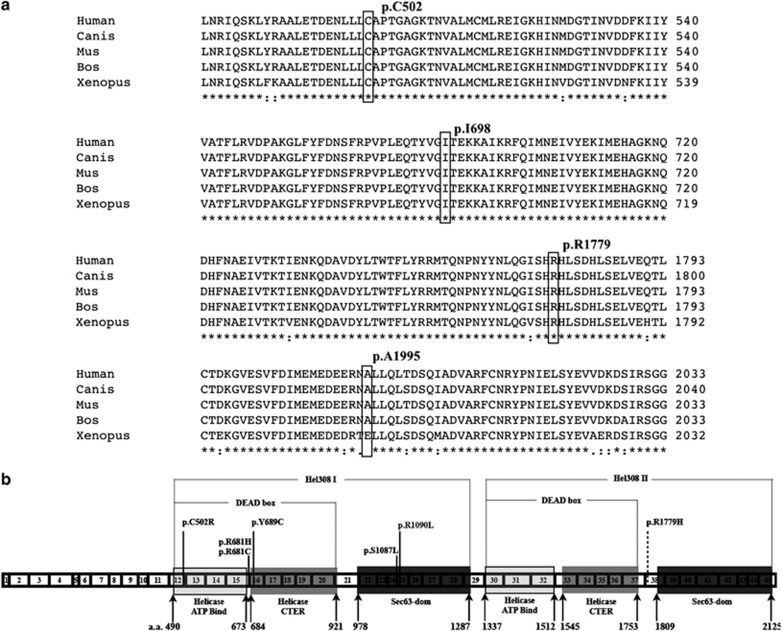
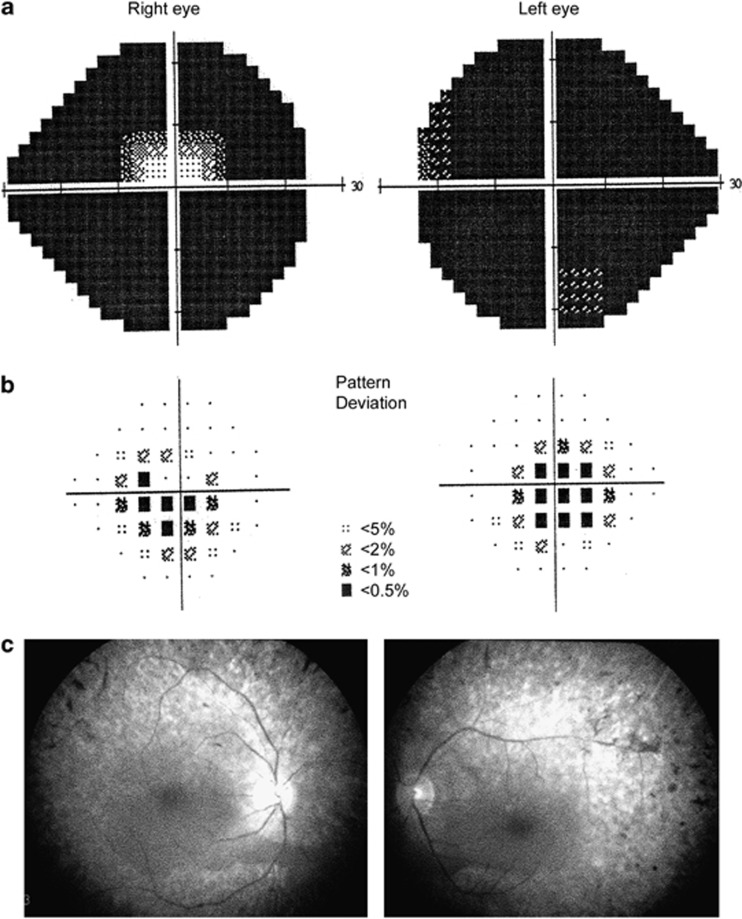
All the four missense variants were tested by using the in silico programs and multiple protein sequences alignment. Three residues (p.C502, p.I698, and p.R1779) were absolutely conserved, whereas the p.A1995 was highly conserved across different species, indicating possible impact on the protein function (Figure 2a). According to the PROSITE and SMART, p.C502 is located in the first helicase ATP-binding domain, p.R1779 is found between the second HELICASE CTR domain and the second sec-63 domain (Figure 2b), p.I698 is in the first helicase CTR domain, whereas p.A1995 is in the second sec-63 domain. The p.A1995T was predicted to be benign by all of the in silico programs and was found in one control subject, suggesting a low chance of being a DCM. The p.C502R, which was found in one sRP patient, was predicted to be pathological, whereas p.I698V was shown to be benign by all five programs (Table 2). For p.R1779H, which was found in another sRP patient, the predictions were inconsistent. As it has been suggested that the Polyphen 2 program could be more reliable,34 p.R1779H is considered pathological. With respect to the carriers of the two potential DCMs, the patient carrying p.C502R is an 82-year-old lady who was diagnosed with RP at the age of 64 years with typical RP fundus changes (Figure 3). Her BCVA was 20/200 and hand movement in her right and left eyes, respectively, with severe visual field damage in both eyes (Figure 3) during her last visit. The patient with p.R1779H had disease onset at the age of 53 years and had typical manifestations of RP (data not shown). These two patients had no other affected family members by the time of recruitment, suggestive of simplex RP. Mutation in some other RP genes, including RHO, RP1, NR2E3, NRL, and BEST1, had all been excluded in these patients.11, 12, 18
Figure 2.
(a) Multiple protein sequence alignment of SNRNP200 (partially), showing the location of p.C502, p.I698, p.R1779, and p.A1995, while the first three residues are absolutely conserved and the last residue are well conserved across different species. The accession numbers of SNRNP200 protein sequences of different species are as follows: Human NP_054733.2, Canis (Canis lupus familiaris) XP_532949.2, Mus (Mus musculus) NP_796188.2, Bos (Bos taurus) NP_001193092.1, Xenopus (Xenopus (Silurana) tropicalis) XP_002932581.1. (b) Protein structure and all reported disease-causing mutations in SNRNP200 with transcript reference No. ENST00000323853. 1–45 demonstrated exon 1–45 with corresponding length. Domain location reference software: PROSITE for helicase ATP-binding and helicase CTER domains; SMART for Sec-63 dom; a.a.: amino acid sequence number of SNRNP200; mutations reference: p.C502R (present study); p.R681H;32 p.R681C;32 p.Y689C;32 p.Q885E;33 p.S1087L;21, 32 p.R1090L;21 p.R1779H (present study, dash line indicated probably be disease causing mutation).
Table 2. Carriers and pathogenic potentials of the four novel SNRNP200 non-synonymous variants.
| Variants | Exon |
Case |
Control | Descriptions |
PolyPhen |
PolyPhen 2 |
SIFT |
SIFT BLink |
PMUT |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | Sex/Age | Prediction | PSIC score | Prediction | HumanVar score | Prediction | Score | Prediction | Score | Prediction | RI | ||||
| c.1504T>C (p.C502R) | 12 | sRP | F/71 | 0/0/178 | Novel | Probably damaging | 4.14 | Probably damaging | 1.00 | Damaging | 0 | Affected Protein Function | 0 | Pathologic | 4 |
| c.2092A>G (p.I698V) | 16 | — | NA | 0/0/178 | Novel | Benign | 0.74 | Benign | 0.00 | Tolerated | 0.12 | Tolerated | 0.23 | Neutral | 9 |
| c.5356G>A (p.R1779H) | 38 | sRP | F/53 | 0/0/178 | Novel | Benign | 0.27 | Probably damaging | 0.95 | Damaging | 0.04 | Affected Protein Function | 0.01 | Neutral | 0 |
| c.5983G>A (p.A1995T) | 42 | — | M/62 | 0/1/178 | Novel | Benign | 0.20 | Benign | 0.00 | Tolerated | 0.14 | Tolerated | 0.31 | Neutral | 5 |
Abbreviations: Age, age (years) at recruitment; PSIC, position-specific independent counts, a scoring system used in PolyPhen; RI, reliability index; sRP, simplex retinitis pigmentosa; Type, type of RP.
Figure 3.
The visual field tests (a and b) and fundus photos (c) of the p.C502R carrier during the last visit with the typical presentations of retinitis pigmentosa.
Discussion
In this study, we have evaluated the mutation profile in 24 exons containing the hotspots in SNRNP200 among a cohort of southern Han Chinese RP patients and controls. A total of 18 novel variants were detected, among which three non-synonymous changes, p.C502R, p.I698V, and p.R1779H, were found exclusively in patients. Moreover, p.C502R and p.R1779H were predicted to be pathological by in silico programs. They are likely to be disease-causing mutations of RP. If they are genuinely pathogenic, the SNRNP200 gene might have accounted for ∼1.1% (2/176, 11 of them from families and 165 were simplex) of overall RP, albeit we only screened the potential mutation hotspots of the gene. In contrast, another non-synonymous variant p.A1995T, which occurred in both patients and controls, was predicted to be benign, suggesting it is unlikely to be pathogenic. Notably, the three DCMs p.Q885E, p.S1087L, and p.R1090L, which were previously identified in three respective adRP pedigrees from northern China,21, 22, 33 were not found in our study. In the first sec-63 domain region, which harbors two of these three mutations (p.S1087L and p.R1090L), only three common SNPs (p.L1184L, p.S1218S, and p.S1230s) and three intronic changes (c.3365+145 G>A, c.3366-25 A>G, and c.3639+53_c.3639+93del) were detected in our study subjects. Likewise, the mutations that were detected in the Caucasian cohort32 were also not identified in our present study. Therefore, results of our study enrich the SNRNP200 mutation spectrum and thus substantiate its contribution to RP.
The spliceosome for pre-mRNA splicing is a specialized RNA and dynamic protein complex. The exons ligation relies on spliceosome splicing out the introns from a transcribed pre-mRNA segment.39 The major spliceosome composes of four small nuclear RNA proteins, called snRNPs, U1, U2, U4/U6, and U5, and a range of non-snRNP splicing factors. The individual snRNP particles participate in the splicing cycle in a highly dynamic manner. Lauber et al.29 demonstrated that human SNRNP200 encoded a U5-specfic 220 kDa protein that is homologous to yeast SNU246 encoding a U5-specific protein Brr2, while both of these two proteins contain two conserved domains being characteristic of the DEXH-box protein family of the putative RNA helicases and RNA-stimulated ATPases. They also showed disruption of SNU246 in yeast is lethal and leads to a splicing defect in vivo.29 Brr2 is a DExD/H-box helicase responsible for U4/U6 unwinding during spliceosomal activation. The two reported mutations, p.R1090L and p.S1087L, are well conserved in many species. The yeast analog of these two mutations (p.R1107L and p.N1104L) were shown to compromise RNA unwinding in budding yeast.21 These indicate that the two mutations in the U5-specfic 220 kDa protein may lead to pathogenesis through impaired RNA unwinding and thus splicing. Notably all five reported mutations are located within hel308-I. The mutations p.R681H, p.R681C, and p.Y689C are located in the boundaries between the helicase ATP-binding and helicase CTR domains, whereas p.S1087L and p.R1089L are located within the sec-63 domain. The mutation p.C502R detected in this study is located in the helicase ATP-binding domain. Hel308-I has the highest sequence conservation among different species, suggesting a critical role in the helicase activity. It has a direct role in the unwinding of the U4/U6 helix.40 Structural defect of Hel308-I may impair helicase/ATPase activity, leading to defects in spliceosome catalysis. In contrast to the unwinding function of Hel308-I, the Hel308-II module of Brr2 does not have ATPase activity and is also unlikely to have helicase activity.40 This may explain the lack of mutation reported on Hel308-II in RP, as the structural defect may not directly affect the spliceosome. Hel308-II has been demonstrated to interact with Prp8 and Snu114 both in vitro and in vivo. Further, the Prp8-CTR facilitates the binding of the Brr2/Prp8–CTR complex to U4/U6 in yeast.41 In this current study, we have identified a potentially pathogenic mutation, p.R1779H, which is located between the second helicase CTR and sec-63 domains. It is the first candidate mutation found in the hel308-II module for RP. Although it does not disrupt the helicase directly, it may have a role in mediating the regulation of Brr2 activity or the interaction with other spliceosomal proteins.
To date, the RNA intron-splicing factors have only been implicated in adRP.21, 32, 42, 43, 44 Although it is clear that the retina requires a relatively high level of RNA splicing activity for optimal tissue-specific physiological function,45 how the defects in this essential macromolecular complex transform into a photoreceptor-specific phenotype is unknown. The underlying reason for the RNA intron-splicing factors to be related to adRP is also unclear. In our current study, two possible mutations were found in sRP patients, suggesting that SNRNP200 may also have a role in other forms of RP. However, as the family members of the two patients were not available for clinical and genetic investigations and these two mutations were heterozygous, it is unclear whether they represent de novo mutations or dominant mutations with reduced penetrance.
In our Chinese study subjects, we did not detect the SNRNP200 mutations that were previously found for adRP,21, 22, 32 suggesting that the mutation spectra of SNRNP200 vary across different populations or even within the same ethnic group. Apart from showing the contribution of SNRNP200 to a proportion of overall RP, our results revealed a highly mutable SNRNP200 in RP. Even in a relatively small number of adRP patients in our cohort from southern China, we detected a large number of SNRNP200 variants that are either potentially causal or associated with RP. These were found within the 24 exons containing known mutable regions. Therefore, screening all the 45 coding exons of SNRNP200 is worthwhile in order to reveal the significance of SNRNP200 in RP. In addition, RP families should also be screened to ascertain segregation of SNRNP200 mutations with RP. As our data indicate possible direct causation of specific SNRNP200 variants on RP, functional analysis of potential disease-causing mutants should be warranted.
Acknowledgments
We express our greatest appreciation to all the participants in the study. The work in this paper was supported in part by the Direct Grants of the Chinese University of Hong Kong (Project codes: 2041770 and 2041771), a block grant of the University Grants Committee Hong Kong, and the Endowment Fund for Lim Por-Yen Eye Genetics Research Centre, Hong Kong.
The authors declare no conflict of interest.
References
- Bundey S, Crews SJ. A study of retinitis pigmentosa in the City of Birmingham. I Prevalence. J Med Genet. 1984;21 (6:417–420. doi: 10.1136/jmg.21.6.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu DN. Genetic aspects of retinitis pigmentosa in China. Am J Med Genet. 1982;12 (1:51–56. doi: 10.1002/ajmg.1320120107. [DOI] [PubMed] [Google Scholar]
- Greenberg J, Bartmann L, Ramesar R, Beighton P. Retinitis pigmentosa in southern Africa. Clin Genet. 1993;44 (5:232–235. doi: 10.1111/j.1399-0004.1993.tb03888.x. [DOI] [PubMed] [Google Scholar]
- Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368 (9549:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- Valentine JA. Night blindness: retinitis pigmentosa sine pigmento. Proc R Soc Med. 1923;16 (Sect Ophthalmol:17. [PMC free article] [PubMed] [Google Scholar]
- Ruther K, Banhart F, Kremmer S, Apfelstedt-Sylla E, Zrenner E. [Social ophthalmologic aspects of retinitis pigmentosa] Ophthalmologe. 1995;92 (5:704–707. [PubMed] [Google Scholar]
- Kaplan J, Bonneau D, Frezal J, Munnich A, Dufier JL. Clinical and genetic heterogeneity in retinitis pigmentosa. Hum Genet. 1990;85 (6:635–642. doi: 10.1007/BF00193589. [DOI] [PubMed] [Google Scholar]
- Kalloniatis M, Fletcher EL. Retinitis pigmentosa: understanding the clinical presentation, mechanisms and treatment options. Clin Exp Optom. 2004;87 (2:65–80. doi: 10.1111/j.1444-0938.2004.tb03152.x. [DOI] [PubMed] [Google Scholar]
- Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis. 2006;1:40. doi: 10.1186/1750-1172-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A. Genetic diseases of vision. Curr Opin Neurol. 1994;7 (1:65–68. doi: 10.1097/00019052-199402000-00012. [DOI] [PubMed] [Google Scholar]
- Zhang X, Chen LJ, Law JP, Lai TY, Chiang SW, Tam PO, et al. Differential pattern of RP1 mutations in retinitis pigmentosa. Molecular vision. 2010;16:1353–1360. [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang X, Chen LJ, Chiang SW, Tam PO, Lai TY, et al. Association of NR2E3 but not NRL mutations with retinitis pigmentosa in the Chinese population. Invest Ophthalmol Vis Sci. 2010;51 (4:2229–2235. doi: 10.1167/iovs.09-4299. [DOI] [PubMed] [Google Scholar]
- Tian Y, Tang L, Cui J, Zhu X. Screening for the carbonic anhydrase IV gene mutations in Chinese retinitis pigmentosa patients. Curr Eye Res. 2010;35 (5:440–444. doi: 10.3109/02713680903503512. [DOI] [PubMed] [Google Scholar]
- Li S, Xiao X, Wang P, Guo X, Zhang Q. Mutation spectrum and frequency of the RHO gene in 248 Chinese families with retinitis pigmentosa. Biochem Biophy Res Commun. 2010;401 (1:42–47. doi: 10.1016/j.bbrc.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li S, Xiao X, Jia X, Guo X. The 208delG mutation in FSCN2 does not associate with retinal degeneration in Chinese individuals. Invest Ophthalmol Vis Sci. 2007;48 (2:530–533. doi: 10.1167/iovs.06-0669. [DOI] [PubMed] [Google Scholar]
- Chiang SW, Wang DY, Chan WM, Tam PO, Chong KK, Lam DS, et al. A novel missense RP1 mutation in retinitis pigmentosa. Eye (Lond) 2006;20 (5:602–605. doi: 10.1038/sj.eye.6701944. [DOI] [PubMed] [Google Scholar]
- Zhang XL, Liu M, Meng XH, Fu WL, Yin ZQ, Zhang X, et al. A complete screen for mutations of the rhodopsin gene in a panel of Chinese patients with autosomal dominant retinitis pigmentosa. Chinese medical sciences journal=Chung-kuo i hsueh k'o hsueh tsa chih/ Chinese Academy of Medical Sciences. 2005;20 (1:30–34. [PubMed] [Google Scholar]
- Chan WM, Yeung KY, Pang CP, Baum L, Lau TC, Kwok AK, et al. Rhodopsin mutations in Chinese patients with retinitis pigmentosa. Br J Ophthalmol. 2001;85 (9:1046–1048. doi: 10.1136/bjo.85.9.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum L, Chan WM, Yeung KY, Lam DS, Kwok AK, Pang CP. RP1 in Chinese: Eight novel variants and evidence that truncation of the extreme C-terminal does not cause retinitis pigmentosa. Human Mut. 2001;17 (5:436. doi: 10.1002/humu.1127. [DOI] [PubMed] [Google Scholar]
- Zhao C, Lu S, Zhou X, Zhang X, Zhao K, Larsson C. A novel locus (RP33) for autosomal dominant retinitis pigmentosa mapping to chromosomal region 2cen-q12.1. Hum Genet. 2006;119 (6:617–623. doi: 10.1007/s00439-006-0168-3. [DOI] [PubMed] [Google Scholar]
- Zhao C, Bellur DL, Lu S, Zhao F, Grassi MA, Bowne SJ, et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet. 2009;85 (5:617–627. doi: 10.1016/j.ajhg.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Mei H, MacDonald IM, Jiao X, Hejtmancik JF. Mutations in ASCC3L1 on 2q11.2 are associated with autosomal dominant retinitis pigmentosa in a Chinese family. Invest Ophthalmol Vis Sci. 51 (2:1036–1043. doi: 10.1167/iovs.09-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. From unwinding to clamping—the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12 (8:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81 (4:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Lyman SK, Schekman R. Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell. 1997;88 (1:85–96. doi: 10.1016/s0092-8674(00)81861-9. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388 (6645:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Feldheim D, Rothblatt J, Schekman R. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol Cell Biol. 1992;12 (7:3288–3296. doi: 10.1128/mcb.12.7.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143 (1:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber J, Fabrizio P, Teigelkamp S, Lane WS, Hartmann E, Luhrmann R. The HeLa 200 kDa U5 snRNP-specific protein and its homologue in Saccharomyces cerevisiae are members of the DEXH-box protein family of putative RNA helicases. Embo J. 1996;15 (15:4001–4015. [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Achsel T, Luhrmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci USA. 1998;95 (8:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Current biology: CB. 1998;8 (15:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- Benaglio P, McGee TL, Capelli LP, Harper S, Berson EL, Rivolta C. Next generation sequencing of pooled samples reveals new SNRNP200 mutations associated with retinitis pigmentosa. Hum Mutat. 2011;32 (6:E2246–E2258. doi: 10.1002/humu.21485. [DOI] [PubMed] [Google Scholar]
- Liu T, Jin X, Zhang X, Yuan H, Cheng J, Lee J, et al. A novel missense snrnp200 mutation associated with autosomal dominant retinitis pigmentosa in a Chinese family. PLoS ONE. 2012;7 (9:e45464. doi: 10.1371/journal.pone.0045464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7 (4:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11 (5:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Lin X, Salzberg SL. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001;29 (5:1185–1190. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40 (Database issue:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39 (Database issue:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 2001;13 (3:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Kim DH, Rossi JJ. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA. 1999;5 (7:959–971. doi: 10.1017/s135583829999012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, et al. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol. 2009;16 (7:731–739. doi: 10.1038/nsmb.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie AB, McHale JC, Keen TJ, Tarttelin EE, Goliath R, van Lith-Verhoeven JJ, et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13) Hum Mol Genet. 2001;10 (15:1555–1562. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]
- Vithana EN, Abu-Safieh L, Allen MJ, Carey A, Papaioannou M, Chakarova C, et al. A human homolog of yeast spre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11) Mol Cell. 2001;8 (2:375–381. doi: 10.1016/s1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- Maita H, Kitaura H, Ariga H, Iguchi-Ariga SM. Association of PAP-1 and Prp3p, the products of causative genes of dominant retinitis pigmentosa, in the tri-snRNP complex. Exp Cell Res. 2005;302 (1:61–68. doi: 10.1016/j.yexcr.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Cao H, Wu J, Lam S, Duan R, Newnham C, Molday RS, et al. Temporal and tissue specific regulation of RP-associated splicing factor genes PRPF3, PRPF31 and PRPC8-implications in the pathogenesis of RP. PLoS One. 2011;6 (1:e15860. doi: 10.1371/journal.pone.0015860. [DOI] [PMC free article] [PubMed] [Google Scholar]